Abstract
The majority of the parasite assessments of New World primates have been conducted through the identification of the eggs found in faeces, though many species of parasites have very similar eggs, leaving uncertainty in the diagnosis. Here, we present the results of a parasite survey of the three species of primates distributed in Mexico, combining non-invasive sampling with molecular techniques via DNA extraction of the eggs found in the faeces. Mitochondrial and ribosomal DNA were employed for species identification and Bayesian phylogenetic analysis. Nine parasite taxa were found in the three primate species: the nematodes Trypanoxyuris minutus, T. multilabiatus, T. pigrae, T. atelis, T. atelophora, Strongyloides sp., unidentified Ancylostomatid, unidentified Ascarid, and the trematode Controrchis biliophilus. We were able to extract and amplify DNA from the eggs of the five species of Trypanoxyuris reported for Mexican primates, two morphologically different trematode eggs, and Strongyloides sp. Phylogenetic analysis confirmed that the two types of trematode eggs belong to Controrchis biliophilus, a member of the family Dicrocoeliidae. For Strongyloides sp., phylogenetic analysis and genetic divergence showed an association between our samples and S. fuelleborni; however, no species could be established due to the lack of more DNA sequences from Strongyloides sp. occurring in Neotropical primates. The use of molecular and phylogenetic methods could help to overcome the limitations imposed by traditional non-invasive sampling because eggs are primarily obtained from the faeces; however, its utility relies on the extant genetic library and the contributions that expand such library. The information presented here could serve as a basis for future research on primate parasitology, allowing a more accurate parasite diagnosis and a more precise evaluation of their zoonotic potential.
Keywords: DNA sequence, Parasite egg, Phylogenetic analysis, Diagnosis
Graphical abstract
Highlights
-
•
Molecular diagnosis of parasites from Mexican primates through non-invasive sampling.
-
•
Phylogenetic analysis identified seven parasite species.
-
•
28S a suitable marker for parasite DNA diagnosis.
-
•
Accurate parasite diagnosis crucial in wildlife conservation and management programs.
1. Introduction
Parasites are important natural components of ecosystems because they actively intervene in the ecological, demographic and life history processes of their hosts, influencing the structure and organization of free-living organism communities (Poulin, 1999, Gómez and Nichols, 2013). The study of parasites provides information not only on host health but also on the evolutionary history and historical biogeography of the host-parasite associations (Brooks and McLennan, 1993), as well as the health of the ecosystem (Lafferty, 1997, Overstreet, 1997, Pérez-Ponce de León, 2014).
Parasites in wildlife vertebrates are challenging to study, and in most occasions the death of the host is required to obtain and identify its parasitic fauna. This has been a major limitation in studying rare and endangered species, such as many Neotropical primates, where sacrifice is unethical or even illegal. For this reason, the majority of the parasitic assessments of New World primates have been conducted via non-invasive sampling techniques. Non-invasive parasitic evaluations rely mostly on egg identification, though many species of parasites have very similar egg morphotypes, making it practically impossible to distinguish species, which results in uncertainty in the diagnosis. Furthermore, while information on human parasites and parasites of veterinary importance is available with detailed guides on parasite species and egg descriptions (Zajac and Conboy, 2006, Ash and Orihel, 2007, Taylor et al., 2015), only a few references are available regarding the parasitic diseases of wildlife mammals (see Samuel et al., 2001). No guides for the diagnosis of parasites in free-ranging primates are currently available, except for the references and diagnostic images compiled by Hasegawa et al. (2009) and the photographs of eggs and larvae presented in different papers on primate parasitology.
Molecular techniques have been mentioned as promising tools for parasitological studies, not only by facilitating species identification regardless of the parasite developmental stage but also by allowing the gathering of data on transmission modes, geographical spreads, ecological dynamics, and evolutionary processes, thus widening the scope of parasitological research (Monis et al., 2002, Gasser, 2006).
