Abstract
Background
Selenium and single-nucleotide-polymorphisms in selenoprotein genes have been associated to diabetes. However, the interaction of selenium with genetic variation in diabetes and oxidative stress-related genes has not been evaluated as a potential determinant of diabetes risk.
Methods
We evaluated the cross-sectional and prospective associations of plasma selenium concentrations with type 2 diabetes, and the interaction of selenium concentrations with genetic variation in candidate polymorphisms, in a representative sample of 1452 men and women aged 18–85 years from Spain.
Results
The geometric mean of plasma selenium levels in the study sample was 84.2 µg/L. 120 participants had diabetes at baseline. Among diabetes-free participants who were not lost during the follow-up (N=1234), 75 developed diabetes over time. The multivariable adjusted odds ratios (95% confidence interval) for diabetes prevalence comparing the second and third to the first tertiles of plasma selenium levels were 1.80 (1.03, 3.14) and 1.97 (1.14, 3.41), respectively. The corresponding hazard ratios (95% CI) for diabetes incidence were 1.76 (0.96, 3.22) and 1.80 (0.98, 3.31), respectively. In addition, we observed significant interactions between selenium and polymorphisms in PPARGC1A, and in genes encoding mitochondrial proteins, such as BCS1L and SDHA, and suggestive interactions of selenium with other genes related to selenoproteins and redox metabolism.
Conclusions
Plasma selenium was positively associated with prevalent and incident diabetes. While the statistical interactions of selenium with polymorphisms involved in regulation of redox and insulin signaling pathways provide biological plausibility to the positive associations of selenium with diabetes, further research is needed to elucidate the causal pathways underlying these associations.
1. Introduction
While early experimental studies suggested a potential role of selenium, an essential nutrient and a component of antioxidant selenoproteins, as an anti-diabetic factor, [[1], [2]] the evidence from human studies has been controversial. Most [3], [4], [5], [6], but not all [7], [8], cross-sectional studies of selenium and type 2 diabetes identified statistically significant positive associations. The prospective ORDET cohort study showed that increased dietary selenium intake was associated with an increased risk of type 2 diabetes [9]. Conversely, selenium at dietary levels of intake was inversely associated with incident diabetes in two separate cohorts of U.S. men (mean toenail selenium 0.84 µg/g) and women (mean toenail selenium 0.77 µg/g) [10]. Other prospective studies have shown null results [6], [11]. More recently, a case-control study in a population from Northern Taiwan found that high serum selenium levels were associated with increased risk for diabetes mellitus [12]. Moreover, positive, although in some cases not statistically significant, associations from clinical trials in selenium-repleted populations supported concerns that high selenium exposure may not be beneficial for type 2 diabetes or insulin resistance [13], [14], [15], [16], [17].
Genetic studies indicate that type 2 diabetes is a highly heritable trait [18]. A number of single-nucleotide-polymorphisms (SNPs) in selenoprotein genes, including SEPS1, SEPP1, GPX1, and GPX4, have been associated to diabetes-related conditions [19], [20], [21], [22], supporting that redox unbalance may be an underlying mechanism in altered glucose metabolism and β-cell dysfunction. Nonetheless, the interaction of selenium status with genetic variation in diabetes and oxidative stress-related genes has not been evaluated as a potential determinant of type 2 diabetes.
The objective of this study was thus to evaluate the gene-environment interactions between plasma levels of selenium and genetic variants related with diabetes or redox metabolic pathways on the prevalence and incidence of diabetes in a population-based study conducted in Spain. Evaluating the interaction of selenium exposure and polymorphisms in candidate genes can potentially provide etiologic insight into the selenium-diabetes relation.
2. Methods
2.1. Study population
The Hortega Study is a population-based survey among adults 15–85 years old residing in the catchment area of the Rio Hortega University Hospital in Valladolid (Spain). In Spain, tertiary hospitals have specific geographic areas for patient referral and integrate the network of primary care centers in each area. The baseline examination visits started in 2001 and were completed in 2003. The study design and data collection methods for the baseline examination have been described elsewhere [23]. Incident health endpoints and deaths that occurred after the baseline examination visit were assessed in 2015 by reviews of health records including primary care, hospitalization and mortality. Records were reviewed by 2 physician reviewers who adjudicated incident events. The study sample consisted of 1502 total participants with stored plasma samples. For the cross-sectional analysis, we excluded 12 participants missing selenium measures, 20 participants missing smoking-related variables, and 17 participants missing data in other relevant covariates, leaving 1452 participants. For prospective analysis we further excluded 120 prevalent cases of diabetes, and 99 individuals who were lost during follow up, leaving 1234 diabetes-free participants at the baseline examination. The research protocol was approved by the Ethics Committee of the Rio Hortega University Hospital and all participants provided written informed consent.
2.2. Plasma selenium levels
We measured plasma selenium levels by inductively coupled-plasma mass spectrometry (ICPMS) on an Agilent 7500CEx ICPMS (Agilent Technologies, United States) following a standardized protocol. The lower and upper detection limits for plasma selenium levels were 29.85 and 205.30 µg/L, respectively. No samples had levels below or above these limits. The inter-assay coefficient of variation for plasma selenium levels was 5.6%. In order to ensure analytical accuracy, a commercially available reference material (Scharlau Standard Solution), which is traceable to the corresponding National Institute of Standards and Technology reference material, was used. Additional internal controls were employed for daily quality control. Acceptable control measures were within two standard deviations of reference means.
2.3. Type 2 diabetes
In the Hortega study, blood samples were obtained after an average of 3 h since the last intake of food (range 0–17). Glucose was measured by the glucose oxidase method with an autoanalyzer (Hitachi 704, Boehringer Mannheim, Germany). A second fasting glucose determination was obtained only in subjects with non-fasting glucose levels equal or higher than 140 mg/dl. Glycosylated hemoglobin (HbA1c) levels were measured from capillary blood samples with a DCA 2000 HbA1c analyzer (Bayer Diagnostics, Tarrytown, NY). Subjects were considered as having prevalent diabetes if, at the time of the baseline examination, they had been previously diagnosed of type 2 diabetes by a physician, if they had a record of use of diabetes medications in the clinical history, if fasting plasma glucose was >126 mg/dl, or if HbA1c was 6.5%. Subjects were considered as having incident diabetes if they were diabetes-free at the time of the baseline examination and they fulfilled the diabetes definition during the follow-up based on the health records.
2.4. DNA isolation, SNP selection and genotyping
We isolated DNA from peripheral blood cells using Chemagic System (Chemagen), and quantified it with PicoGreen dsDNA Quantification Reagent (Invitrogen, Carlsbad, CA, USA). DNA was diluted to a final concentration of 100 ng/µl. We used bibliography searches and the SYSNPS program [24], based on public data sources including Ensembl and HapMap (GRCh36), to identify 597 single nucleotide polymorphisms (SNPs) from 155 candidate genes implicated in diabetes metabolic pathways such as inflammation, redox homeostasis and response to insulin or gluconeogenesis. We included SNPs previously reported to be related to health outcomes in humans or to have functional implications.
The SNPs were genotyped using an oligo-ligation-assay (SNPlex, Applied Biosystems, Foster City) following the manufacturer's guidelines. The polymorphisms nomenclature was based in Den Dunnen JT et al. recommendations. The mean genotyping coverage across all genotyped SNPs was 95%. We excluded 42 SNPs because of a genotyping coverage <90%, 59 SNPs because they had less than 3 genotypes and 41 SNPs because they did not meet a Hardy-Weinberg equilibrium p-value <0.01. 101 SNPs were further excluded because they had a MAF<0.05 or a minor genotype frequency <20 in the study population. The final number of SNPs included in gene-environment interaction analyses was 354.
