Abstract
BACKGROUND: Blood-based proteomic profiling may aid and expand our understanding of diseases and their different phenotypes. The aim of the presented study was to profile serum samples from patients with malignant melanoma using affinity proteomic assays to describe proteins in the blood stream that are associated to stage or recurrence of melanoma. MATERIAL AND METHODS: Multiplexed protein analysis was conducted using antibody suspension bead arrays. A total of 232 antibodies against 132 proteins were selected from (i) a screening with 4595 antibodies and 32 serum samples from melanoma patients and controls, (ii) antibodies used for immunohistochemistry, (iii) protein targets previously related with melanoma. The analysis was performed with 149 serum samples from patients with malignant melanoma. Antibody selectivity was then assessed by Western blot, immunocapture mass spectrometry, and epitope mapping. Lastly, indicative antibodies were applied for IHC analysis of melanoma tissues. RESULTS: Serum levels of regucalcin (RGN) and syntaxin 7 (STX7) were found to be lower in patients with both recurring tumors and a high Breslow's thickness (T-stage 3/4) compared to low thickness (T-stage 1/2) without disease recurrence. Serum levels of methylenetetrahydrofolate dehydrogenase 1-like (MTHFD1L) were instead elevated in sera of T3/4 patients with recurrence. The analysis of tissue sections with S100A6 and MTHFD1L showed positive staining in a majority of patients with melanoma, and S100A6 was significantly associated to T-stage. CONCLUSIONS: Our findings provide a starting point to further study RGN, STX7, MTHFD1L and S100A6 in serum to elucidate their involvement in melanoma progression and to assess a possible contribution to support clinical indications.
Introduction
Cutaneous malignant melanoma is one of the most aggressive forms of skin cancer for which incidence rates have steadily increased over the last decades. Despite a very favorable prognosis associated to early stage localized melanomas, patients with distant metastases have an overall poor prognosis with 5-year survival rates <5% [1]. The American Joint Committee on Cancer (AJCC) melanoma staging system [2] is used to predict disease progression and takes into account different clinical and histopathological variables. Despite a continuous refinement of covariates included in the staging system (Breslow's thickness, ulceration, mitotic index, lymph node status, the presence of distant metastases, and concentration of serum lactate dehydrogenase), improvements for classifying patients into high or low risk for recurrence or metastatic disease are still needed. One of the most important determinants of prognosis and treatment for clinically localized melanoma is Breslow's thickness (T-stage), which measures the thickness of the primary tumor in millimeters. There is still a lack of molecular understanding about why thick tumors are more prone to spread and metastasize compared to thin ones. Rather than promoting metastasis, Breslow's thickness is likely an indicator of the disease biology within tumor cells [3]. Thus, finding proteins that correlate to T-stage may provide novel insights about the mechanisms that regulate disease progression. The current edition of AJCC staging includes measuring serum lactate dehydrogenate (LDH) to classify late stage melanomas, and also S100B has been linked to clinical stage and tumor progression. However, adding additional serum markers would be of great value to improve the more widespread poor clinical sensitivity and specificity that are currently limiting the use of LDH and S100B in a clinical setting.
To find additional proteins associated to the disease from serum or plasma analysis, proteomics approaches by either mass spectrometry (MS) [4] or multiplexed immunoassays [5] can be used. Commonly, MS allows for the analysis of many proteins in a limited set of samples at a time, while the affinity-based assays offer to analyze a larger set of samples with a selected set of defined protein targets. There has been a lack of affinity reagents for a more extensive analysis of proteins, but resources with binders have become available to extend the opportunities for protein analysis with immunoassays [6]. One initiative is the Human Protein Atlas (HPA) [7] that since its first release in 2005 has published data from more than 25,000 antibodies on protein expression of cell lines and various types of normal and cancer tissues.
In this study, we used the HPA resource of antibodies to screen serum samples with suspension bead array assays [8] for proteins associated to melanoma progression, recurrence or survival. The indicative antibodies were validated by using several proteomics approaches acknowledging their potential application dependent performance [9]. Lastly, we evaluated the selected antibodies for immunohistochemical analysis of melanoma tissues.
Material and Methods
Samples
Serum samples were collected at the Department of Oncology, Uppsala University Hospital between 1995 and 2004, at first admittance after primary surgery due to malignant melanoma. Serum samples from 20 anonymous individuals used as controls for the screening were collected at the same hospital. Due to logistical reasons, sera were collected by experienced staff within 1–100 days after primary removal of the tumor. The study was reviewed and approved by the Research Ethics Committee (Dnr 2005/232) at Uppsala University, Uppsala, Sweden. All patients were staged according to UICC 2002 (TNM system). Disease specific survival was defined as survival with end-point death due to malignant melanoma. Disease recurrence was defined by local recurrence of tumor growth in the skin or development of metastases. Among 63 patients with recurrence, 25 and 10 were recorded to have positive or negative lymph node status respectively, while data on lymph node status was missing for 28 patients. Patients were followed until the time of death or until August 2007. The follow-up period ranged from 7.3 to 241 months with a median follow-up time of 80 months and during this time, 63 patients (42%) relapsed. The sample set of 149 patients used for the main study is summarized in Table 1, while the subset of 16 patient samples used during the screening is found in S-Table 1.
Table 1.
Sample Demographics
| Description | N | Gender (%Female) | Mean Age (min-max) | |
|---|---|---|---|---|
| T-stage | M1 disease at dx | 5 | 40 | 66 (44–85) |
| T1 < =1.00 mm | 60 | 42 | 55 (28–84) | |
| T2 = 1.01–2.00 mm | 32 | 66 | 62 (31–88) | |
| T3 = 2.01–4.00 mm | 29 | 41 | 63 (24–88) | |
| T4> 4.00 mm | 17 | 29 | 63 (37–81) | |
| Unclassified | 6 | |||
| Recurrence | Yes | 86 | 50 | 57 (28–84) |
| No | 63 | 40 | 62 (24–88) | |
| T-stage/Recurrence | T1-T2/No recurrence | 74 | 51 | 56 (28–84) |
| T3-T4/Recurrence | 34 | 34 | 63 (24–88) | |
| DFS 5 y | Yes | 91 | 49 | 58 (24–88) |
| No | 58 | 40 | 61 (30–88) | |
| Metastasis status at follow-up | M1 at diagnosis | 5 | 40 | 66 (44–85) |
| Metastasis | 43 | 44 | 56 (24–82) | |
| No metastasis | 101 | 47 | 60 (28–88) | |
| Location of the tumor | Face | 17 | 53 | 65 (31–88) |
| Lower part of body | 41 | 73 | 57 (30–85) | |
| Trunk | 5 | 20 | 56 (29–84) | |
| Upper part of body | 82 | 32 | 60 (24–88) | |
| Unclassified | 4 | |||
| Ulceration | Yes | 27 | 41 | 61 (30–84) |
| No | 37 | 43 | 58 (28–88) | |
| Unclassified | 85 |
Sample demographics of the melanoma patients. DSF; Disease-free survival (5 years).
Antibody Suspension Bead Arrays
To create multiplexed bead arrays in suspension, antibodies form the Human Protein Atlas (HPA) were covalently coupled to color-coded magnetic microspheres (MagPlex®, Luminex Corp.) as previously described [10]. The coupling efficiency for each antibody was confirmed via R-phycoerythrin-labeled anti-rabbit IgG antibody (anti-rabbit RPE, Jackson ImmunoResearch Laboratories). Samples were diluted, biotinylated and incubated with beads in accordance to previous protocols [11] and with minor modifications. Briefly, crude serum stored at −80 °C were distributed in randomized layouts across two 96-well plates, centrifuged (3000 rpm for 3 min) and diluted 1:10 in PBS prior addition of activated biotin (Pierce) at a 10-fold molar excess. After 2 h of incubation at 4 °C, the labeling reaction was quenched by Tris–HCl, pH 8.0, and stored at −20 °C. For the proceeding assay procedure, biotinylated samples were diluted 1:50 (SELMA, CyBio) and heat-treated at 56 °C in a water bath for 30 min before incubating with the suspension bead array (~150 beads per ID) overnight on a 384-well assay plate (Greiner). Beads were washed on a plate washer (EL406, Biotek), antibody-target interactions were fixated by 0.4% paraformaldehyde and florescent readout was enabled by the addition of R-phycoerythrin-labeled streptavidin (Invitrogen). Binding events were measured using a dedicated flow cytometer (FlexMap 3D, Luminex Corp.).
