Abstract
Members of the genus Baylisascaris utilize omnivores or carnivores as their definitive hosts. The best known member of this genus is Baylisascaris procyonis, which is an intestinal parasite of raccoons. The closest relative of B. procyonis is B. columnaris, which utilizes the common skunk as its definitive host. Although B. procyonis has been extensively studied, relatively little is known of B. columnaris. For example, the mitochondrial genome of B. procyonis has been sequenced in its entirety. Conversely, the mitochondrial genome of B. columnaris remains largely unexplored. Likewise, the prevalence of this parasite in its wild host has not been documented. In this study, we collected parasites from a wild population of skunks in the state of Utah, United States. The cytochrome c oxidase subunit 1 and 2 genes, NADH dehydrogenase 2 and several tRNA genes were sequenced from the mitochondrial genome of these parasites. We also determined the prevalence of B. columnaris in a wild population of skunks. In this work we identify several novel polymorphic genetic loci between B. procyonis and B. columnaris. These findings provide additional molecular targets for the differentiation of Baylisascaris species through clarification of genetic differences between B. columnaris and B. procyonis.
Graphical abstract

Highlights
-
•
Prevalence of B. columnaris infection in wild striped skunks ∼60%.
-
•
Several mtDNA genes from B. columnaris sequenced.
-
•
Mitochondrial gene sequences show several species specific SNPs in B. columnaris.
1. Introduction
The Baylisascaris genus of nematodes (family: Ascarididae) is comprised of nine recognized species, each parasitizing distinct definitive hosts and a vast array of intermediate hosts (Bauer, 2013, Kazacos, 2001, Sorvillo et al., 2002). The most widely studied of these is B. procyonis, which primarily utilizes the common raccoon (Procyon lotor) as its principal definitive host (Kazacos, 2001). Embryonated eggs of Baylisascaris species are infectious to over 90 species of mammals and birds including humans. Larval infections with Baylisascaris species can lead to irreversible neural, optical and visceral damage, in both wild animals as well as humans (Gavin et al., 2002, Gavin et al., 2005).
The severity of pathology of Baylisascaris infection in the intermediate host is known to vary depending on the species of the infecting parasite (Tiner, 1953). Understanding which species of Baylisascaris a host is infected with may provide valuable information concerning prognosis and treatment, as well as the source of the infection. As with other nematodes, Baylisascaris species have been traditionally identified through morphometric data. However, distinguishing larval nematodes to the species level is problematic (Graeff-Teixeira et al., 2016). With the current widespread availability of molecular tools, the use of genetic analysis to rapidly and accurately identify organisms to the species level is increasingly standard. However, differentiation of closely related species such as B. procyonis and B. columnaris has proven difficult (Dangoudoubiyam et al., 2009, Gatcombe et al., 2010).
Extensive previous research has documented the life-cycle, distribution and prevalence of B. procyonis (Graeff-Teixeira et al., 2016, Kazacos and Boyce, 1989, Roussere et al., 2003). Additionally, the complete mt genome of B. procyonis, has been sequenced (Xie et al., 2011a). In contrast to the relative abundance of studies on B. procyonis, little is known of its closest relative, B. columnaris. Previous research using copromicroscopic detection has shown B. columnaris infection prevalence of ∼25% in a population of Eastern Spotted skunks (Spilogale putorius) (Lesmeister et al., 2008) and 25% prevalence in captive Striped skunks in Europe (d'Ovidio et al., 2017). To our knowledge, the prevalence of B. columnaris in wild populations of Striped skunks has not previously been reported. Recently, Franssen et al. (2013) reported the cloning and sequencing of partial sequences of B. columnaris for the Cox1 (413 bp) and Cox2 (483 bp) genes. Additionally, in this work Franssen et al. identified three single nucleotide polymorphisms (SNPs) in the partial Cox1 sequence and one SNP found in the partial Cox2 sequence which are useful in differentiating B. procyonis from B. columnaris. The partial sequence of these mitochondrial genes has provided a reference which has facilitated the differentiation of B. procyonis from B. columnaris by DNA sequencing. In addition to the four SNPs which allow for the molecular differentiation of B. procyonis and B. columnaris, Franssen et al. also reported a number of intragenic differences in B. columnaris mitochondrial sequences suggesting there is a high degree of genetic diversity in this species.