2.5. Other variables
Information on age, sex, education, smoking status and alcohol intake was based on self-report. Body mass index (BMI) was calculated using measured height and weight. Urine cotinine was measured by enzyme-linked immunosorbent assay (ELISA) (Kit “Análisis DRI® Cotinina”, Microgenics laboratories). 1121 individuals had cotinine concentrations below the limit of detection (34 ng/mL).
2.6. Statistical methods
We used the survey package in R software (version 3.30; R Development Core Team 2016) to account for the sampling design and survey weights. Plasma selenium concentrations were log-transformed for analyses. Cut-offs for plasma selenium tertiles were based on weighted distributions in the study sample. For the cross-sectional analysis, we estimated multi-adjusted odd ratios of type 2 diabetes (with 95% CIs) by selenium levels using logistic regression models. For the prospective analysis, we used Cox proportional hazard models with age as time scale and age at baseline treated as staggered entries. Follow up time was calculated in years from the date of baseline visit to the date of newly diagnosed diabetes for patients with incident diabetes and, for those without incident diabetes, to the date of the administrative censoring (November 30, 2015) or the date of death. Plasma selenium concentrations were introduced in the models as tertiles comparing each of the 2 highest tertiles of plasma selenium with the lowest tertile, or as a log-transformed (continuous) variable to compare the prevalence and incidence of type 2 diabetes for a doubling of selenium concentrations.
Statistical models were initially unadjusted (Model 1) and then they were adjusted for age, sex and education (<high school, ≥ high school) (Model 2). We additionally adjusted for urine cotinine levels (<34, 34–500, and >500 ng/mL), smoking status (never, former, and current) and alcohol status (never, former, current) (Model 3). As a sensitivity analysis we further adjusted the statistical models for BMI. We did not consider BMI in the final model because the number of missing values was high and the results did not change. We also modeled selenium models without using the logarithmic transformation, with essentially similar results (data not shown). P-values for linear and non-linear trend were obtained from Wald tests for log-transformed plasma selenium and plasma selenium spline terms in regression models.
Gene-environment interaction analyses were evaluated from interaction terms for log-transformed plasma selenium levels with indicator variables for genotypes. For each SNP we estimated three models for the interaction with selenium in separate models assuming dominant, recessive and additive inheritance, and calculated the P-values for interaction in each model by using Wald tests for the corresponding interaction terms. We calculated a Bonferroni-corrected p-value of 0.0002 (estimated as 0.05 divided by 242 effective number of SNPs. The effective number of SNPs in our study population was obtained using Plink software [25] based on the LD structure). For polymorphisms with the top-10 interaction p-values, we reported the associations of selenium and diabetes among subgroups of participants carrying the specific genotypes of interest. If more than 1 inheritance model showed statistically significant SNP-selenium interactions at the nominal p-value of 0.05, we reported the best fitting inheritance model selected by comparing a general model that included separate dummy variables for the heterozygote and minor allele homozygote (reference major allele homozygote) with the models assuming dominant (minor allele homozygote and heterozygote versus major allele homozygote), recessive (minor allele homozygote versus heterozygote and major allele homozygote) or additive inheritance (0, 1, or 2, minor allele dosage).
3. Results
3.1. Description of the study population
The overall geometric mean concentration of plasma selenium was 84.2 µg/L (Table 1). The weighted prevalence of diabetes was 5.6% (n=120). The incidence rate of diabetes over the study period was 5.0 per 1000 person-years. The median follow-up time was 13.2 years. Participants with prevalent diabetes compared to participants without prevalent diabetes were older, more likely to be men and former smokers, and to have lower education and serum cotinine levels (Table 1). Participants with incident diabetes showed similar characteristics compared to participants with prevalent diabetes, except for alcohol intake (Supplemental Table 1).
Table 1.
Participant characteristics by prevalent diabetes status.
| Characteristics | No diabetes | Diabetes | Overall |
|---|---|---|---|
| (n=1332) | (n=120) | (n=1452) | |
| Age, mean (SE) | 47.8 (0.22) | 66.6 (1.4) | 49.0 (0.2) |
| Gender, % male (SE) | 48.3 (0.3) | 60.0 (4.6) | 49.0 (0.2) |
| Education, % < high school (SE) | 20.8 (1.0) | 45.0 (4.8) | 22.3 (1.0) |
| Urine cotinine (ng/mL), geometric mean (95% CI) | 8.8 (7.3, 10.5) | 3.6 (2.2, 5.8) | 8.3 (7.0, 9.7) |
| Smoking status, | |||
| Former, % (SE) | 28.1 (1.2) | 39.3 (4.8) | 28.8 (1.2) |
| Current, % (SE) | 27.5 (1.3) | 14.3 (3.9) | 26.7 (1.2) |
| Alcohol intake (g/day); mean (SE) | 11.1 (0.6) | 11.3 (2.9) | 11.2 (0.6) |
| Glucose lowering medication; % yes (SE) | – – – | 50.0 (4.9) | 3.1 (0.4) |
| Plasma selenium (µg/L), geometric mean (95% CI) | 83.9 (82.8, 85.1) | 88.9 (84.1, 93.8) | 84.2 (83.1, 85.4) |
Abbreviations: SE, standard error; CI, confidence interval.
3.2. The association of selenium and Type 2 Diabetes
In multi-adjusted models, plasma selenium concentrations were linearly and positively associated with the prevalence of diabetes (p-linear trend 0.03) (Table 2). The odds ratios for prevalent diabetes comparing the second and third to the first tertiles of selenium distribution were, respectively, 1.80 (95% confidence interval [CI], 1.03, 3.14) and 1.97 (1.14, 3.41). (Table 2). For incident diabetes, we observed consistent, although borderline statistically significant, positive associations (hazard ratio comparing the second and third to the first selenium tertiles were 1.76 [0.96, 3.22] and 1.80 [0.98, 3.31], respectively. The hazard ratio for a doubling of selenium levels was 2.06 (1.07, 3.95) for diabetes prevalence and 1.55 [0.85, 2.81]) for diabetes incidence models (Table 2). The P-values for non-linear spline terms were not statistically significant in both cross-sectional and prospective analyses (non-linearity p-values in fully adjusted models were 0.55 and 0.66, respectively) (data not shown).
Table 2.