Immunocapture Mass Spectrometry
Immunocapture mass spectrometry (IC-MS) was used to validate on-target binding as previously described [12]. For each investigation, 1.6 μg from anti-MTHFD1L HPA029040, anti-RGN HPA029104, anti-S100A6 HPA007575, anti-STX7 HPA001467, or normal rabbit IgG were coupled to 500,000 beads, respectively, and coupling efficiency was confirmed as described above. A pool of sera (anti- MTHFD1L, RGN HPA029104) or plasma (anti-MTHFD1L and anti-STX7 HPA001467, assayed in a plate together with other unrelated antibodies) from four healthy individuals of both mixed genders (Seralab) was diluted 1:10 in assay buffer (see above) and heat-treated at 56 °C for 30 min. Duplicate IC-MS experiments were performed for anti-STX7 and anti-MTHFD1L, while a single incubation was performed with anti-RGN and anti-S100A6. The sample volumes were split into 1 ml aliquots and 500,000 coupled beads with either anti-MTHFD1L HPA029040, anti-RGN HPA029104, anti-S100A6 or anti-STX7 HPA001467 were added. As a reference, normal rabbit IgG coupled beads were assayed alongside the specific immune-captures of an experiment. Samples were incubated with beads overnight, washed 3× in PBS/Chaps 0.03% and re-suspended in 100 μl of ammonium bicarbonate (50 mM) with 2.5% sodium deoxycholate. Captured proteins were reduced with 1 mM dithiothreitol (DTT), alkylated by 4 mM iodoacetamide (IAA) and digested by trypsin/Lys-C Mix (Promega) overnight at 37 °C. Next, 5 μl TFA (10%) was added to the digested sample and incubated for 30 min to allow SDC to precipitate. The supernatants containing the peptides were then separated from the beads, dried and re-suspended in Buffer A (97% water, 3% acetonitrile (ACN), 0.1% formic acid (FA)). Chromatography separation was performed using a 50 cm x 75 μm ID Easy spray analytical column (PepMap RSLC C18) installed on Ultimate 3000 RSLC nanosystem (Thermo Scientific). Peptide ions were then analyzed on a Q-Exactive HF (Thermo) operated in a data dependent mode. Raw data files were processed using the software MaxQuant under the following parameters: human protein sequences from Uniprot (http://www.uniprot.org, Swiss-prot Reviewed, 20,198 hits on 03/17/2016)); two missing cleavages were allowed; carbamidomethylation on cysteine was selected as fixed modification and oxidation of methionine and N-term acetylation as variable modification, match between runs applied. Proteins identified were analyzed within a large analysis including 276 IC-MS experiments performed under the same experimental conditions in order to exclude proteins representing common contaminants for the immunoprecipitation procedure (data not shown). Protein intensities were averaged before the z-score calculation, for experiments performed in duplicate. Also, in this case, proteins not obtained in duplicate were filtered out before analysis. For each protein identified, z-scores were calculated considering all the 276 experiments as a population. A protein was considered specifically captured by the antibody when associated to z-score higher than 3. Scatter plots representing z-scores (x-axes) and protein intensities (y-axes) were generated as representation of antibody specificity.
Epitope Mapping
Antibodies were epitope mapped on high-density peptide arrays (Roche NimbleGen) as previously described [13]. Each antibody was pooled with 11 other antibodies prior incubation at 1 μg antibody/ml with arrays containing 16-mer peptides with a 15-residue overlap. The peptides covered the sequences corresponding to the protein fragments used for the generation of HPA008060, HPA007575, HPA029104 and HPA029040 antibodies.
Western Blotting
For Western blot analysis, a serum pool from 20 patients with disease recurrence (9 female, 11 male, median age 61.5) and a pool from 16 patients with no recurrence of melanoma (9 female, 7 male, median age 58.5) were created. These pools of serum were thawed on ice, diluted 1:50 in LDS sample buffer, sample reducing agent (both from NuPAGE Invitrogen) and Milli-Q water. Samples were heated at 95 °C for 5 min and loaded on a Bis-Tris 5–12% gel (Invitrogen) that was subjected to electrophoresis at 200 V (XCell SurLock mini-cell). Following transfer onto a PVDF-membrane and subsequent blocking (blocking reagent for ELISA with 0.1% Tween20, Roche) overnight, primary antibodies were incubated at 1 μg/ml for 24 hours at 4 °C. Blots were developed using HRP-labeled anti-rabbit IgG (Dako) with a chemiluminescent substrate (Immun-Star Western C Kit, Biorad). Images were acquired with a Chemidoc scanner (Bio-Rad Laboratories) using ImageJ software.
Sandwich Assays
To measure S100A6 in the full set of melanoma samples, HPA008060 and a commercial S100A6 antibody (AF4584, R&D Systems) were used as capture reagents together with a monoclonal antibody (CPTC-Calcyclin-3) for detection. Capture reagents were coupled together with four additional anti-S100A6 antibodies (S-Table 2) to magnetic beads and pooled into one bead mixture as above, and efficient coupling was confirmed by both anti-rabbit R-PE and R-PE-labeled anti-mouse (Moss Inc.). One bare bead (empty bead) and normal rabbit IgG were also included in the bead mixture. The detection antibody CPTC-Calcyclin-3 was biotinylated in accordance with previously described protocols [14]. Serum samples were centrifuged (3000 rpm for 3 min), diluted 1:30 in assay buffer (10% normal rIgG in PVXC) and a pipetting robot (Selma, CyBio) was used to add 45 μl of diluted samples to 5 μl beads in two separate 96-well plates (Greiner). Following an overnight incubation, beads were washed 3× PBST on a liquid handler (EL406, Biotek) and 25 μl of labeled detection antibody was added at 0.5 μg/ml in assay buffer for 40 min. After 3x washing of beads, 0.5 μg/ml R-phycoerythring-labeled streptavidin (Invitrogen) in PBST was incubated for 20 min and beads were finally washed and measured in PBST (Lx200, Luminex Corp.).