In Mexico, there are three native species of primates: the mantled howler monkey (Alouatta palliata), the black howler monkey (Alouatta pigra), and the spider monkey (Ateles geoffroyi). These primates are all considered to be endangered species by Mexican law (SEMARNAT, 2010) and are threatened mainly by habitat loss and the illegal pet trade (Duarte-Quiroga and Estrada, 2003, Rodríguez-Luna et al., 2009). As habitat fragmentation and landscape anthropogenization increases, encounters between primates and domestic fauna and humans have become more common, and a clear parasitological diagnosis is critical to evaluate the possibilities of cross-infections and the risks that this could have for primate conservation and human health. The proper identification of parasite species is essential to addressing this issue.
We present the results of a parasite survey of these three Mexican primates along their distribution range in Mexico. Non-invasive sampling methods were combined with molecular techniques to enhance parasite species identification via DNA extraction of the eggs found in the primate faeces and by inferring their phylogenetic position. In addition, a list of all the helminths parasitizing primates in Mexico was summarized from available bibliographical sources with the aim of generating a checklist of the helminths in this group of mammals. This information could serve as a basis for future research on primate parasitology, assisting with a more accurate identification of parasite species. This could provide a more precise evaluation of their zoonotic potential, the implications for primate conservation and management and for public health.
2. Methods
2.1. Sample collection and parasitological examinations
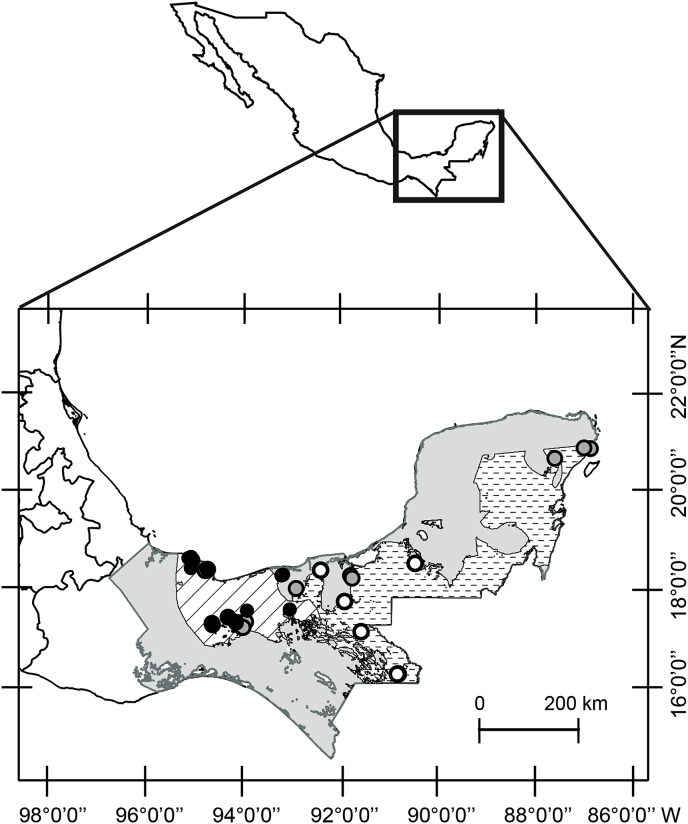
The study area comprises the tropical rainforests of southeastern Mexico, including fragmented and continuous forests, protected and unprotected areas, and agroforestry lands across the primates distribution range in Mexico. A total of 420 samples were collected between 2013 and 2015 from 68 primate troops inhabiting 52 localities (Fig. 1). All samples correspond to free-ranging populations, except those in Villahermosa and Palenque, which correspond to captive populations in zoos. In most localities, more than one forest location was surveyed.
Fig. 1.
Surveyed sites for parasites in Mexican primates. Dots indicate sampling sites, black: Alouatta palliata; white: A. pigra; and grey: Atetes geoffroyi. Polygons indicate the primate distribution range in Mexico, diagonal lines: A. palliata; dashes: A. pigra; and grey: A. geoffroyi.