Odds ratios (95% confidence intervals) of prevalent diabetes and hazard ratios (95% confidence intervals) of incident diabetes by plasma selenium levels.
|
Prevalent diabetes (N=1452) |
Incident diabetes (N=1234) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Plasma Selenium, µg/L | Cases/ Non cases | Model 1 OR (95% CI) | Model 2 OR (95% CI) | Model 3 OR (95% CI) | Cases/ Non cases | Model 1 HR (95% CI) | Model 2 HR (95% CI) | Model 3 HR (95% CI) |
| <76.3 | 30 / 462 | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) | 19 / 412 | 1.00 (Reference) | 1.00 (Reference) | 1.00 (Reference) |
| 76.3–94.2 | 42 / 444 | 1.63 | 1.78 | 1.80 | 30 / 376 | 1.78 | 1.83 | 1.76 |
| (0.96, 2.76) | (1.02, 3.11) | (1.03, 3.14) | (0.98, 3.26) | (1.00, 3.35) | (0.96, 3.22) | |||
| ≥94.2 | 48 / 426 | 1.78 | 1.97 | 1.97 | 26 / 371 | 1.78 | 1.86 | 1.80 |
| (1.06, 2.97) | (1.15, 3.39) | (1.14, 3.41) | (0.97, 3.28) | (1.01, 3.41) | (0.98, 3.31) | |||
| Per doubling of the dose | 120 / 1332 | 1.81 | 2.07 | 2.06 | 75 / 1159 | 1.53 | 1.59 | 1.55 |
| (0.99, 3.29) | (1.08, 3.98) | (1.07, 3.95) | (0.84, 2.77) | (0.87, 2.91) | (0.85, 2.81) | |||
| P lineal | 0.05 | 0.03 | 0.03 | 0.16 | 0.13 | 0.15 | ||
Model 1 is unadjusted. Model 2 is adjusted for age (years), gender (male, female), education (<high school, ≥high school). Model 3 is further adjusted for urine cotinine levels (<34, 34–500, and >500 ng/mL), smoking status (never, former and current smoker) and alcohol intake (g/day). In cox proportional hazard models for the estimation of hazard ratios, the time scale was age and age at baseline was treated as staggered entries in all adjustment models. Abbreviations: OR, odds ratio; CI, confidence interval; HR, hazard ratio.
3.3. Gene-environment interaction
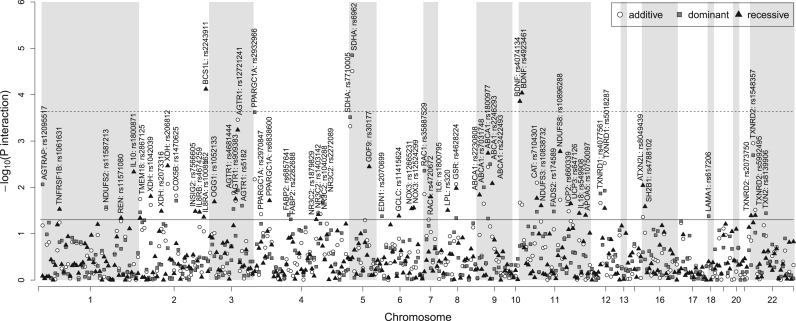
At the Bonferroni-corrected level of statistical significance, the following candidate SNPs showed significant evidence of a differential association of plasma selenium levels and prevalent diabetes: rs6962 in SDHA (p-interaction=0.00002) and rs2243911 in BCS1L (p-interaction=0.00008), which are related to mitochondrial enzymes; and rs4923461 and rs4074134 in BDNF (p-interaction=0.00009 and 0.0001, respectively) and rs2932966 in PPARGC1A (p-interaction=0.0002), which are involved in signaling pathways (Fig. 1, Table 3). The statistical interactions of selenium with SNPs in BDNF from recessive models, however, must be taken with caution because the number of participants with prevalent diabetes among homozygous carriers of the minor allele was low (the number of cases/non cases was 116/1237 and 3/71 among carriers of the AA+AG and GG genotypes, respectively, in rs4923461, and 115/1243 and 4/73 among carriers of the GG+GA and AA genotypes, respectively, in rs4074134 [data not shown]). For rs7710005 in SDHA (p-interaction=0.0003) and rs12721241 in AGTR1 (p-interaction=0.0003), the evidence was suggestive (Fig. 1, Table 3). In addition, among top-10 interaction p-values, we found statistical interactions of selenium with polymorphisms from selenoprotein genes such as rs1548357 in TXNRD2 (p-interaction=0.001) and rs5018287 in TXNRD1 (p-interaction=0.003) and other genes encoding mitochondrial enzymes such as rs10896288 in NDUFS8 (p-interaction =0.002) or involved in lipid transport (rs1800977 [p-interaction =0.002] and rs2246293 [p-interaction =0.003] in ABCA1) (Fig. 1, Table 3).
Fig. 1.
Candidate gene-selenium interaction -log10 P-values for adjusted odds ratios of prevalent diabetes. P-values for the interactions of log-transformed plasma selenium levels with 354 SNPs on prevalent diabetes derived from logistic regression models (dominant, recessive and additive model) adjusted for age, sex, education, urine cotinine levels (<34, 34–500, and >500 ng/mL), smoking status (never, former and current smokers) and alcohol intake (g/day) are presented (Y axis) according to the position of the SNPs on the chromosome (X axis). The horizontal solid and dashed lines correspond to a nominal significance level of 0.05 and to the effective SNP number significance level of 0.0002, respectively.
Table 3.
Odds ratio (95% confidence interval) of prevalent diabetes per a two-fold increase in selenium concentrations by top-10 interaction p-values, and associated hazard ratio (95% confidence interval) of incident diabetes.
| Prevalent diabetes | Incident diabetes* | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Model | Genotype | Cases/Non cases | Odds ratio (95% CI) | P- int. | Cases/Non cases | Hazard ratio (95% CI) | P- int. | Consequence |
| SDHA | rs6962 | DOM | G/G | 84/966 | 1.01 (0.55, 1.87) | 0.00002 | 59/833 | 1.43 (0.70, 2.92) | 0.87 | Non synonymous |
| G/A+A/A | 36/345 | 15.06 (5.08, 44.64) | 14/307 | 1.65 (0.38, 7.18) | ||||||
| rs7710005 | DOM | A/A | 78/918 | 1.23 (0.65, 2.33) | 0.0003 | 57/790 | 1.24 (0.65, 2.38) | 0.81 | Intronic | |
| A/G+G/G | 37/356 | 11.70 (4.02, 34.07) | 14/317 | 1.52 (0.34, 6.73) | ||||||
| BCS1L | rs2243911 | REC | C/C+C/T | 94/1025 | 3.82 (1.99, 7.33) | 0.00008 | 57/889 | 1.65 (0.81, 3.39) | 0.59 | Intronic |
| T/T | 24/278 | 0.25 (0.07, 0.83) | 15/244 | 1.06 (0.26, 4.31) | ||||||
| PPARGC1A | rs2932966 | DOM | A/A | 80/767 | 1.10 (0.57, 2.12) | 0.0002 | 45/665 | 2.02 (0.87, 4.68) | 0.40 | Intronic |
| AC + C/C | 39/521 | 11.13 (3.73, 33.24) | 27/454 | 1.18 (0.47, 2.95) | ||||||
| AGTR1 | rs12721241 | ADD | G/G | 75/850 | 1.10 (0.58, 2.07) | 0.0003 | 49/734 | 1.47 (0.72, 2.98) | 0.70 | Intronic |
| G/A | 34/404 | 6.37 (2.68, 15.12) | 22/358 | 1.82 (0.67, 4.89) | ||||||
| A/A | 8/53 | 36.96 (6.58, 207.77) | 3/46 | 2.24 (0.31, 16.09) | ||||||
| TXNDR2 | rs1548357 | ADD | T/T | 59/627 | 0.93 (0.44, 1.98) | 0.001 | 31/550 | 2.15 (0.77, 5.95) | 0.45 | Intronic |
| T/C | 47/556 | 4.12 (2.04, 8.33) | 32/480 | 1.39 (0.69, 2.80) | ||||||
| C/C | 12/117 | 18.17 (4.26, 77.47) | 8/103 | 0.90 (0.18, 4.35) | ||||||
| NDUFS8 | rs10896288 | REC | GC+G/G | 100/1051 | 3.22 (1.52, 6.83) | 0.002 | 60/907 | 1.71 (0.84, 3.50) | 0.29 | 5´UTR |
| C/C | 18/244 | 0.15 (0.02, 0.90) | 12/219 | 0.74 (0.19, 2.88) | ||||||
| ABCA1 | rs1800977 | REC | C/T+C/C | 96/1129 | 1.46 (0.79, 2.70) | 0.002 | 59/984 | 1.67 (0.78, 3.55) | 0.43 | 5´ UTR |
| T/T | 21/162 | 20.55 (4.01, 105.43) | 12/142 | 0.83 (0.17, 3.92) | ||||||
| rs2246293 | DOM | C/C | 29/346 | 10.54 (2.97, 37.35) | 0.003 | 21/301 | 1.18 (0.46, 3.06) | 0.62 | 5´ UTR | |
| C/G+G/G | 77/875 | 1.27 (0.64, 2.54) | 48/763 | 1.64 (0.69, 3.88) | ||||||
| TXNRD1 | rs5018287 | ADD | G/G | 41/426 | 0.81 (0.36, 1.81) | 0.003 | 20/370 | 1.39 (0.43, 4.47) | 0.76 | Intronic |
| G/A | 57/661 | 2.75 (1.52, 4.97) | 42/571 | 1.63 (0.87, 3.03) | ||||||
| A/A | 22/240 | 9.42 (2.93, 30.24) | 13/213 | 1.90 (0.59, 6.17) | ||||||
The Bonferroni-corrected significance level was 0.0002.