Tissue Microarray, IHC and Slide Scanning
The generation and characteristics of the melanoma tissue microarray (TMA) cohort has been described elsewhere [15], [16]. In brief, 108 cases of primary malignant melanoma cases were collected as triplicate or duplicate 0.6 mm cores onto three TMA-blocks and used for further studies after review and approval by the Research Ethics Committee (Dnr 2005/232) at Uppsala University, Uppsala, Sweden. IHC and slide scanning were performed at the Swedish SciLifeLab facilities in accordance with protocols described elsewhere [17]. In brief, 4-μm TMA sections collected on SuperFrost Plus slides were deparaffinized in xylene, re-hydrated in graded alcohols, blocked for endogenous peroxidase, and subjected to heat-induced antigen retrieval using PT module buffer 1 (pH = 6, ThermoScientific) in a Decloaking Chamber (Biocare Medical) at 125 °C for 4 minutes. Automated IHC was performed using a LabVision Autostainer 480S (Thermo Fisher Scientific, Runcorn, UK). Primary antibodies used were: S100A6 (HPA007575, Atlas Antibodies, Stockholm, Sweden 1:750) and MTHFD1L (HPA029040, Atlas Antibodies, Stockholm, Sweden 1:250). Antibodies were diluted in UltraAb Diluent (Thermo Fisher Scientific, Fremont, CA, USA). Primary antibodies were applied to the slides for 30 min at room temperature. The slides were subsequently washed and incubated with UltraVision LP kit for 30 min at room temperature (secondary reagent containing anti-rabbit/anti-mouse antibodies conjugated to horseradish peroxidase-labeled polymers, #TL-125 HL, Thermo Fisher Scientific, Runcorn, UK). Following washing steps, the slides were developed for 5 min using 3,3-diaminobenzidine as the substrate (DAB Quanto, Thermo Fisher Scientific, Runcorn, UK). Slides were counterstained in Mayers hematoxylin (01820, Histolab) for 5 minutes using the Autostainer XL (Leica), and then rinsed in lithium carbonate water (diluted 1:5 from saturated solution) for 1 minute. The slides were dehydrated in graded ethanol and lastly coverlipped (PERTEX, Histolab) using an automated glass coverslipper (CV5030, Leica). The slides were scanned using the automated scanning system Aperio XT (Aperio Technologies).
Experimental Study Design
A screening was performed using 4595 antibodies and 32 serum samples from late-stage melanoma patients (N = 16) and non-diseased controls (N = 16) (S-Table 1). This analysis was performed alongside >500 other serum and plasma samples from patients with other types of cancer, neurodegenerative and cardiovascular diseases (data not shown). As a criterion to include antibodies for further analysis, at least two different antibodies raised against the same protein had to reveal a significant difference (P < .01) from Wilcoxon rank-sum test analysis. We also added antibodies based on manual annotations of profiles. This included antibodies that were significant for melanoma case–control comparison but not for any of the other types of cancers when comparing to the respective non-diseased controls (P < .01). To this list, targets were added based on literature surveys and from antibodies used for IHC analysis of tissues from these melanoma patients.
The selected targets were assembled for the generation of a targeted suspension bead array (SBA1). A second SBA (SBA2) was created later in order to expand our analysis and to assess the reproducibility of indicative profiles. SBA2 included multiple antibodies against proteins that showed indicative profiles in SBA1 assays. It also included antibodies against proteins with RNA expression ≥1 FPKM in melanoma tissue and at least 10 times higher RNA expression levels compared to 11 other tumor types (unpublished data), antibodies used for IHC analysis of tissue from these melanoma patients, as well as antibodies that were suggested by plasma Western blot assays.
All serum assays were designed by randomizing samples, replicates of a pooled sample and sample-free controls across microtiter plates. Repeated assays provided three experimental datasets (Assay 1, 2 and 3) that were 2 months and 2 years apart. Assays 1 and 2 use samples from one preparation, while a new sample preparation was made for Assay 3.
Data Analysis
Statistical analysis and visualization of data were performed in R [18] by using the median fluorescent intensities (MFI) for >50 events per bead identity to represent relative protein abundances. Outlying samples were identified and excluded from analysis via robust principal components analysis [19]. All statistical and technical evaluations were based on normalized data and the shown MFI values refer to these.
Normalization of the screening data was performed by probabilistic quotient normalization (PQN) [20] of log-transformed data, followed by MA-loess normalization to adjust for between-plate effects [21]. Data from the 149 samples was normalized by PQN and secondly by linear regression normalization of log-transformed data to adjust for age- and gender effects. The gender covariate was added to the intensity ~ age normalization only if it significantly (ANOVA p-value< 0.05) contributed to the model. Coefficients of variation (CV) were calculated per bead ID to estimate the technical variation across sample replicates.
For comparative analysis, Wilcoxon signed-rank test was used for the two group comparisons recurrence (yes/no), metastasis under follow-up (yes/no), 5-year disease-free survival (yes/no), and gender (female/male). Linear regression models were computed for linear trends in signals over T-stage and age, using log-transformed data. Unsupervised hierarchical clustering was based on Pearson correlations for normalized data that were pre-processed by log-transformation, centering and unit variance scaling.
The immunohistochemically stained melanoma samples were annotated by evaluating all available cores per individual to generate an overall score comprised of an intensity level (1 = Negative, 2 = Weak or 3 = Distinct) and the fraction of stained tumor cells (0 = Negative, 1 = <25%, 2 = 25–75%, or 3 = >75%). The combined intensity and fraction scores for each individual were calculated by multiplying the intensity level (1–3, or) by fraction (1–3, or) and used for statistical analysis of S100A6 in relation to T-stage. All two-group comparisons between clinical variables and data from immunohistochemistry (IHC) stainings and RNAseq expression values were analyzed by Wilcoxon signed-rank tests, and the association from tissue IHC annotations in relation to T-stage by Kendal's rank correlations.
Results
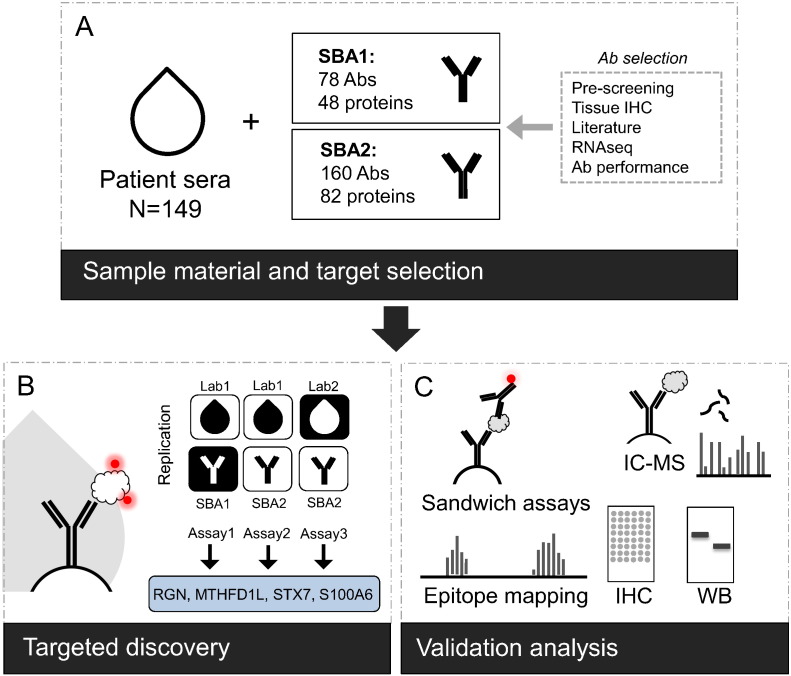
We performed an antibody-based protein profiling of serum samples in context of malignant melanoma. Starting from results of a screening with 16 late-stage melanoma patients versus 16 non-diseased controls, we selected protein candidates to be subsequently assessed for association to melanoma in 149 patient sera. We then compared protein profiles between patients with different stages and outcomes of melanoma to identify protein signatures associated to melanoma in different stages of disease. Antibodies indicating significant associations were further investigated for their binding selectivity, followed by tissue profiling on matched sample material (Figure 1).
Figure 1.
Study overview.
(A) From an initial study employing 4595 antibodies on 16 melanoma cases and 16 non-diseased controls, antibodies against 48 and 82 protein targets were selected for further analysis in 149 sera from melanoma patients. Antibody selection was based on results from the initial study, as well as candidates from immunohistochemical staining on tissue material from matched individuals, literature searches, RNAseq data on melanoma tissue or antibody performance in Western blots with plasma. (B) The 149 patients were analyzed using a direct-labeling assay with 78 antibodies on suspension bead array (SBA1, Assay 1). The performance of the most significant set of 25 antibodies was studied further in a new bead array (SBA2, 160-plex) for repeated analysis of the previously prepared 149 samples (Assay 2). In addition, a new sample labeling and analysis with SBA2 was performed two years later (Assay 3). Antibody profiles for candidate proteins RGN, S100A6, STX7 and MTHFD1L displayed reproducible associations to T-stage or recurrence. (C) Candidate proteins were further evaluated for binding selectivity of antibodies using epitope mapping, immuno-capture mass spectrometry, Western blot, sandwich immunoassays and immunohistochemistry.