In this study we sought to determine the prevalence of B. columnaris in a wild population of Striped skunks (Mephitis mephitis) in Salt Lake County, Utah, USA. In addition to determining the prevalence of these parasites in a native skunk population we report the complete sequence of 11 mitochondrial genes (Cox1, Cox2, ND2 and 8 tRNA genes) comprising 3638 bp of the B. columnaris mitochondrial genome. Results revealed several novel SNPs in these genes, which further facilitates and improves the molecular distinction between B. procyonis and B. columnaris. Additionally, several intragenic SNPs were identified in B. columnaris worms. In summary, these results demonstrate the prevalence of B. columnaris in a wild population of skunks and extend the number of known genetic differences between B. procyonis and B. columnaris.
2. Methods
2.1. Animals
Skunks were acquired through nuisance animal calls from the public in Salt Lake County, Utah, USA during the fall and winter of 2013. Institutional Animal Care and Use Committee (IACUC) or ethics committee approval was not necessary, as animals were not sacrificed for research purposes. All skunks were collected by U.S. Department of Agriculture employees as part of their routine duties. No animals were trapped or euthanized for the purposes of this study. Skunks were captured in Live traps (Tru-Catch, Belle Fourche, South Dakota, USA). Skunks were euthanized by chemical immobilization with 5/1 ketamine/xylazine followed by intracardiac injection of potassium chloride. The presence of nematode parasites was determined by emptying the small intestinal contents through manual extrusion using finger/grip pressure and the visual examination of intestinal contents.
2.2. Parasite identification
Species determination of collected worms was performed by extracting DNA from one or more worms from each skunk and performing DNA sequencing of the Cox1 gene. Sequences were aligned with the previously published Cox1 to determine worm genus and species (GenBank: KC543474.1) (https://blast.ncbi.nlm.nih.gov). A total of 34 worms were collected and the Cox1 gene of 22 of these worms was sequenced. All sequenced worms were identified by DNA sequence as B. columnaris. A single worm from several animals was then used for more extensive DNA sequencing of a number of genes as described below.
2.3. Primer design
PCR primers were designed based on the mitochondrial genome of B. procyonis (ac. No. JF951366), using Primer 3 software (Untergasser et al., 2012). In order to ensure amplification of the gene of interest, primers were constructed to be within highly conserved regions of the mitochondrial genome. Primers were designed to flank the gene of interest by ∼100–300 bp from the 5′ and 3′ ends. This primer design resulted in the amplification of segments of DNA which contained the complete gene of interest (Cox1, Cox2, ND2) as well as several small tRNA genes contained at the 5′ and 3’ ends of each amplicon (Table 1).
Table 1.
Amplicon primers.
| Primer Name | Target genes | Forward Primer | Reverse Primer |
|---|---|---|---|
| Amplicon 1 | Q, R, I tRNA ND2, & S, L, K tRNA | CCGTTGGCCTTAACTGTTCG | GGCAACCCAACAACCATAG |
| Amplicon 2 | COX1 | CGGGTTTTCTGTTCCTGTGG | ACCTGATTGGAAGTCAGGTG |
| Amplicon 3 | D, G tRNA COX2, & H tRNA | GTGGATAAGGGGCCTTGTG | ATAAACCCCCGCCAGTTCTC |
Three separate amplicons were generated with Amplicon 1 containing the sequences for the tRNA genes Q, R, I, S, and L, as well as the complete ND2 gene. Amplicon 2 contained the complete Cox1 gene and Amplicon 3 contained the tRNA genes D, G, and H as well as the complete Cox2 gene. Primer sequences for each amplicon are contained in Table 1.