Odds ratios of prevalent diabetes per a doubling of selenium dose and associated test for interaction were obtained from logistic regression models with log-transformed selenium as a continuous variable. Hazard ratios of incident diabetes per a doubling of selenium dose and associated test for interaction were obtained from cox proportional hazard regression models with log-transformed selenium as a continuous variable. Models were adjusted by age (years), sex (male, female), education (<high school, ≥high school), smoking status (never, former and current smoker), urine cotinine levels (<34, 34–500, and >500 ng/mL), and alcohol intake (g/day). *In cox proportional hazard models for the estimation of hazard ratios, the time scale was age and age at baseline was treated as staggered entries in all adjustment models. Abbreviations: SNP, single nucleotide polymorphism; CI, confidence interval.
The corresponding associations of selenium with incident diabetes among carriers of these polymorphisms were directionally consistent for most genotypes, although the interaction p-values were not statistically significant (Table 3). However, the number of incident diabetes cases was markedly lower compared to the number of prevalent diabetes cases, making the estimation of gene-environment interactions in incident diabetes models difficult. Supplemental Table 2 shows the associations of selenium and incident diabetes by genotype for polymorphisms with the top-10 interaction p-values and at least 11 incident diabetes cases among carriers of the minor genotype, which include polymorphisms in genes related to oxidative metabolism such as rs206812 in XDH (p-interaction =0.004) and rs2297520 in NOS2A (p-interaction=0.02), genes encoding proteins related to selenoproteins (rs3761144 in GSS [p-interaction =0.02] and rs740603 in TXNRD2 [p-interaction =0.02]), genes encoding mitochondrial enzymes (rs9435739 in SDHB [p-interaction =0.02], rs10504961 in UQCRB [p-interaction =0.006] and rs1053517 in NDUFS1 [p-interaction =0.02]); and genes related to lipid synthesis and transport (rs7566605 in INSIG2 [p-interaction =0.01], rs2246293 in ABCA1 [p-interaction 0.02] and rs619054 in APOA5 [p-interaction 0.02]) (Supplemental Table 2).
4. Discussion
This study evaluated the cross-sectional association of baseline plasma selenium concentrations with prevalent diabetes, the prospective association of baseline plasma selenium concentrations with incident diabetes among participants free of diabetes at baseline, and the interaction of selenium and diabetes-related candidate polymorphisms, using a representative sample of the general population. At selenium exposure concentrations that were similar to other European countries, plasma selenium was linearly and positively associated with both the prevalence and incidence of diabetes. We found significant interactions of selenium levels with genetic variation in BCS1L, SDHA and PPARGC1A, and suggestive interactions in genes related to oxidative metabolism including genes related to selenoproteins and mitochondrial proteins. Findings from our gene-environment interaction analysis are, thus, consistent with a potential dysfunction of redox mechanisms associated with altered selenium levels in diabetes.
The observed positive associations of selenium with diabetes were largely consistent with findings from most, but not all, population-based studies [2], [12], [16], [26]. Interestingly, prospective studies mostly reported null or inverse associations [2], [6], [10], [11]. In the Olivetti Heart Study population (mean plasma selenium 77.5 ng/mL), the cross-sectional, positive, association of selenium with diabetes was not confirmed in prospective analysis [6]. In our data, selenium exposure, as measured in plasma, consistently showed a positive association with both prevalent and incident diabetes. Random sampling and, differences in exposure, genetic background, sample size, and residual confounding, are plausible explanations for the inconsistent findings across different observational studies. In a recent meta-analysis of randomized controlled trials (N=20,294 participants) the combined relative risks (RRs) for the association between selenium and the risk of type 2 diabetes was 1.09 (95% CI: 0.99–1.20). The authors concluded that the accumulated evidence does not support the use of selenium supplementation for type 2 diabetes prevention [26].
The biological mechanisms underlying the association of plasma selenium with type 2 diabetes are not well understood and experimental evidence is, so far, somewhat contradictory. Some in vivo and in vitro studies suggested that selenium might have insulin-like actions including stimulation of glucose uptake, glycolysis, gluconeogenesis and fatty acid synthesis [1], [27]. Other in vitro studies in pancreatic cells, showed increased insulin secretion in response to selenium treatment [28]. Furthermore, selenate treatment conferred protection to rat insulinoma cells INS1 against streptozotocin-induced cell death [29], suggesting that the antioxidant properties of selenium may be protective for pancreatic beta-cells. Selenium, however, has also shown potential adverse effects on glucose metabolism, such as hyperglycemia in animal models with high-selenium diets [30]. Interestingly, some animal studies suggest that both selenium over-supplementation and deficiency could be associated with type 2 diabetes in a U-shaped dose-response relationship [31]. In a previous work in our study population [23], we observed non-linear dose-responses of selenium exposure with oxidative stress biomarkers. However, in the present analysis the association of selenium and diabetes was linear, with no evidence for a threshold of potential selenium diabetogenic effects.
Mechanistic studies suggest that elevated Se intake induces hepatic insulin resistance through changes in reactive oxygen species (ROS) levels [32], [33]. Mice with elevated gene expression of glutathione peroxidase 1 (GPx1), a major antioxidant selenoprotein, developed insulin resistance and obesity [34]. Increased GPX1 activity interfered with the maintenance of normal insulin function by overquenching intracellular ROS, which are required for sensitizing insulin signaling [35], [36]. Consistently, low expression and activity of major antioxidant enzymes, including selenoproteins such as glutathione peroxidases (GPxs), rendered β-cells sensitive to signaling [37]. In addition, the administration of Selenoprotein P (SeP), which promotes selenium accumulation in the body and is involved in selenium distribution from the liver to extra-hepatic tissues [38], aggravated insulin resistance in hepatocytes and myocytes [39]. Moreover, both genetic deletion and RNA interference-mediated knockdown of SeP improved systemic insulin sensitivity and glucose tolerance in mice [39]. Circulating selenoprotein P levels were positively associated with fasting plasma glucose levels in humans [40], [41]. Consistently, individuals with type 2 diabetes showed higher concentrations of SeP compared to individuals without type 2 diabetes, although the sample size was small (n=100) [41]. Unfortunately, we did not have genotyped polymorphisms in SEPP1, the gene encoding SeP.