Antibody Screening and Target Selection
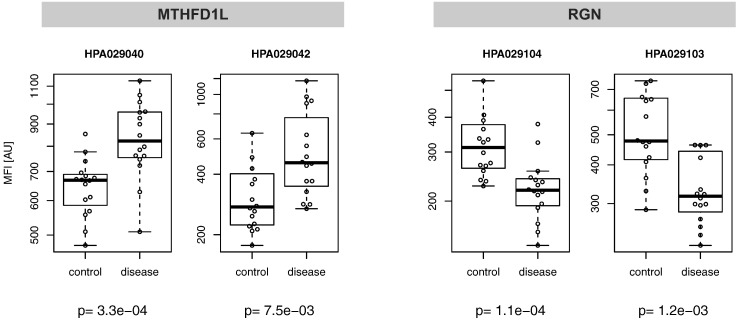
From the initial analysis of 32 serum samples with 4595 antibodies, a set of 31 antibodies passed the selection criterion of P < .01. This included paired antibodies against the proteins regucalcin (RGN, HPA029104; P = 1.1E-4, HPA029103; P = 1.3E-3) and methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like (MTHFD1L, HPA029040; P = 3.3E-4, HPA029042; P = 7.5E-5). The two antibodies against RGN revealed decreased while anti-MTHFD1L binders showed elevated levels in melanoma patients compared to non-diseased controls (Figure 2). In addition to the above, 12 other targets were included for which only one single binder revealed a significant difference (P < .01). This resulted in a list of 43 antibodies targeting 26 unique proteins. To this list, we added 15 proteins (24 antibodies) proposed by literature [22], [23], [24] and 8 candidate proteins (8 antibodies) previously identified in matched tissue material from the same 149 patients as was analyzed within this study [15], [16], [25], including Syntaxin 7 (STX7). Subsequently, 75 antibodies and 3 controls were combined in a suspension bead array (SBA1, 78-plex) that targeted 48 unique proteins. We also generated a second SBA (SBA2, 160-plex) that comprised 67 antibodies against targets selected from SBA1 data, 44 from RNA expression, 30 suggested by plasma Western blot assays, 16 by IHC analysis of melanoma patients (unpublished data), and 3 controls. Thus, SBA2 comprised 25 antibodies against proteins that showed indicative profiles in SBA1 data.
Figure 2.
Candidates from screening analysis.
The initial screening was performed with 32 sera and 4595 antibodies. The main candidates that showed differential detection levels between late-stage melanoma patients and non-diseased controls were antibodies towards the proteins RGN and MTHFD1L.
Assessment of Data and Comparative Analysis
Antibody bead arrays offer high throughput assays to study proteins in serum samples. We developed a study design to confirm the initial findings through the replication of analysis over time and with different antibody compositions of the bead arrays. We utilized SBA1 to initially analyze 149 serum samples from patients with malignant melanoma (Assay 1). SBA2 was then used to repeat the screening of the 149 samples, first by applying the same labeled samples as above (Assay 2) 2 months later and secondly by freeze-thawing and re-labeling the complete sample set another 2 years later (Assay 3), thus allowing for an assessment of the reproducibility of candidates. In all data sets, we observed that a proportion (20–26%) of the antibody profiles were associated to age (P < .05) and thus normalized the data on age prior statistical analysis. A set of 25 antibodies were included in both bead arrays and the median Spearman correlation coefficient for these 25 antibody profiles was rho = 0.64 (−0.11–0.95) and 0.60 (−0.39–0.95) for Assay 1–2 and 1–3 respectively. Among the 160 antibodies applied in Assay 2 and 3, the median correlation was 0.75 (0.22–0.98). As a reference, the distribution of correlation coefficients of random antibody pairs was found to be centered around zero, with median |rho| < 0.1 in all three repeated assays (S-Figure 1). The median technical variation (%CVs) considering all antibodies was <5% in replicated assays, while the median biological variation (%CV for all samples but the replicates) was 20–35%. This assessment deemed the quality of the data for further analysis.
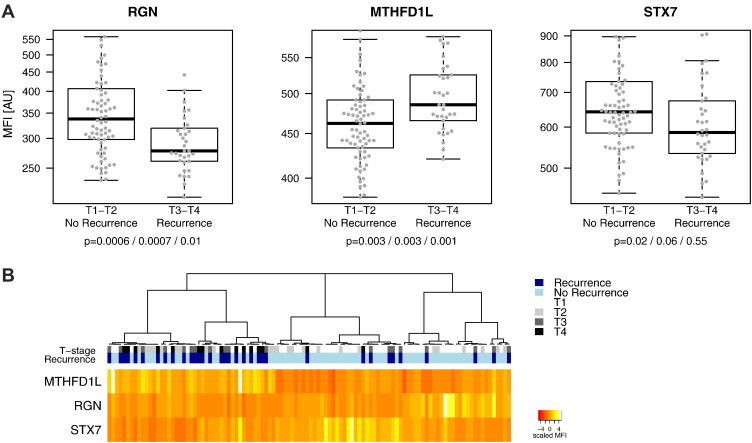
The data from Assays 1, 2, and 3 was analyzed for associations to the clinical parameters T-stage, disease recurrence, metastasis (under follow-up), ulceration and disease-free survival. Ranking of the targets based on P values from Wilcoxon rank sum tests revealed three protein profiles (P < .05) that differed with T-stage (RGN, S100A6, STX7) and two with disease recurrence and metastasis (MTHFD1L and RGN) (Table 2, S-Figure 2). The technical performance of antibodies towards these targets is summarized in Table 3. anti-RGN antibody HPA020104 showed linear and significant decrease over the four T-stages (Assay 1; lm P = .0002, Assay 2; lm P = .002). In case of S100A6, serum levels were significantly lower in T2 compared to patients with other T-stages (S-Figure 2). This was confirmed by a developed sandwich immunoassay that showed a correlation of rho = 0.88 to the SBA data (S-Figures 3 and 4). The levels for RGN, MTHFD1L and STX7 differed in between individuals grouped into patients staged as T1 and T2 (“early stage”) with no recurrence (N = 74) versus those patients staged as T3 and T4 (“late stage”) with disease recurrence (N = 34) (Figure 3A). Unsupervised hierarchical clustering of the multivariate RGN, MTHFD1L, and STX7 profiles for these patients lead to an enrichment of the recurrent and non-recurrent patients within three main clusters (Figure 3B). In one cluster, 70% (21/34) of patients with recurrence were represented, while 72% (53/74) of non-recurrent patients were found in the other two clusters. Cluster analysis within the complete sample of set is presented in S-Figure 5. No notable effects related to age, gender or sampling date were observed (Table 2, S-Figure 6). Associations to disease-free survival or ulceration were not reproducible.
Table 2.