Non-invasive sampling techniques were employed, collecting faecal samples immediately after defecation to avoid contamination. In general, a single monkey troop was surveyed in one day, starting the collection at dawn and moving along with the troop to gather as many samples as possible, avoiding repeatedly sampling the same individual. On occasions where the monkey troop was too small (<10 individuals) or there were many troops nearby, more than one troop was surveyed in a day. Faecal samples were placed in 50 ml falcon tubes, and stored at −4 °C until transported to the laboratory, where they were preserved at −20 °C. Preserved samples were examined for parasite eggs under direct light microscopy (10xm 40x, 100x) using flotation in saturated sodium chloride solution and simple sedimentation techniques (Greiner and McIntosh, 2009). Both methods were performed for each collected sample using 2.5 g of faeces and examining 6 drops in each procedure, in order to avoid missing parasites with different egg densities. The initial identification of the parasites was based on egg morphology, shape, size and colour. The percentage of infected hosts was estimated for each parasite taxa in each host species; in addition, we also quantified the number of hosts that were infected by at least one helminth species.
When a drop was found positive for any type of parasite, the entire drop was transferred to a new slide and observed under the stereoscope, where eggs with different appearances were individually separated with the aid of a 0.5–10 μl micropipette and sited in a drop of distilled water (5 μl) on a new slide. The eggs were rinsed several times in fresh drops of distilled water to remove the concentrated solution and then placed in 0.5-ml Eppendorf tubes with 7 μl of distilled water and kept at −20 °C until DNA extraction. Each egg morphotype was measured (length and width) and photographed to characterize its shape.
DNA was successfully extracted from a pool of 5 eggs of the same general appearance using the SIGMA REDExtract-N-Amp Tissue PCR Kit (St. Louis, MO, USA) and the Chelex® 100 (Bio-Rad, Richmond, CA, USA) chelating resin method. Whenever possible, two molecular markers were used for species identification: a fragment of the mitochondrial cytochrome c oxidase subunit 1 gene (cox1) and a fragment of the nuclear ribosomal large subunit gene (28S). For cox1, two sets of primers amplifying adjacent regions were used: pr-a: 5′-TGGTTTTTTGTGCATCCTGAGGTTTA-3′, pr-b: 5′-AGAAAGAACGTAATGAAAATGAGCAAC-3' (Nakano et al., 2006), and LCO1490: 5′-GGTCAACAAATCATAAAGATATTGG-3′, HC02198: 5′-TAAACTTCAGGGTGACCAAAAAATCA-3' (Folmer et al., 1994). PCR conditions for cox1 were as follows: initial denaturation at 94 °C for 1 min, followed by 30 cycles at 94 °C for 1 min, 40 °C for 1 min, 72 °C for 2 min, and post-amplification extension for 7 min at 72 °C.
The 28S primers included 502: 5′-CAAGTACCGTGAGGGAAAGTTGC-3′, and 536: 5′-CAGCTATCCTGAGGGAAAC-3' (García-Varela and Nadler, 2005). PCR conditions for 28S were as follows: 94 °C for 4 min, followed by 34 cycles at 94 °C for 0:30 min, 54 °C for 0:50 min, 72 °C for 1:30 min, and a post-amplification extension for 7 min at 72 °C. PCR products were treated with Exo-SAP (Thermo scientific) according to the manufacturer's instructions and were sequenced at the Instituto de Biología, Universidad Nacional Autónoma de México. Sequences obtained in this study were deposited in GenBank (Supplementary material S1).
2.2. Phylogenetic analyses
To accomplish species identification, at least one molecular marker was used for each parasite taxa. The 28S sequences were used for all egg morphotypes, since it has been mentioned that ribosomal DNA performs better for diagnostic proposes than mitochondrial DNA (Blouin, 2002). In few cases, two molecular markers were used for phylogenetic analyses, as in the case of Strongyloides spp.
DNA sequences were aligned using CLUSTAL W and MESQUITE v. 2.75. For cox1, no gaps were required to align the nucleotide sequences. To infer the phylogenetic position of the different eggs within the phylogeny of the major helminth group they belong to (usually at the level of order or family), we used a set of DNA sequences available in GenBank, using the closest identifiable egg species as a proxy by conducting a nucleotide blast (BLASTN) (Supplementary material S1). Phylogenetic analyses were conducted by Bayesian Inference (BI) employing Monte Carlo Markov Chain analysis in the program MrBayes v. 3.2.2 (Ronquist and Huelsenbeck, 2003) as implemented in the CIPRES Science Gateway (Miller et al., 2010). MrModeltest v. 2.3 (Nylander, 2004) was used to select the best model of evolution for each gene for each egg species using the Akaike information criterion. The Bayesian analyses included two simultaneous runs of Markov chain Monte Carlo, each for four million generations, sampling trees every 4000 generations, with a heating parameter value of 0.2 and a “burn-in” of 25%. A 50% majority-rule consensus tree was constructed from the post burn-in trees. Genetic divergence (p-distance) was calculated using MEGA v. 6 (Tamura et al., 2013); standard error of the distances was estimated by bootstrap resampling with 100 replications.