While we did not find statistically significant interactions of selenium with available polymorphisms in selenoproteins such as GPx1 and GPx4, we found statistically suggestive interactions with polymorphisms in GSS, the gene encoding the glutathione synthetase, which participates in the synthesis of the GPx substrate glutathione, and in genes encoding other selenoproteins from the thioredoxin reductase family, which have been related to oxidative metabolism in diabetes [42], [43]. Alternatively, experimental evidence support that other selenoproteins are involved in endoplasmic reticulum (ER) function [44], [45], [46], [47], [48], [49], [50]. Alterations of ER selenoproteins participate in the accumulation of misfolded proteins and associated ER stress [44], [47], [51], [52], [53]. Interestingly, ER in the pancreatic beta cells has an important role in insulin secretion [54]. In our data, carriers of the GA or AA genotypes in rs7566605 from the insulin induced gene 2 (INSIG2), which encodes an ER protein involved in the transport of sterol regulatory element binding proteins to the Golgi, showed increased incidence of diabetes with increased selenium levels compared to carriers of the GG genotype.
Our gene-environment interaction results also point to other redox pathways. For instance, we identified differential associations of selenium and diabetes by polymorphisms in genes encoding mitochondrial chain enzymes such as subunits A and B of the succinate dehydrogenase complex, which are major catalytic subunits of the succionate-ubiquinone oxidoreductase, the NADH-ubiquinone oxidoreductase, and BCS1, a chaperone involved in the assembly of the ubiquinol-cytochrome c reductase. These genes are expressed in pancreas and liver, and the presence of altered gene expression could translate in an unbalance of mitochondrial ROS resulting in increased cell damage. Indeed, increased ROS in the mitochondria compartment resulted in chronic oxidative stress and B-cell dysfunction in pancreatic cell lines [55]. In addition, rs2243911 in BCS1L, is close to RNF25, a gene encoding a protein that interacts with NFkb, a transcription factor involved in immune-mediated pancreatic beta-cells damage [56].
Interestingly, in diabetes prevalence models, individuals with AC or CC genotypes in rs2932966 from the peroxisome proliferator-activated receptor-γ coactivator 1 alpha (PGC-1α) gene (PPARGC1A) showed increased prevalence of diabetes with increased selenium levels. Associations of PPARGC1A variants with a range of metabolic traits including type 2 diabetes have been reported in human studies [57], [58], [59], [60], [61], [62]. Selenium preserved mitochondrial functional performance by increasing levels of PGC-1α, which is a signaling protein in energy metabolism pathways, potentially including insulin signaling pathways [63], in neuronal cell lines and prevented ischemic neuronal damage in mice [64]. Other lines of evidence suggest that elevated PGC-1α levels might, in turn, increase hepatic selenoprotein P biosynthesis and insulin resistance [65].
Our study needs to be viewed within its strengths and limitations. First, one weakness was the lack of non-fasting plasma glucose determinations in the whole study population. Only individuals with suggestive evidence of altered non-fasting glucose levels underwent a second measurement in fasting conditions. This strategy allows the analysis of diabetes status, but not glycaemia as a continuous measure. Second, a limitation to study gene by environment interactions was the moderate sample size. While we could detect some differential associations by carriers of different genotypes in candidate genes, even after applying a conservative alpha of significance threshold, larger studies are needed for replication of findings. Third, it is unclear whether the evaluated polymorphisms can induce a change of gene expression or a functional change in the corresponding encoded proteins. The main goal of our study was the identification of differential associations of selenium and diabetes by genetic variants pointing to genes and pathways known to be related to glucose metabolism. Additional functional studies of the identified interacting polymorphisms are needed. Importantly, this is an observational study and we cannot exclude that alterations in both selenium metabolism and transport and glucose metabolism have a common cause (for instance, potentially hepatic damage) or the presence of unmeasured confounders that may partly explain the observed associations. However, the ability to compare cross-sectional and prospective associations with consistent findings is a major strength of the study and makes these alternate explanations unlikely. In addition, the strength of the estimated associations including extensive adjustment for potential confounders, the dose-response relationship, and the biological plausibility of the reported interactions given the involvement of the candidate genes on diabetes pathways, make that our study adds to the body of suggestive evidence supporting of a role of selenium on diabetes risk. Other strengths of our study include the representative sampling design that allows our findings to be generalized to a general population framework.
In conclusion, we observed linear associations of plasma selenium with prevalent and incident diabetes and found evidence that these associations may be modified by alteration of redox and glucose metabolism, including insulin-signaling pathways. While our results add to the concern about the potential adverse effects of selenium, future studies are needed to confirm the observed interactions and to establish the biological and clinical implications of our findings under different levels of selenium exposure.
Funding
This work was supported by the Strategic Action for Research in Health Sciences [CP12/03080, PI15/00071, PI10/0082, PI13/01848, PI14/00874 and PI11/00726], GRUPOS 03/101; PROMETEO/2009/029 and ACOMP/2013/039 from the Valencia Government, GRS/279/A/08 from Castilla-Leon Government and European Network of Excellence Ingenious Hypercare (EPSS- 037093) from the European Commission; CIBER Fisiopatología Obesidad y Nutrición (CIBERobn) [CIBER-02–08–2009, CB06/03 and CB12/03/30016] and CIBER de Diabetes y Enfermedades Metabólicas Relacionadas (CIBERDEM). The Strategic Action for Research in Health sciences, CIBEROBN and CIBERDEM are initiatives from Carlos III Health Institute Madrid and the Spanish Ministry of Economy and Competitiveness and co-funded with European Funds for Regional Development (FEDER).