Associations to Clinical Parameters
| Assay | MTHFD1L HPA029040 | RGN HPA029104 | S100A6 HPA008060 | S100A6 HPA007575 | STX7 HPA001467 | |
|---|---|---|---|---|---|---|
| T-stage | 1 | 0.03 | 0.005 | 0.006 | 0.01 | |
| 2 | 0.007 | 0.01 | 0.01 | 0.006 | ||
| 3 | 0.01 | 0.01 | 0.02 | 0.02 | ||
| Recurrence | 1 | 0.0007 | 0.0003 | 0.004 | ||
| 2 | 0.003 | 0.005 | 0.06 | |||
| 3 | 0.0006 | 0.008 | 0.3 | |||
| Metastasis | 1 | 0.007 | 0.0006 | |||
| 2 | 0.004 | 0.01 | ||||
| 3 | 0.002 | 0.08 | ||||
| Age | 1 | 1.0 | 0.6 | 0.7 | 0.5 | 0.5 |
| 2 | 1.0 | 0.1 | 0.7 | 0.4 | 0.8 | |
| 3 | 0.4 | 0.7 | 0.2 | 0.3 | 0.2 | |
| Gender | 1 | 0.9 | 0.1 | 0.1 | 0.2 | 0.04 |
| 2 | 0.4 | 0.2 | 0.4 | 0.4 | 0.4 | |
| 3 | 0.9 | 0.02 | 0.06 | 0.06 | 0.5 |
The table summarizes the association analysis of the candidate profiles to selected clinical parameters in form of p-values. A1: Assay 1 with SBA1; A2: Assay 2 with SBA2; A3: Assay 3 with SBA2. For T-stage Kruskal-Wallis tests were used. Wilcoxon signed-rank test were used for recurrence; Yes/No, Metastasis under follow-up; Yes/No, Age; >60/<60, and Gender (Female/Male). Spearman correlations (rho) represent inter-assay reproducibility.
Table 3.
Antibody Performance
| Assay | MTHFD1L HPA029040 | RGN HPA029104 | S100A6 HPA008060 | S100A6 HPA007575 | STX7 HPA001467 | |
|---|---|---|---|---|---|---|
| CV (%) | 1 | 2.5 | 1.3 | 2.3 | 1.7 | 2.8 |
| 2 | 4.1 | 5.1 | 3.9 | 2.6 | 2 | |
| 3 | 3.1 | 3 | 3.7 | 2 | 1.1 | |
| Correlation (rho) | 1–2 | 0.69 | 0.81 | 0.92 | 0.94 | 0.63 |
| 2–3 | 0.87 | 0.82 | 0.98 | 0.97 | 0.78 | |
| 1–3 | 0.72 | 0.74 | 0.93 | 0.94 | 0.45 |
The performance of the antibodies in the SBA assays are summarized as: Technical variation between replicated samples (%CV) and inter-assay reproducibility represented by Spearman's rho between replicated measurements in Assay 1, 2, and 3.
Figure 3.
Candidate profiles from targeted analysis.
(A) The boxplots illustrate the protein targets with differential serum levels in patients with early stage (T1 and T2) melanoma without recurrence (N = 74) versus late stage (T3 and T4) melanoma patients that had a relapse during the time of follow-up (N = 34). These were detected by anti-RGN (HPA029104), anti-MTHFD1L (HPA029040) and anti-STX7 (HPA001467). Respective p-values are shown for each two-group comparison referring to the repeated analyses (Assay 1, 2 and 3). (B) The heat map visualize unsupervised hierarchical clustering of candidate profiles for the 108 patients in (A), showing three main clusters, of which one was enriched for late T-stage/relapsing (darker blue/gray colors) and the other two for early T-stage/non-relapsing (lighter blue/gray colors) patients respectively.
Antibody Validation
In order to assure that the highlighted antibodies were binding the intended targets, a series of validation assessments were considered and performed (see Supplementary information for further details). An overview of the outcome of the antibody validation assays is given in Table 4 indicating that antibody performance is indeed application dependent [9].
Table 4.
Antibody Validation Assays
| Method | MTHFD1L HPA029040 | RGN HPA029104 | S100A6 HPA008060 | S100A6 HPA007575 | STX7 HPA001467 |
|---|---|---|---|---|---|
| Epitope Mapping | Supportive | Supportive | Supportive | Supportive | - |
| IC-MS | uncertain | Supportive | - | uncertain | Supportive |
| Western Blot | Supportive | uncertain | - | - | - |
| Sandwich immunoassay | uncertain | uncertain | Supportive | Supportive | - |
| IHC | Supportive | - | - | Supportive | Supportive* |
The results of the antibody validation assays are summarized for candidate antibody profiles: Annotation is based on “supportive” and “uncertain”, where the latter implies that one assay was conducted but required further optimization. The annotation “– “refers to that the antibody was not tested in this assay. Indicated by the asterisk (*), the IHC analysis for STX7 has been published elsewhere [16].
First, we checked the data from the replicated SBA assays and the performance of paired antibodies binders to the targets of interest. Candidate profiles were supportively reproducible between the three assays (median rho = 0.81, Table 3). They also demonstrated uniqueness by the low (rho< 0.1) pair-wise correlations between profiles (S-Figure 7). For RGN, the indicative antibody (HPA029104) was directed towards the N-terminal part of RGN and a paired antibody (HPA029102) was included that was raised against the internal region of the protein. The latter binder confirmed the observations of HPA029104 in Assay 1 (P = .02, S-Figure 8), however not in Assay 2 or 3 (P > .05). Also for S100A6, two antibodies (HPA008060 and HPA007575) were both available that were raised against a 72-residue region of S100A6. These binders showed a high correlation with each other in different assays (Assay 1; rho = 0.91, Assay 2; rho = 0.92). No paired antibodies were supportive for the other targets primarily due to signal intensities in the background range. For the two anti-MTHFD1L binders, HPA029040 and HPA029042 that were both found during the initial screening (Figure 2), HPA029042 did not reveal a consistence association to disease in Assays 1, 2 and 3. Secondly, we applied high-density peptide arrays to describe and narrow down the region of antigen binding of antibodies into distinct epitopes, and investigated if these were homologous to sequences of known high abundant serum proteins. The reactivity from anti-MTHFD1L, anti-S100A6 and anti-RGN antibodies is presented in S-Figure 9. A homology search (BLASTp) of all identified epitope regions returned only the targeted proteins MTHFD1L (E-values of 1e-6, 2e-9 and 8e-25), S100A6 (HPA008060; E = 8e-4 and E = 3e-13, HPA007575; E = 0.05) and RGN (E = 0.02) with 100% protein sequence identity at 100% query coverage. The highest scoring alignments to sequences that represent other proteins than the intended targets are presented in S-Table 3. No sequence homology was found to other known serum proteins that could indicate off-target binding.
Next, we performed immunocapture followed by mass spectrometry (IC-MS) in an un-depleted pool of serum (RGN, S100A6) or plasma (STX7, MTHFD1L) from healthy donors. As shown in S-Figure 10, anti-RGN and anti-STX7 specifically captured the cognate target proteins and with enrichment of z-scores 16.6 and 11.5 respectively (S-Table 4, Supplementary data). Anti-S100A6 showed a less pronounced level of enrichment (z-score 2.4). The anti-MTHFD1L binder HPA029040 generated no matching spectra for any expected target peptide (Supplementary data).
Furthermore, we performed Western blot analysis using the anti-MTHFD1L and anti-RGN antibody on sera from melanoma patients. In contrast to the data obtained from IC-MS analysis, the tested RGN antibody did not provide any bands with these sera. However, for anti-MTHFD1, one band close to the molecular weight of transcript variant 8 of MTHFD1L (36.3 kDa) was detected that showed stronger intensity in the pool from patients with disease recurrence (S-Figure 11). Western blot analysis with the other anti-MTHFD1L binder (HPA029042) indicated binding to a protein of different molecular weight.
In summary, we performed a series of antibody validation assays that indicate the challenge of finding a suitable approach for confirming the screening data. However, we found supportive evidence for protein detection of RGN and STX7 using IC-MS, MTHFD1L using Western blot, as well as paired antibodies and sandwich assays for S100A6.