2.4. DNA collection and PCR conditions
Upon collection of parasites (described above), individual specimens were preserved by immediately freezing each worm at −80 C. For genetic analysis, specimens were thawed and a ∼2.0 mm portion of each worm macerated with a razor blade. The macerated tissue samples were then placed in a 1.5 mL microcentrifuge tube and DNA extracted using the Qiagen DNeasy Blood & Tissue Kit (Qiagen Inc., Valencia, CA, USA) per manufacturer instructions.
PCR reactions were carried out in a final reaction volume of 50 μl containing 2 μl of DNA, 25 μl of OneTaq DNA polymerase (New England Biolabs, Ipswich, MA USA), 0.5 μl of each primer with 22 μl of molecular grade water. Polymerase chain reaction conditions used to amplify the Cox1 and Cox2 containing amplicons (Amplicons 2 and 3), involved a 30 s initial denaturation at 94 °C followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and elongation at 68 °C for 80 s, followed by a 10-min final extension at 68 °C. The PCR amplification of the ND2 target region (Amplicon 1) was identical to the aforementioned profile, with the exception of the annealing temperature being 63 °C.
2.5. DNA sequencing
PCR reaction products were electrophoresed in 1.5% agarose gel for 45 min. The PCR amplified products of interest for each amplicon were then extracted and gel purified using the Wizard SV Gel and PCR Clean-Up System (Promega Madison, WI). The sequencing of each amplicon was performed using primers designed to bind approximately every 500 bp (Supplemental Fig. 1). Cycle sequencing was performed by the Brigham Young University DNA Sequencing Center using an ABI 3730xl automated sequencer. Primers were designed using the forward DNA strand and sequencing was performed on a single strand. Contigs for each sample were assembled by mapping sample reads to the mitochondrial genome of B. procyonis (ac. No. JF951366). Sequence reads were determined using Geneious software (Biomatters Limited, San Francisco, CA, USA). Overlapping regions of contigs for each sample were aligned to generate consensus reads. SNP analysis, multiple alignments, and prediction of transmembrane and cytoplasmic regions were performed using Geneious software.
3. Results
3.1. B. columnaris prevalence in a wild population of skunks
We first sought to determine the prevalence of B. columnaris in a wild population of skunks. The intestinal tracts of 16 skunks were collected and the prevalence of nematode infection determined by gross examination of the intestinal contents. Ten of the 16 skunks examined were found to be infected with roundworms, with numbers of worms ranging from 1 to 10 in infected animals (Table 2). All worms were identified as B. columnaris based on gross morphology and confirmed by detailed sequence homology to the published partial sequence of B. columnaris Cox1 (GenBank: KC543474.1).
Table 2.
Parasite prevalence.
| Host Name |
Worm sequenced |
|
|---|---|---|
| A | yes | 1 |
| Z | no | 0 |
| Y | no | 0 |
| B | yes | 1 |
| C | yes | 4 |
| D | yes | 2 |
| X | no | 0 |
| E | yes | 4 |
| W | no | 0 |
| F | yes | 8 |
| G | yes | 1 |
| H | yes | 10 |
| V | no | 0 |
| I | yes | 2 |
| J | yes | 1 |
| U | no | 0 |
| Total | 10 | 34 |
3.2. DNA sequence heterogeneity between B. procyonis and B. columnaris
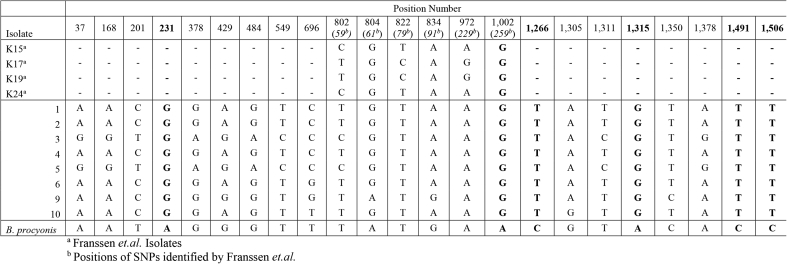
Due to the essential function of Cox1, this gene is present in a wide variety of organisms yet has sufficient variation to distinguish closely related species (Hebert et al., 2003). Consequently, the sequence of the Cox1 gene is a frequently used marker for population genetic and phylogenetic studies (Ai et al., 2011, Xie et al., 2011b). In this study, the complete Cox1 gene (1578 bp) of eight Baylisascaris worms isolated from eight different hosts was determined (Table 3). Our analysis revealed 11 novel loci, which consistently distinguished B. columnaris from B. procyonis, including the three previously reported by Franssen et al. (Fig. 1). The majority of these species specific SNPs are found in the 3’ portion of the gene with 8 of the 11 being found between nucleotides 1002 and 1506 of the Cox1 gene. Previously published work has shown a relatively high degree of intraspecies heterogeneity in the Cox1 gene of B. columnaris (Franssen et al., 2013). In our analysis of Cox1, 11 total intraspecies SNPs were identified 10 of these having not previously been reported (Fig. 1).