Duality of interest
No potential conflicts of interest relevant to this article were reported.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.04.022.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Stapleton S.R. Selenium: an insulin-mimetic. Cell Mol. Life Sci. 2000;57(13–14):1874–1879. doi: 10.1007/PL00000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rayman M.P., Stranges S. Epidemiology of selenium and type 2 diabetes: can we make sense of it? Free Radic. Biol. Med. 2013;65:1557–1564. doi: 10.1016/j.freeradbiomed.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Bleys J., Navas-Acien A., Guallar E. Serum selenium and diabetes in U.S. adults. Diabetes Care. 2007;30(4):829–834. doi: 10.2337/dc06-1726. [DOI] [PubMed] [Google Scholar]
- 4.Gao S., Jin Y., Hall K.S., Liang C., Unverzagt F.W., Ji R., Murrell J.R., Cao J., Shen J., Ma F., Matesan J., Ying B., Cheng Y., Bian J., Li P., Hendrie H.C. Selenium level and cognitive function in rural elderly Chinese. Am. J. Epidemiol. 2007;165(8):955–965. doi: 10.1093/aje/kwk073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laclaustra M., Navas-Acien A., Stranges S., Ordovas J.M., Guallar E. Serum selenium concentrations and diabetes in U.S. adults: national Health and Nutrition Examination Survey (NHANES) 2003–2004. Environ. Health Perspect. 2009;117(9):1409–1413. doi: 10.1289/ehp.0900704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stranges S., Galletti F., Farinaro E., D'Elia L., Russo O., Iacone R., Capasso C., Carginale V., De Luca V., Della Valle E., Cappuccio F.P., Strazzullo P. Associations of selenium status with cardiometabolic risk factors: an 8-year follow-up analysis of the Olivetti Heart study. Atherosclerosis. 2011;217(1):274–278. doi: 10.1016/j.atherosclerosis.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 7.Hughes K., Aw T.C., Kuperan P., Choo M. Central obesity, insulin resistance, syndrome X, lipoprotein(a), and cardiovascular risk in Indians, Malays, and Chinese in Singapore. J. Epidemiol. Community Health. 1997;51(4):394–399. doi: 10.1136/jech.51.4.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coudray C., Roussel A.M., Mainard F., Arnaud J., Favier A. Lipid peroxidation level and antioxidant micronutrient status in a pre-aging population; correlation with chronic disease prevalence in a French epidemiological study (Nantes, France) J. Am. Coll. Nutr. 1997;16(6):584–591. [PubMed] [Google Scholar]
- 9.Stranges S., Sieri S., Vinceti M., Grioni S., Guallar E., Laclaustra M., Muti P., Berrino F., Krogh V. A prospective study of dietary selenium intake and risk of type 2 diabetes. BMC Public Health. 2010;10:564. doi: 10.1186/1471-2458-10-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park K., Rimm E.B., Siscovick D.S., Spiegelman D., Manson J.E., Morris J.S., Hu F.B., Mozaffarian D. Toenail selenium and incidence of type 2 diabetes in U.S. men and women. Diabetes Care. 2012;35(7):1544–1551. doi: 10.2337/dc11-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akbaraly T.N., Arnaud J., Rayman M.P., Hininger-Favier I., Roussel A.M., Berr C., Fontbonne A. Plasma selenium and risk of dysglycemia in an elderly French population: results from the prospective Epidemiology of Vascular Ageing Study. Nutr. Metab. (Lond.) 2010;7:21. doi: 10.1186/1743-7075-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu C.W., Chang H.H., Yang K.C., Kuo C.S., Lee L.T., Huang K.C. High serum selenium levels are associated with increased risk for diabetes mellitus independent of central obesity and insulin resistance. BMJ Open Diabetes Res. Care. 2016;4(1):e000253. doi: 10.1136/bmjdrc-2016-000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein E.A., Thompson I.M., Jr, Tangen C.M., Crowley J.J., Lucia M.S., Goodman P.J., Minasian L.M., Ford L.G., Parnes H.L., Gaziano J.M., Karp D.D., Lieber M.M., Walther P.J., Klotz L., Parsons J.K., Chin J.L., Darke A.K., Lippman S.M., Goodman G.E., Meyskens F.L., Jr., Baker L.H. Vitamin E and the risk of prostate cancer: the selenium and vitamin E cancer prevention trial (SELECT) JAMA. 2011;306(14):1549–1556. doi: 10.1001/jama.2011.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lippman S.M., Klein E.A., Goodman P.J., Lucia M.S., Thompson I.M., Ford L.G., Parnes H.L., Minasian L.M., Gaziano J.M., Hartline J.A., Parsons J.K., Bearden J.D., 3rd, Crawford E.D., Goodman G.E., Claudio J., Winquist E., Cook E.D., Karp D.D., Walther P., Lieber M.M., Kristal A.R., Darke A.K., Arnold K.B., Ganz P.A., Santella R.M., Albanes D., Taylor P.R., Probstfield J.L., Jagpal T.J., Crowley J.J., Meyskens F.L., Jr, Baker L.H., Coltman C.A., Jr. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA. 2009;301(1):39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stranges S., Marshall J.R., Natarajan R., Donahue R.P., Trevisan M., Combs G.F., Cappuccio F.P., Ceriello A., Reid M.E. Effects of long-term selenium supplementation on the incidence of type 2 diabetes: a randomized trial. Ann. Int. Med. 2007;147(4):217–223. doi: 10.7326/0003-4819-147-4-200708210-00175. [DOI] [PubMed] [Google Scholar]
- 16.Thompson P.A., Ashbeck E.L., Roe D.J., Fales L., Buckmeier J., Wang F., Bhattacharyya A., Hsu C.H., Chow H.H., Ahnen D.J., Boland C.R., Heigh R.I., Fay D.E., Hamilton S.R., Jacobs E.T., Martinez M.E., Alberts D.S., Lance P. Selenium supplementation for prevention of colorectal adenomas and risk of associated type 2 diabetes. J. Natl. Cancer Inst. 2016;108(12) doi: 10.1093/jnci/djw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Czernichow S., Couthouis A., Bertrais S., Vergnaud A.C., Dauchet L., Galan P., Hercberg S. Antioxidant supplementation does not affect fasting plasma glucose in the Supplementation with Antioxidant Vitamins and Minerals (SU.VI.MAX) study in France: association with dietary intake and plasma concentrations. Am. J. Clin. Nutr. 2006;84(2):395–399. doi: 10.1093/ajcn/84.1.394. [DOI] [PubMed] [Google Scholar]
- 18.Gjesing A.P., Hornbak M., Allin K.H., Ekstrom C.T., Urhammer S.A., Eiberg H., Pedersen O., Hansen T. High heritability and genetic correlation of intravenous glucose- and tolbutamide-induced insulin secretion among non-diabetic family members of type 2 diabetic patients. Diabetologia. 2014;57(6):1173–1181. doi: 10.1007/s00125-014-3207-y. [DOI] [PubMed] [Google Scholar]
- 19.Alanne M., Kristiansson K., Auro K., Silander K., Kuulasmaa K., Peltonen L., Salomaa V., Perola M. Variation in the selenoprotein S gene locus is associated with coronary heart disease and ischemic stroke in two independent Finnish cohorts. Hum. Genet. 2007;122(3–4):355–365. doi: 10.1007/s00439-007-0402-7. [DOI] [PubMed] [Google Scholar]