Tissue Profiling
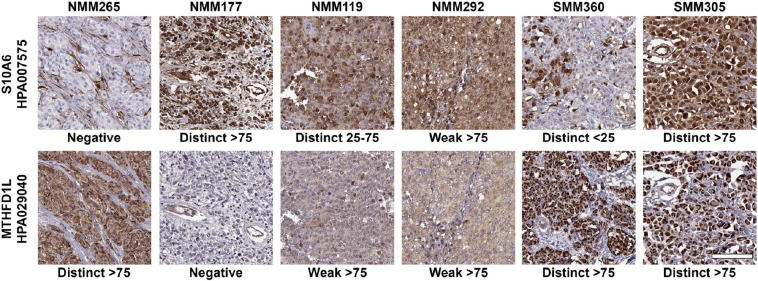
Lastly, we performed immunohistochemistry (IHC) with MTHFD1L (HPA029040) and S100A6 (HPA007575) to stain tissue microarrays built from a subset of 108 individuals among the 149 that had been screened on antibody bead arrays. Figure 4 shows representative images for the observed staining patterns for each of the two proteins. For both for MTHFD1L and S100A6, only 3 patients (2.8%) showed a negative staining (S-Figure 12). The staining intensity of MTHFD1L showed a trend of lower intensity in patients with recurrence (Wilcoxon's P = .065) and lower T-stage (Kendal's tau = 0.069), although there was no association between these two clinical parameters and the fraction of positive cells. In contrast, for S100A6, the fraction of positive cells showed a significant association to T-stage (Kendal's tau = −0.25, P = .008), with a higher fraction of positive cells for the lower T-stages. Also, by combining the S100A6 staining intensity and fraction of positive cells into a continuous score, there was a negative association between score and T-stage (Kendal's tau = −0.20, P = .023). The majority (82%, N = 89) of individuals with positive S100A6 expression showed a distinct staining pattern and of these, most samples (78%, N = 69) revealed >75% positive cells. In this group of 69 patients, the difference in the distribution of patients across T-stages were as follows: T1; 22 patients, T2; 17 patients; T3; 14 patients; T4; 9 patients (S-Figure 13). We did not observe any significant association between tissue staining patterns and serum profiles for S100A6 and MTHFD1L.
Figure 4.
Immunohistochemistry analysis of S100A6 and MTHFD1L in melanoma.
Representative images of S100A6 (top row) and MTHFD1L (bottom row) show the various appearances of immunohistochemical expression patterns in consecutive sections from six different tumors. Staining intensity were scored as negative, weak or distinct and fraction of stained tumor cells was scored as negative, >25%, 25–75% or >75%. Scale bar is 100 μm and valid for all images.
Discussion
An affinity-based approach was used to screen for proteins associated to melanoma in serum samples. A screening of more than 4500 antibodies on 32 serum samples from melanoma patients and controls lead to build a targeted bead array for the analysis of 149 serum samples. In this study, we identified associations for the proteins RGN, MTHFD1L, and STX7 with melanoma recurrence and tumor size (T1-T2 vs. T3-T4). We also identified S100A6 to be differentially abundant in T2 compared to other T-stages in the analyzed serum sample set. Following antibody validation by ultra-dense peptide arrays, immunocapture mass spectrometry, Western blot and sandwich immunoassays to validate the selectivity of the antibodies, we applied the identified antibodies to immunohistochemistry analysis of skin tissue.
This study was initiated by using an unbiased discovery approach to screen serum samples for indicative antibodies. The set of binders was used without any prior knowledge about the relation of their target proteins with disease nor if the antibodies would be able to detect respective targets in the chosen assay system and sample type. In other words, the assay was not designed for known markers of disease and consequently such candidates might have been missed in our analysis (false negatives). For this first screening, we used a set of 32 samples (16 with malignant melanoma diagnosis and 16 controls). At the time of study, 4595 antibodies were available and included due to a concentration suitable for immobilization onto beads (>0.05 mg/ml). This study was performed in parallel to previously presented investigations for multiple sclerosis [21] and osteoporosis [26], [27]. The melanoma-associated profiles have not been highlighted as main candidates in any of the other studies. Further investigations in 149 serum samples then revealed four candidates with discriminative power between T-stage or recurrence of melanoma patients in this sample set. Two of the four highlighted antibodies, namely RGN (Regucalcin), and MTHFD1L (methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1) were proposed by our screening efforts. In addition, antibodies to proteins previously studied in the context of melanoma, namely STX7 (Syntaxin 7) and S100A6 (S100 calcium binding protein A6) were included and highlighted. The negative correlation between STX7 expression and T-stage within this set of tissue samples has previously been described [16]. The trends observed for STX7 in serum were in agreement with these observations.
In the presented study, we observed no correlation between S100A6 serum levels and expression in tissue and immunohistochemistry data showed that lower T-stages were associated with a higher fraction of positive cells. Although data on S100A6 protein expression in cutaneous melanoma tissue is limited, S100A6 has been demonstrated to be expressed in a variety of mesenchymal tumors that arise in the skin [28], [29]. Ribe et al. suggested S100A6 as a potential marker for distinguishing melanoma from the benign melanocytic lesion Spitz nevi [30]. The authors showed that S100A6 was expressed in significantly fewer melanomas than Spitz nevi cases, and that there was a lack of protein expression in normal intraepidermal melanocytes. In another study, Böni et al. demonstrated IHC expression of S100A6 in all examined 39 primary cutaneous melanomas. The study showed that metastases were associated with a weaker S100A6 staining, but only for patients with Breslow thickness <0.75 mm [31].
One novel candidate associated with melanoma was RGN, also known as senescence marker protein 30 (SMP30) and mainly expressed by adrenal glands, liver, kidney and cerebellum. This protein is Ca2+ binding and functions to maintain intracellular Ca2+ homeostasis [32], [33]. Several studies have also suggested that it plays an essential role in the development of carcinogenesis in several tissues and cell types, by its multifunctional and potentially regulatory role on e.g. signal transduction, cell proliferation and apoptosis [34], [35]. In a very recent study, increased mRNA expression levels of RGN were associated to a prolonged relapse-free survival when analyzed in 87 breast cancer patients, which also contributed to the conclusion that RGN may exhibit anticancer properties in human breast cancer [36]. As later discussed for mRNA, our study showed low protein levels of RGN in skin tissue (of melanoma patients and non-disease individuals), which points towards RGN being a potential cancer-related systemic marker and not necessarily specific for malignant melanoma [37].
The other protein was MTHFD1L, a mitochondrial enzyme involved in the synthesis of tetrahydrofolate (THF) in the mitochondrion. Up-regulation of this protein has been suggested to participate in colorectal cancer progression [38] and, being a target of a potential microRNA tumor suppressor, expression of MTHFD1L and other mitochondrial enzymes has been related with tumor maintenance [39], [40]. Interestingly, metabolic reprogramming – one of the hallmarks of cancer – has been proposed to lead to higher expression of mitochondrial folate enzymes such as MTHFD1L. Jain et al. analyzed microarray gene expression data on 60 primary human cancer cell lines and showed that MTHFD1L and other mitochondrial glycine biosynthesis enzymes were expressed at higher levels in rapidly proliferating cell lines [41]. In the same study, MTHFD1L and two additional glycine synthesis enzymes were also shown to be associated with greater mortality in 1300 patients with early stage breast cancer.
In addition to the antibody centric data, we related the mRNA expression levels of the selected candidates by using data available from Cancer Genome Atlas [42]. In a set of 103 tumors from patients with cutaneous melanoma (SKCM), three of the candidate proteins (STX7, MTHFD1L, S100A6) showed medium to high mRNA expression in tumor tissue (median FPKM values; 9.0, 11.7, 930 for STX7, MTHFD1L and S100A6 respectively), whereas RGN expression was negligible (median FPKM = 0.05; S-Figure 14). In this SKCM cohort, data on disease recurrence was missing and samples were mainly classified as T-stage 2 (N = 66) or T-stage 3 (N = 27). The mRNA expression levels of RGN and STX7 were in general lower in the later T-stages (T3 vs. T2: P = .059 and P = .054 respectively, S-Figure 15). We found no association (P > .3) between mRNA expression and age or gender for any of the four candidates.