Table 3.
Sequence data.
Fig. 1.
Single nucleotide polymorphisms in the Cox1 gene of B. columnaris, compared to B. procyonis. Nucleotide position numbers are shown at the top of the figure. Italicized numbers represent the position number from a previously published partial sequence of the B. columnaris Cox1 gene (Franssen et al., 2013). Species-specific SNPs are shown in bold.
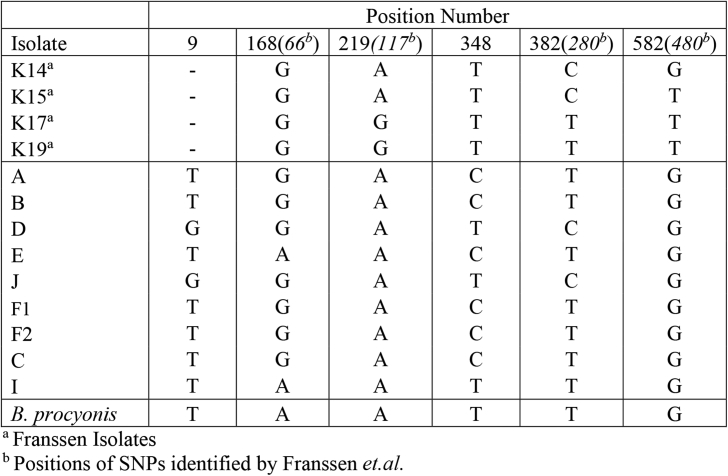
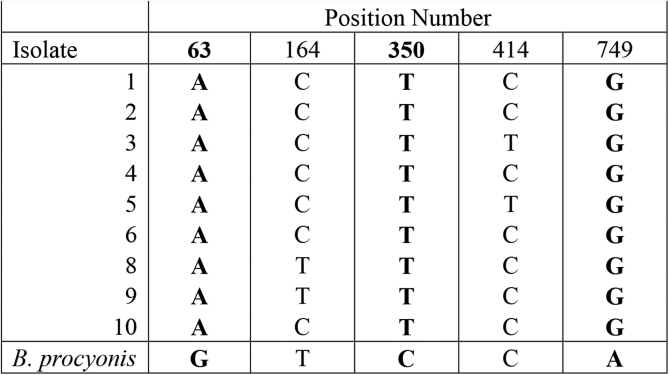
Previous sequencing of a partial sequence of the B. columnaris Cox2 gene has shown one specific SNP which was useful in differentiating B. procyonis from B. columnaris as well as three intragenic SNPs (Franssen et al., 2013). These previous studies were done in a European population of skunks. In our sequencing analysis of a North American population, we found six intragenic SNPs within the Cox2 gene. Four of these SNPs were previously identified by Franssen et al. (2013). Importantly, our data showed that none of these nucleotide variations were species specific, whereas previous analysis had suggested the SNP at position 168 was useful in differentiating B. columnaris from B. procyonis (Fig. 2). There have been no previous reports of sequencing of the B. columnaris ND2 gene. In our sequencing of this gene and subsequent analysis, three SNPs were identified which consistently differentiated B. procyonis from B. columnaris. In addition, two intraspecies SNP were identified in the ND2 gene of B. columnaris (Fig. 3).