- 20.Katunga L.A., Gudimella P., Efird J.T., Abernathy S., Mattox T.A., Beatty C., Darden T.M., Thayne K.A., Alwair H., Kypson A.P., Virag J.A., Anderson E.J. Obesity in a model of gpx4 haploinsufficiency uncovers a causal role for lipid-derived aldehydes in human metabolic disease and cardiomyopathy. Mol. Metab. 2015;4(6):493–506. doi: 10.1016/j.molmet.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuzuya M., Ando F., Iguchi A., Shimokata H. Glutathione peroxidase 1 Pro198Leu variant contributes to the metabolic syndrome in men in a large Japanese cohort. Am. J. Clin. Nutr. 2008;87(6):1939–1944. doi: 10.1093/ajcn/87.6.1939. [DOI] [PubMed] [Google Scholar]
- 22.Hellwege J.N., Palmer N.D., Ziegler J.T., Langefeld C.D., Lorenzo C., Norris J.M., Takamura T., Bowden D.W. Genetic variants in selenoprotein P plasma 1 gene (SEPP1) are associated with fasting insulin and first phase insulin response in Hispanics. Gene. 2014;534(1):33–39. doi: 10.1016/j.gene.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galan-Chilet I., Tellez-Plaza M., Guallar E., De Marco G., Lopez-Izquierdo R., Gonzalez-Manzano I., Carmen Tormos M., Martin-Nunez G.M., Rojo-Martinez G., Saez G.T., Martin-Escudero J.C., Redon J., Javier Chaves F. Plasma selenium levels and oxidative stress biomarkers: a gene-environment interaction population-based study. Free Radic. Biol. Med. 2014;74:229–236. doi: 10.1016/j.freeradbiomed.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Lorente-Galdos B., Medina I., Morcillo-Suarez C., Heredia T., Carreno-Torres A., Sangros R., Alegre J., Pita G., Vellalta G., Malats N., Pisano D.G., Dopazo J., Navarro A. Select your SNPs (SYSNPs): a web tool for automatic and massive selection of SNPs. Int. J. Data Min. Bioinform. 2012;6(3):324–334. doi: 10.1504/ijdmb.2012.049249. [DOI] [PubMed] [Google Scholar]
- 25.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao S., Zhang A., Huang S. Selenium supplementation and the risk of type 2 diabetes mellitus: a meta-analysis of randomized controlled trials. Endocrine. 2014;47(3):758–763. doi: 10.1007/s12020-014-0298-7. [DOI] [PubMed] [Google Scholar]
- 27.Mueller A.S., Pallauf J. Compendium of the antidiabetic effects of supranutritional selenate doses. In vivo and in vitro investigations with type II diabetic db/db mice. J. Nutr. Biochem. 2006;17(8):548–560. doi: 10.1016/j.jnutbio.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Campbell S.C., Aldibbiat A., Marriott C.E., Landy C., Ali T., Ferris W.F., Butler C.S., Shaw J.A., Macfarlane W.M. Selenium stimulates pancreatic beta-cell gene expression and enhances islet function. FEBS Lett. 2008;582(15):2333–2337. doi: 10.1016/j.febslet.2008.05.038. [DOI] [PubMed] [Google Scholar]
- 29.Steinbrenner H., Hotze A.L., Speckmann B., Pinto A., Sies H., Schott M., Ehlers M., Scherbaum W.A., Schinner S. Localization and regulation of pancreatic selenoprotein P. J. Mol. Endocrinol. 2013;50(1):31–42. doi: 10.1530/JME-12-0105. [DOI] [PubMed] [Google Scholar]
- 30.Satyanarayana S., Sekhar J.R., Kumar K.E., Shannika L.B., Rajanna B., Rajanna S. Influence of selenium (antioxidant) on gliclazide induced hypoglycaemia/anti hyperglycaemia in normal/alloxan-induced diabetic rats. Mol. Cell Biochem. 2006;283(1–2):123–127. doi: 10.1007/s11010-006-2387-2. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa-Wong A.N., Berry M.J., Seale L.A. Selenium and metabolic disorders: an emphasis on type 2 diabetes risk. Nutrients. 2016;8(2) doi: 10.3390/nu8020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Zhang W., Chen H., Liao N., Wang Z., Zhang X., Hai C. High selenium impairs hepatic insulin sensitivity through opposite regulation of ROS. Toxicol. Lett. 2014;224(1):16–23. doi: 10.1016/j.toxlet.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Steinbrenner H. Interference of selenium and selenoproteins with the insulin-regulated carbohydrate and lipid metabolism. Free Radic. Biol. Med. 2013;65:1538–1547. doi: 10.1016/j.freeradbiomed.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 34.McClung J.P., Roneker C.A., Mu W., Lisk D.J., Langlais P., Liu F., Lei X.G. Development of insulin resistance and obesity in mice overexpressing cellular glutathione peroxidase. Proc. Natl. Acad. Sci. USA. 2004;101(24):8852–8857. doi: 10.1073/pnas.0308096101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahadev K., Zilbering A., Zhu L., Goldstein B.J. Insulin-stimulated hydrogen peroxide reversibly inhibits protein-tyrosine phosphatase 1b in vivo and enhances the early insulin action cascade. J. Biol. Chem. 2001;276(24):21938–21942. doi: 10.1074/jbc.C100109200. [DOI] [PubMed] [Google Scholar]
- 36.Hansen L.L., Ikeda Y., Olsen G.S., Busch A.K., Mosthaf L. Insulin signaling is inhibited by micromolar concentrations of H(2)O(2). Evidence for a role of H(2)O(2) in tumor necrosis factor alpha-mediated insulin resistance. J. Biol. Chem. 1999;274(35):25078–25084. doi: 10.1074/jbc.274.35.25078. [DOI] [PubMed] [Google Scholar]
- 37.Lei X.G., Vatamaniuk M.Z. Two tales of antioxidant enzymes on beta cells and diabetes. Antioxid. Redox Signal. 2011;14(3):489–503. doi: 10.1089/ars.2010.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill K.E., Wu S., Motley A.K., Stevenson T.D., Winfrey V.P., Capecchi M.R., Atkins J.F., Burk R.F. Production of selenoprotein P (Sepp1) by hepatocytes is central to selenium homeostasis. J. Biol. Chem. 2012;287(48):40414–40424. doi: 10.1074/jbc.M112.421404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misu H., Takamura T., Takayama H., Hayashi H., Matsuzawa-Nagata N., Kurita S., Ishikura K., Ando H., Takeshita Y., Ota T., Sakurai M., Yamashita T., Mizukoshi E., Yamashita T., Honda M., Miyamoto K., Kubota T., Kubota N., Kadowaki T., Kim H.J., Lee I.K., Minokoshi Y., Saito Y., Takahashi K., Yamada Y., Takakura N., Kaneko S. A liver-derived secretory protein, selenoprotein P, causes insulin resistance. Cell Metab. 2010;12(5):483–495. doi: 10.1016/j.cmet.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 40.Misu H., Ishikura K., Kurita S., Takeshita Y., Ota T., Saito Y., Takahashi K., Kaneko S., Takamura T. Inverse correlation between serum levels of selenoprotein P and adiponectin in patients with type 2 diabetes. PLoS One. 2012;7(4):e34952. doi: 10.1371/journal.pone.0034952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang S.J., Hwang S.Y., Choi H.Y., Yoo H.J., Seo J.A., Kim S.G., Kim N.H., Baik S.H., Choi D.S., Choi K.M. Serum selenoprotein P levels in patients with type 2 diabetes and prediabetes: implications for insulin resistance, inflammation, and atherosclerosis. J. Clin. Endocrinol. Metab. 2011;96(8):E1325–E1329. doi: 10.1210/jc.2011-0620. [DOI] [PubMed] [Google Scholar]
- 42.Grankvist K., Holmgren A., Luthman M., Taljedal I.B. Thioredoxin and thioredoxin reductase in pancreatic islets may participate in diabetogenic free-radical production. Biochem Biophys. Res. Commun. 1982;107(4):1412–1418. doi: 10.1016/s0006-291x(82)80156-3. [DOI] [PubMed] [Google Scholar]