Besides the availability of binders for the whole proteome, one of the major challenges of affinity based assays is the need for validation [43]. Very recently, guidelines were proposed to use different approaches for confirming that the observed data reports abundance from the intended target of interest [44]. Compared to many other studies found in the literature, we here used a series of assessments to increase the certainty of on-target binding. By including data from epitope mapping, immune-capture mass spectrometry, Western blot and sandwich assays, we aimed to provide detailed information about the antibodies used in the screening. A remaining hurdle for such efforts is that antibodies perform differently well in the different assays. In addition, each type of assay provides unique performance characteristics in terms of analytical sensitivity and selectivity. Each assay indeed has its own requirements when it comes to the preparation, consumption and throughput of samples, as well as antibody consumption and formulation. For example, the anti-MTHFD1L binder HPA029040 revealed supportive data from Western blot analysis while no peptides were identified by mass spectrometry. Similarly, although we were not able to verify antibody specificity for anti-S100A6 using a single IC-MS experiment, an in-house developed sandwich immunoassay verified the selectivity of this antibody when applied for bead-based serum assays. Unfortunately, the study of body fluids presently does not enable overexpression or knock-down of the target protein of interest, thus relies on sensitive methods for the analysis. Even though tremendous advances have been made in the field of mass spectrometry [45], remaining challenges are related to sensitivity, speed and sample consumption of serum analysis [46].
Our study has also some weaknesses and we acknowledge the lack of replication in additional and independent sample sets. This would be needed to confirm that the found associations are truly linked to the disease and not only to the phenotypes of the sample set used here. In this context, other diseases should be evaluated, such as we did during the initial screening efforts. This would allow further assessing the disease specificity and weather changes in protein expression of the melanoma tissue causes altered serum levels. Secondly, the preferred outcome of the discovery effort would include a sandwich assay for the targets, as demonstrated for S100A6. Such assays would offer an attractive endpoint to integrate and translate our findings into further studies. To achieve this, however, new antibodies may have to be generated and tested for their functionality focusing on sandwich assays with serum or plasma samples.
In conclusion, we used antibody based assays for serum analysis of melanoma and found two novel as well as two previously known proteins in the context of the disease. Further and independent studies are though needed to confirm our observations and to provide an extended understanding about the proteins, such as in relation to the pathophysiology, disease progression and disease specificity. The antibodies highlighted though our screening effort may have the potential to contribute to a better understanding of melanoma and could be shortlisted as candidate targets for the development of diagnostic assays for serum samples.
Conflict of Interest Statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Acknowledgements
We thank the entire staff of the Human Protein Atlas and the whole Biobank Profiling group at SciLifeLab in Stockholm for their great efforts. Mun-Gwan Hong is acknowledged for support with statistical analysis. Björn Forsström is acknowledged for support with epitope mapping. We also thank the clinical teams and biobank staffs who helped with sample collection. Adil Mardinoglu and Cheng Zhang are acknowledged for their support for RNA expression analysis. The authors would like to thank Gordon Whiteley and Henry Rodriguez, CPTAC/NCI for kindly providing CPTC antibodies. The KTH Center for Applied Precision Medicine (KCAP) funded by the Erling-Persson Family Foundation is acknowledged for financial support. This work was supported by grants for Science for Life Laboratory, the Knut and Alice Wallenberg Foundation, U-CAN and by the SRA grant from the Swedish Government (CancerUU).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.tranon.2017.03.002.
Appendix A. Supplementary material
Supplementary Information.
Supplementary Data.
References
- 1.Mansfield AS, Markovic SN. Novel therapeutics for the treatment of metastatic melanoma. Future Oncol. 2009;5:543–557. doi: 10.2217/fon.09.15. [DOI] [PubMed] [Google Scholar]
- 2.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG, Ding S. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redpath M, van Kempen L, Robert C, Spatz A. Springer; New York: 2014. Molecular testing in Cutaneous Melanoma Molecular Testing in Cancer; pp. 363–374. [Google Scholar]
- 4.Pernemalm M, Lehtiö J. Mass spectrometry-based plasma proteomics: state of the art and future outlook. Expert Rev Proteomics. 2014;11:431–448. doi: 10.1586/14789450.2014.901157. [DOI] [PubMed] [Google Scholar]
- 5.Ayoglu B, Häggmark A, Neiman M, Igel U, Uhlén M, Schwenk JM, Nilsson P. Systematic antibody and antigen-based proteomic profiling with microarrays. Expert Rev Mol Diagn. 2011;11:219–234. doi: 10.1586/erm.10.110. [DOI] [PubMed] [Google Scholar]
- 6.Taussig MJ, Schmidt R, Cook EA, Stoevesandt O. Development of proteome-wide binding reagents for research and diagnostics. Proteomics Clin Appl. 2013;7:756–766. doi: 10.1002/prca.201300060. [DOI] [PubMed] [Google Scholar]
- 7.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 8.Drobin K, Nilsson P, Schwenk JM. Highly multiplexed antibody suspension bead arrays for plasma protein profiling. Methods Mol Biol. 2013;1023:137–145. doi: 10.1007/978-1-4614-7209-4_8. [DOI] [PubMed] [Google Scholar]
- 9.Fredolini C, Byström S, Pin E, Edfors F, Tamburro D, Iglesias MJ, Häggmark A, Hong MG, Uhlén M, Nilsson P. Immunocapture strategies in translational proteomics. Expert Rev Proteomics. 2016;13:83–98. doi: 10.1586/14789450.2016.1111141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayoglu B, Chaouch A, Lochmüller H, Politano L, Bertini E, Spitali P, Hille M, Niks EH, Gualandi F, Pontén F. Affinity proteomics within rare diseases: a BIO-NMD study for blood biomarkers of muscular dystrophies. EMBO Mol Med. 2014;6:918–936. doi: 10.15252/emmm.201303724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwenk JM, Igel U, Neiman M, Langen H, Becker C, Bjartell A, Pontén F, Wiklund F, Grönberg H, Nilsson P. Toward next generation plasma profiling via heat-induced epitope retrieval and array-based assays. Mol Cell Proteomics. 2010;9:2497–2507. doi: 10.1074/mcp.M110.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Remnestål J, Just D, Mitsios N, Fredolini C, Mulder J, Schwenk JM, Uhlén M, Kultima K, Ingelsson M, Kilander L. CSF profiling of the human brain-enriched proteome reveals associations of neuromodulin and neurogranin to Alzheimer's disease. Proteomics Clin Appl. 2016 doi: 10.1002/prca.201500150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsström B, Axnäs BB, Stengele KP, Bühler J, Albert TJ, Richmond TA, Hu FJ, Nilsson P, Hudson EP, Rockberg J. Proteome-wide epitope mapping of antibodies using ultra-dense peptide arrays. Mol Cell Proteomics. 2014;13:1585–1597. doi: 10.1074/mcp.M113.033308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dezfouli M, Vickovic S, Iglesias MJ, Nilsson P, Schwenk JM, Ahmadian A. Magnetic bead assisted labeling of antibodies at nanogram scale. Proteomics. 2014;14:14–18. doi: 10.1002/pmic.201300283. [DOI] [PubMed] [Google Scholar]