Fig. 2.
Single nucleotide polymorphisms in the Cox2 gene of B. columnaris, compared to B. procyonis. Nucleotide position numbers are shown at the top of the figure. Italicized numbers represent the position number from a previously published partial sequence of the B. columnaris Cox2 gene (Franssen et al., 2013).
Fig. 3.
Single nucleotide polymorphisms in the ND2 gene of B. columnaris, compared to B. procyonis. Nucleotide position numbers are shown at the top of the figure. Species-specific SNPs are shown in bold.
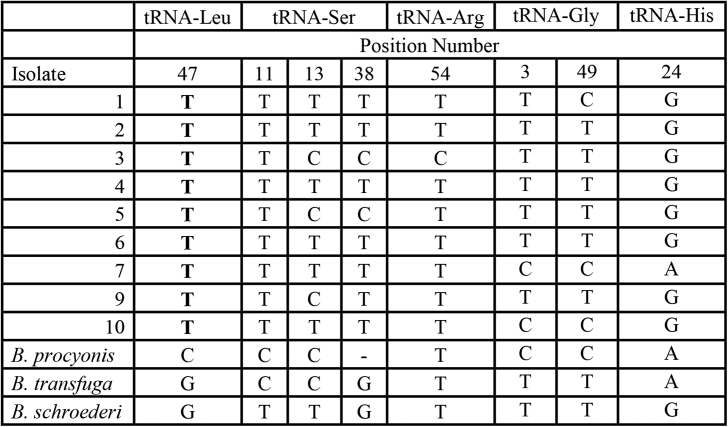
Alignment of eight of the B. columnaris concatenated sequences and homologous regions of B. procyonis were used to determine the sequence of several tRNA genes from B. columnaris. This analysis revealed two SNPs and an indel within mt-tRNA genes which differentiated B. procyonis from B. columnaris. In addition, five intragenic SNPs were identified (one of them being in the same position as the indel of tRNA S (Fig. 4)). Based on the high degree of similarity between these organisms, we were initially skeptical of this degree of variation in tRNA genes. Therefore, we next compared the tRNA genes from all mt-DNA genomes of sequenced Baylisascaris species. This comparison demonstrated a relatively high degree of sequence variation in the tRNA genes of several closely related Baylisascaris species (Fig. 4). This level of heterogeneity in tRNA sequences from numerous Baylisascaris species lends confidence in this unanticipated level of heterogeneity if tRNA genes. Additionally, concatenated sequences were used to generate maximum likelihood relationships of B. columnaris with other Ascarid species. Results of these analyses demonstrated maximum likelihood relationships in agreement with previous findings (Franssen et al., 2013).
Fig. 4.
Single nucleotide polymorphisms in several tRNA genes of B. columnaris, compared to B. procyonis, B. transfuga and B. schroederi. Nucleotide position numbers are shown at the top of the figure. SNPs which distinguish B. columnaris from other Baylisascaris species are shown in bold.
4. Discussion
Both skunks as well as raccoons commonly live in urban areas facilitating human contact and potential ingestion of embryonated eggs which can cause visceral, ocular and neural larval migrans (Roussere et al., 2003, Sorvillo et al., 2002). In the USA several cases of debilitating as well as fatal larval migrans has been attributed to B. procyonis. Until recently, the molecular identification of B. columnaris was not possible due to the lack of any DNA sequences in public databases. In 2009, a partial (529 bp) sequence of the B. columnaris Cox2 gene was generated (Dangoudoubiyam et al., 2009). In 2013 work by Franssen et al. resulted in partial sequences for B. columnaris Cox1 (413 bp) and Cox2 (483 bp) genes (Franssen et al., 2013). The availability of these sequences has facilitated the molecular identification of these parasites for us as well as other researchers (d'Ovidio et al., 2017).