- 43.Calabrese V., Cornelius C., Leso V., Trovato-Salinaro A., Ventimiglia B., Cavallaro M., Scuto M., Rizza S., Zanoli L., Neri S., Castellino P. Oxidative stress, glutathione status, sirtuin and cellular stress response in type 2 diabetes. Biochim. Biophys. Acta. 2012;1822(5):729–736. doi: 10.1016/j.bbadis.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Rueli R.H., Torres D.J., Dewing A.S., Kiyohara A.C., Barayuga S.M., Bellinger M.T., Uyehara-Lock J.H., White L.R., Moreira P.I., Berry M.J., Perry G., Bellinger F.P. Selenoprotein S reduces endoplasmic reticulum stress-induced phosphorylation of tau: potential role in selenate mitigation of tau pathology. J. Alzheimers Dis. 2017;55(2):749–762. doi: 10.3233/JAD-151208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai J., Liu H., Zhou J., Huang K. Selenoprotein R protects human lens epithelial cells against D-galactose-induced apoptosis by regulating oxidative stress and endoplasmic reticulum stress. Int. J. Mol. Sci. 2016;17(2):231. doi: 10.3390/ijms17020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., Jiang L., Li Y., Luo X., He J. Excessive selenium supplementation induced oxidative stress and endoplasmic reticulum stress in chicken spleen. Biol. Trace Elem. Res. 2016;172(2):481–487. doi: 10.1007/s12011-015-0596-9. [DOI] [PubMed] [Google Scholar]
- 47.Lee J.H., Park K.J., Jang J.K., Jeon Y.H., Ko K.Y., Kwon J.H., Lee S.R., Kim I.Y., Selenoprotein S-dependent Selenoprotein K binding to p97(VCP) protein is essential for endoplasmic reticulum-associated degradation. J. Biol. Chem. 2015;290(50):29941–29952. doi: 10.1074/jbc.M115.680215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J.H., Kwon J.H., Jeon Y.H., Ko K.Y., Lee S.R., Kim I.Y. Pro178 and Pro183 of selenoprotein S are essential residues for interaction with p97(VCP) during endoplasmic reticulum-associated degradation. J. Biol. Chem. 2014;289(20):13758–13768. doi: 10.1074/jbc.M113.534529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hwang D., Seo S., Kim Y., Kim C., Shim S., Jee S., Lee S., Jang M., Kim M., Yim S., Lee S.K., Kang B., Jang I., Cho J. Selenium acts as an insulin-like molecule for the down-regulation of diabetic symptoms via endoplasmic reticulum stress and insulin signalling proteins in diabetes-induced non-obese diabetic mice. J. Biosci. 2007;32(4):723–735. doi: 10.1007/s12038-007-0072-6. [DOI] [PubMed] [Google Scholar]
- 50.Wu Y., Zhang H., Dong Y., Park Y.M., Ip C. Endoplasmic reticulum stress signal mediators are targets of selenium action. Cancer Res. 2005;65(19):9073–9079. doi: 10.1158/0008-5472.CAN-05-2016. [DOI] [PubMed] [Google Scholar]
- 51.Jang J.K., Park K.J., Lee J.H., Ko K.Y., Kang S., Kim I.Y. Selenoprotein S is required for clearance of C99 through endoplasmic reticulum-associated degradation. Biochem. Biophys. Res. Commun. 2017 doi: 10.1016/j.bbrc.2017.03.060. [DOI] [PubMed] [Google Scholar]
- 52.Qin H.S., Yu P.P., Sun Y., Wang D.F., Deng X.F., Bao Y.L., Song J., Sun L.G., Song Z.B., Li Y.X. Paclitaxel inhibits selenoprotein S expression and attenuates endoplasmic reticulum stress. Mol. Med. Rep. 2016;13(6):5118–5124. doi: 10.3892/mmr.2016.5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turanov A.A., Shchedrina V.A., Everley R.A., Lobanov A.V., Yim S.H., Marino S.M., Gygi S.P., Hatfield D.L., Gladyshev V.N. Selenoprotein S is involved in maintenance and transport of multiprotein complexes. Biochem. J. 2014;462(3):555–565. doi: 10.1042/BJ20140076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Araki E., Oyadomari S., Mori M. Endoplasmic reticulum stress and diabetes mellitus. Int. Med. 2003;42(1):7–14. doi: 10.2169/internalmedicine.42.7. [DOI] [PubMed] [Google Scholar]
- 55.Pi J., Bai Y., Zhang Q., Wong V., Floering L.M., Daniel K., Reece J.M., Deeney J.T., Andersen M.E., Corkey B.E., Collins S. Reactive oxygen species as a signal in glucose-stimulated insulin secretion. Diabetes. 2007;56(7):1783–1791. doi: 10.2337/db06-1601. [DOI] [PubMed] [Google Scholar]
- 56.Salem H.H., Trojanowski B., Fiedler K., Maier H.J., Schirmbeck R., Wagner M., Boehm B.O., Wirth T., Baumann B. Long-term IKK2/NF-kappaB signaling in pancreatic beta-cells induces immune-mediated diabetes. Diabetes. 2014;63(3):960–975. doi: 10.2337/db13-1037. [DOI] [PubMed] [Google Scholar]
- 57.Kim J.W., Yoon K.H. Glucolipotoxicity in pancreatic beta-cells. Diabetes Metab. J. 2011;35(5):444–450. doi: 10.4093/dmj.2011.35.5.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oberkofler H., Linnemayr V., Weitgasser R., Klein K., Xie M., Iglseder B., Krempler F., Paulweber B., Patsch W. Complex haplotypes of the PGC-1alpha gene are associated with carbohydrate metabolism and type 2 diabetes. Diabetes. 2004;53(5):1385–1393. doi: 10.2337/diabetes.53.5.1385. [DOI] [PubMed] [Google Scholar]
- 59.Jemaa Z., Kallel A., Sleimi C., Mahjoubi I., Feki M., Ftouhi B., Slimane H., Jemaa R., Kaabachi N. The Gly482Ser polymorphism of the peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) is associated with type 2 diabetes in Tunisian population. Diabetes Metab. Syndr. 2015;9(4):316–319. doi: 10.1016/j.dsx.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 60.Ruchat S.M., Weisnagel S.J., Vohl M.C., Rankinen T., Bouchard C., Perusse L. Evidence for interaction between PPARG Pro12Ala and PPARGC1A Gly482Ser polymorphisms in determining type 2 diabetes intermediate phenotypes in overweight subjects. Exp. Clin. Endocrinol. Diabetes. 2009;117(9):455–459. doi: 10.1055/s-0029-1216352. [DOI] [PubMed] [Google Scholar]
- 61.Franks P.W., Ekelund U., Brage S., Luan J., Schafer A.J., O'Rahilly S., Barroso I., Wareham N.J. PPARGC1A coding variation may initiate impaired NEFA clearance during glucose challenge. Diabetologia. 2007;50(3):569–573. doi: 10.1007/s00125-006-0580-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jablonski K.A., McAteer J.B., de Bakker P.I., Franks P.W., Pollin T.I., Hanson R.L., Saxena R., Fowler S., Shuldiner A.R., Knowler W.C., Altshuler D., Florez J.C., Diabetes Prevention Program Research G. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 2010;59(10):2672–2681. doi: 10.2337/db10-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.GeneCards HumanGene Database. PPARGC1A Gene. 〈http://www.genecards.org/cgi-bin/carddisp.pl?Gene=PPARGC1A#pathways_interactions〉 (accessed 24 March)th, 2017.
- 64.Mehta S.L., Kumari S., Mendelev N., Li P.A. Selenium preserves mitochondrial function, stimulates mitochondrial biogenesis, and reduces infarct volume after focal cerebral ischemia. BMC Neurosci. 2012;13:79. doi: 10.1186/1471-2202-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Speckmann B., Walter P.L., Alili L., Reinehr R., Sies H., Klotz L.O., Steinbrenner H. Selenoprotein P expression is controlled through interaction of the coactivator PGC-1alpha with FoxO1a and hepatocyte nuclear factor 4alpha transcription factors. Hepatology. 2008;48(6):1998–2006. doi: 10.1002/hep.22526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material