- 15.Bolander A, Agnarsdóttir M, Wagenius G, Strömberg S, Pontén F, Ekman S, Brattström D, Larsson A, Einarsson R, Ullenhag G. Serological and immunohistochemical analysis of S100 and new derivatives as markers for prognosis in patients with malignant melanoma. Melanoma Res. 2008:412–419. doi: 10.1097/CMR.0b013e328315c690. [DOI] [PubMed] [Google Scholar]
- 16.Strömberg S, Agnarsdóttir M, Magnusson K, Rexhepaj E, Bolander A, Lundberg E, Asplund A, Ryan D, Rafferty M, Gallagher WM. Selective expression of Syntaxin-7 protein in benign melanocytes and malignant melanoma. J Proteome Res. 2009:1639–1646. doi: 10.1021/pr800745e. [DOI] [PubMed] [Google Scholar]
- 17.Kampf C, Olsson I, Ryberg U, Sjöstedt E, Pontén F. Production of Tissue Microarrays, Immunohistochemistry Staining and Digitalization Within the Human Protein Atlas. J Vis Exp. 2012;63 doi: 10.3791/3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 19.Hubert M, Rousseeuw PJ, Vanden Branden K. ROBPCA: A new approach to robust principal component analysis. Dent Tech. 2005;47:64–79. [Google Scholar]
- 20.Kato BS, Nicholson G, Neiman M, Rantalainen M, Holmes CC, Barrett A, Uhlén M, Nilsson P, Spector TD, Schwenk JM. Variance decomposition of protein profiles from antibody arrays using a longitudinal twin model. Proteome Sci. 2011;9:73. doi: 10.1186/1477-5956-9-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong MG, Lee W, Nilsson P, Pawitan Y, Schwenk JM. Multidimensional Normalization to Minimize Plate Effects of Suspension Bead Array Data. J Proteome Res. 2016 doi: 10.1021/acs.jproteome.5b01131. [DOI] [PubMed] [Google Scholar]
- 22.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer SR, Erickson LA, Ichetovkin I, Knauer DJ, Markovic SN. Circulating serologic and molecular biomarkers in malignant melanoma. Mayo Clin Proc. 2011;86:981–990. doi: 10.4065/mcp.2011.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma P, Wagner K, Wolchok JD, Allison JP. Novel cancer immunotherapy agents with survival benefit: recent successes and next steps. Nat Rev Cancer. 2011;11:805–812. doi: 10.1038/nrc3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agnarsdóttir M, Sooman L, Bolander A, Strömberg S, Rexhepaj E, Bergqvist M, Pontén F, Gallagher W, Lennartsson J, Ekman S. SOX10 expression in superficial spreading and nodular malignant melanomas. Melanoma Res. 2010;20:468–478. doi: 10.1097/CMR.0b013e3283403ccd. [DOI] [PubMed] [Google Scholar]
- 26.Qundos U, Drobin K, Mattsson C, Hong MG, Sjöberg R, Forsström B, Solomon D, Uhlén M, Nilsson P, Michaëlsson K. Affinity proteomics discovers decreased levels of AMFR in plasma from Osteoporosis patients. Proteomics Clin Appl. 2015 doi: 10.1002/prca.201400167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byström S, Ayoglu B, Häggmark A, Mitsios N, Hong MG, Drobin K, Forsström B, Fredolini C, Khademi M, Amor S. Affinity proteomic profiling of plasma, cerebrospinal fluid, and brain tissue within multiple sclerosis. J Proteome Res. 2014:4607–4619. doi: 10.1021/pr500609e. [DOI] [PubMed] [Google Scholar]
- 28.Fullen DR, Garrisi AJ, Sanders D, Thomas D. Expression of S100A6 protein in a broad spectrum of cutaneous tumors using tissue microarrays. J Cutan Pathol. 2008;35(Suppl. 2):28–34. doi: 10.1111/j.1600-0560.2007.00866.x. [DOI] [PubMed] [Google Scholar]
- 29.Lesniak W, Slomnicki LP, Filipek A. S100A6 - new facts and features. Biochem Biophys Res Commun. 2009;390:1087–1092. doi: 10.1016/j.bbrc.2009.10.150. [DOI] [PubMed] [Google Scholar]
- 30.Ribe A, McNutt NS. S100A6 protein expression is different in Spitz nevi and melanomas. Mod Pathol. 2003;16:505–511. doi: 10.1097/01.MP.0000071128.67149.FD. [DOI] [PubMed] [Google Scholar]
- 31.Böni R, Heizmann CW, Doguoglu A, Ilg EC, Schäfer BW, Dummer R, Burg G. Ca(2+)-binding proteins S100A6 and S100B in primary cutaneous melanoma. J Cutan Pathol. 1997;24:76–80. doi: 10.1111/j.1600-0560.1997.tb01100.x. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi M. Role of regucalcin in maintaining cell homeostasis and function (review) Int J Mol Med. 2005;15:371–389. [PubMed] [Google Scholar]
- 33.Arun P, Aleti V, Parikh K, Manne V, Chilukuri N. Senescence marker protein 30 (SMP30) expression in eukaryotic cells: existence of multiple species and membrane localization. PLoS One. 2011;6:e16545. doi: 10.1371/journal.pone.0016545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamaguchi M. The anti-apoptotic effect of regucalcin is mediated through multisignaling pathways. Apoptosis. 2013;18:1145–1153. doi: 10.1007/s10495-013-0859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi M. Regucalcin, an inhibitor of cell signaling and transcription activity, is involved in suppression of human carcinogenesis. Curr Res Cancer. 2016;1:1–8. [Google Scholar]
- 36.Yamaguchi M, Osuka S, Weitzmann MN, Shoji M, Murata T. Increased regucalcin gene expression extends survival in breast cancer patients: Overexpression of regucalcin suppresses the proliferation and metastatic bone activity in MDA-MB-231 human breast cancer cells in vitro. Int J Oncol. 2016;49:812–822. doi: 10.3892/ijo.2016.3538. [DOI] [PubMed] [Google Scholar]
- 37.Yamaguchi M, Isogai M, Shimada N. Potential sensitivity of hepatic specific protein regucalcin as a marker of chronic liver injury. Mol Cell Biochem. 1997;167:187–190. doi: 10.1023/a:1006859121897. [DOI] [PubMed] [Google Scholar]
- 38.Sugiura T, Nagano Y, Inoue T, Hirotani K. A novel mitochondrial C1-tetrahydrofolate synthetase is upregulated in human colon adenocarcinoma. Biochem Biophys Res Commun. 2004;315:204–211. doi: 10.1016/j.bbrc.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 39.Selcuklu SD, Donoghue MT, Rehmet K, de Souza Gomes M, Fort A, Kovvuru P, Muniyappa MK, Kerin MJ, Enright AJ, Spillane C. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J Biol Chem. 2012;287:29516–29528. doi: 10.1074/jbc.M111.335943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vazquez A, Tedeschi PM, Bertino JR. Overexpression of the mitochondrial folate and glycine-serine pathway: a new determinant of methotrexate selectivity in tumors. Cancer Res. 2013;73:478–482. doi: 10.1158/0008-5472.CAN-12-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas Research N Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker M. Reproducibility crisis: Blame it on the antibodies. Nature. 2015;521:274–276. doi: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- 44.Uhlen M, Bandrowski A, Carr S, Edwards A, Ellenberg J, Lundberg E, Rimm DL, Rodriguez H, Hiltke T, Snyder M. A proposal for validation of antibodies. Nat Methods. 2016;13:823–827. doi: 10.1038/nmeth.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timms JF, Hale OJ, Cramer R. Advances in mass spectrometry-based cancer research and analysis: from cancer proteomics to clinical diagnostics. Expert Rev Proteomics. 2016;13:593–607. doi: 10.1080/14789450.2016.1182431. [DOI] [PubMed] [Google Scholar]
- 46.Fredolini C, Byström S, Sanchez-Rivera L, Tamburro D, Ioannou M, Branca R, Uhlén M, Lehtio J, Nilsson P, Schwenk JM. 2017. Mass spectrometry based validation of antibodies for protein assays in plasma. [Unpublished results] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information.
Supplementary Data.