In this study, we utilized gene sequencing to identify roundworms as B. columnaris. In this population of wild skunks we show the incidence of B. columnaris infection to be ∼60%. We then extended earlier findings by sequencing a total of 3638 bp of the mitochondrial genome of B. columnaris. In so doing several novel SNPs were identified, facilitating the molecular discrimination of B. columnaris from B. procyonis. Additionally, several intragenic SNPs in B. columnaris isolates were identified. These SNPs provide additional species specific targets for the molecular differentiation of B. columnaris from B. procyonis. Importantly this data demonstrate that three previously reported species specific SNPs do not accurately differentiate B. procyonis from B. columnaris (168 of Cox2 and 804 and 834 of Cox1). All of these SNPs involved transitions between the purines G and A. This unique finding, compared to previously published work, is likely due to the previous study being done in The Netherlands, while skunks (and by extension parasites of skunks) are native to the Americas. Parasites infecting European skunks are likely from a relatively small founder population of nematodes infecting skunks transported outside of North America. It is logical to assume that the nematodes (as well as the skunks) would have less genetic diversity than a native, free roaming population of animals.
A recent study on the prevalence of B. columnaris in captive European skunks found that ∼25% of skunks tested were infected with B. columnaris (d'Ovidio et al., 2017). Data indicates that in our study area there is a much higher infection rate of Striped skunks than reported in studies of captive Striped skunks in Europe. This is likely due to wild animals encountering other wild skunks as well as infected intermediate hosts more commonly than captive skunks. As expected, more intragenic variation of parasites was observed from this population of native wild skunks compared to the imported non-native population studied previously (Franssen et al., 2013). The higher prevalence of parasites in our study compared to a previous study of Spotted skunks (Lesmeister et al., 2008) is likely due to the difficulty in accurately identifying infected animals through fecal analysis compared to our method of visual inspection of the small intestine. Additionally, several of the animals in our study had very few worms infecting them. Animals infected with a single male worm would not be detected by microscopic analysis of feces but would easily be identified through visual inspection on the intestinal contents. Differences in the susceptibility to Baylisascaris infection by these two types of skunks, as well as other environmental factors may also play a role in variations in infection prevalence.
In summary, in this study several novel intragenic SNPs were identified. Additionally, nine novel polymorphisms are identified which aid in the molecular differentiation of B. procyonis from B. columnaris. Three polymorphisms, which were previously thought to differentiate these two species were shown to in fact be intragenic rather than species specific SNPs. These findings provide for more accurate molecular differentiation of these closely related parasites.
Acknowledgments
The authors acknowledge the help of Cameron Sargent and Elliot Moss for their technical assistance in isolating DNA.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ijppaw.2017.03.009.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Ai L., Chen M.X., Alasaad S., Elsheikha H.M., Li J., Li H.L., Lin R.Q., Zou F.C., Zhu X.Q., Chen J.X. Genetic characterization, species differentiation and detection of Fasciola spp. by molecular approaches. Parasit. Vectors. 2011;4:101. doi: 10.1186/1756-3305-4-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C. Baylisascariosis–infections of animals and humans with 'unusual' roundworms. Vet. Parasitol. 2013;193:404–412. doi: 10.1016/j.vetpar.2012.12.036. [DOI] [PubMed] [Google Scholar]
- d'Ovidio D., Pantchev N., Noviello E., Del Prete L., Maurelli M.P., Cringoli G., Rinaldi L. Survey of Baylisascaris spp. in captive striped skunks (Mephitis mephitis) in some European areas. Parasitol. Res. 2017;116:483–486. doi: 10.1007/s00436-016-5307-8. [DOI] [PubMed] [Google Scholar]
- Dangoudoubiyam S., Vemulapalli R., Kazacos K.R. PCR assays for detection of Baylisascaris procyonis eggs and larvae. J. Parasitol. 2009;95:571–577. doi: 10.1645/GE-1905.1. [DOI] [PubMed] [Google Scholar]
- Franssen F., Xie K., Sprong H., van der Giessen J. Molecular analysis of Baylisascaris columnaris revealed mitochondrial and nuclear polymorphisms. Parasit. Vectors. 2013;6:124. doi: 10.1186/1756-3305-6-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatcombe R.R., Jothikumar N., Dangoudoubiyam S., Kazacos K.R., Hill V.R. Evaluation of a molecular beacon real-time PCR assay for detection of Baylisascaris procyonis in different soil types and water samples. Parasitol. Res. 2010;106:499–504. doi: 10.1007/s00436-009-1692-6. [DOI] [PubMed] [Google Scholar]
- Gavin P.J., Kazacos K.R., Shulman S.T. Baylisascariasis. Clin. Microbiol. Rev. 2005;18:703–718. doi: 10.1128/CMR.18.4.703-718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin P.J., Kazacos K.R., Tan T.Q., Brinkman W.B., Byrd S.E., Davis A.T., Mets M.B., Shulman S.T. Neural larva migrans caused by the raccoon roundworm Baylisascaris procyonis. Pediatr. Infect. Dis. J. 2002;21:971–975. doi: 10.1097/00006454-200210000-00017. [DOI] [PubMed] [Google Scholar]
- Graeff-Teixeira C., Morassutti A.L., Kazacos K.R. Update on Baylisascariasis, a highly pathogenic zoonotic infection. Clin. Microbiol. Rev. 2016;29:375–399. doi: 10.1128/CMR.00044-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D., Ratnasingham S., deWaard J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. Biol. Sci. 2003;270(Suppl. 1):S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazacos K.R. Baylisascaris procyonis and related species. In: Samuel W.M., Pybus M.J., Kocan A.A., editors. Parasitic Diseases of Wild Mammals. second ed. 2001. pp. 301–341. [Google Scholar]
- Kazacos K.R., Boyce W.M. Baylisascaris larva migrans. J. Am. Vet. Med. Assoc. 1989;195:894–903. [PubMed] [Google Scholar]
- Lesmeister D.B., Millspaugh J.J., Wade S.E., Gompper M.E. A survey of parasites identified in the feces of eastern spotted skunks (Spilogale putorius) in western Arkansas. J. Wildl. Dis. 2008;44:1041–1044. doi: 10.7589/0090-3558-44.4.1041. [DOI] [PubMed] [Google Scholar]
- Roussere G.P., Murray W.J., Raudenbush C.B., Kutilek M.J., Levee D.J., Kazacos K.R. Raccoon roundworm eggs near homes and risk for larva migrans disease, California communities. Emerg. Infect. Dis. 2003;9:1516–1522. doi: 10.3201/eid0912.030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorvillo F., Ash L.R., Berlin O.G., Morse S.A. Baylisascaris procyonis: an emerging helminthic zoonosis. Emerg. Infect. Dis. 2002;8:355–359. doi: 10.3201/eid0804.010273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiner J.D. The migration, distribution in the brain, and growth of ascarid larvae in rodents. J. Infect. Dis. 1953;92:105–113. doi: 10.1093/infdis/92.2.105. [DOI] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Zhang Z., Niu L., Wang Q., Wang C., Lan J., Deng J., Fu Y., Nie H., Yan N., Yang D., Hao G., Gu X., Wang S., Peng X., Yang G. The mitochondrial genome of Baylisascaris procyonis. PLoS One. 2011;6:e27066. doi: 10.1371/journal.pone.0027066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Zhang Z., Wang C., Lan J., Li Y., Chen Z., Fu Y., Nie H., Yan N., Gu X., Wang S., Peng X., Yang G. Complete mitochondrial genomes of Baylisascaris schroederi, Baylisascaris ailuri and Baylisascaris transfuga from giant panda, red panda and polar bear. Gene. 2011;482:59–67. doi: 10.1016/j.gene.2011.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.