3. Results
3.1. Parasite diversity and percentage of infected hosts
Alouatta palliata contained the highest number of samples infected with at least one parasite species (97/126), followed by Ateles geoffroyi (124/248) and Alouatta pigra (19/46). Nine parasite taxa were found in the three primate species, the majority of which were nematodes, along with one species of trematode (Table 1, Fig. 2). Parasite species richness was similar in the two species of Alouatta, with three taxa per howler monkey species, while seven taxa of parasites were found in A. geoffroyi.
Table 1.
Percentage of infection of parasites and sampling effort for each Mexican primate species.
| Parasite Phylum | Parasite taxa | A. palliata | A. pigra | A. geoffroyi |
|---|---|---|---|---|
| Platyhelminthes | Controrchis biliophilus | 10.3% | 2.0% | |
| Nematoda | Ancylostomatid | 1.6% | ||
| Ascarid | 0.8% | |||
| Strongyloides sp. | 2.2% | 13.3% | ||
| Trypanoxyuris sp. | 7.1% | 15.2% | 14.5% | |
| T. atelis | 17.7% | |||
| T. atelophora | 9.7% | |||
| T. minutus | 57.9% | 2.2% | ||
| T. multilabiatus | 10.3% | |||
| T. pigrae | 23.9% | |||
| Sample size | 126 | 46 | 248 | |
| Localities sampled | 9 | 6 | 15 | |
| Forest fragments sampled | 17 | 9 | 26 | |
| Troops sampled | 22 | 10 | 36 | |
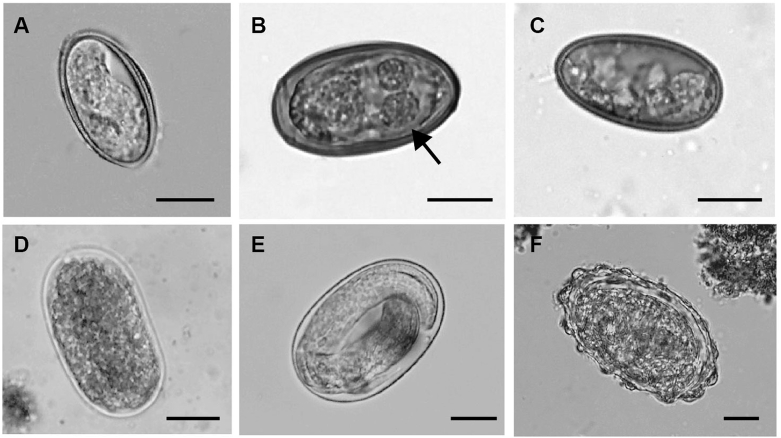
Fig. 2.
Egg morphotypes found in the faeces of Mexican primates. A) Trypanoxyuris sp., B) Controrchis biliophilus, arrow pointing to the two eyespot remnants; C) trematode, diagnosed as C. biliophilus by molecular data; D) unidentified ancylostomatid; E) Strongyloides sp.; F) unidentified ascarid. Scale bar is equal to 15 μm.
In general, nematodes of the genus Trypanoxyuris reached the highest percentage of infection in all primates. The eggs of these pinworms are morphologically undistinguishable among species (Solórzano-García et al., 2015, Solórzano-García et al., 2016); fortunately, adult pinworms were present in most of the faeces, making it possible to identify them at the species level. However, this was not the case for the other nematodes, i.e., Strongyloides sp., the ancylostomid, and the ascarid, for which egg morphology is not a reliable method to establish species identification.
The helminth parasite fauna of the three species of primates in Mexico is composed of 23 species, based on the information available in different bibliographical sources and the information provided by our field survey of the last two years. Of the 23 species, there are 3 platyhelminthes (2 trematodes and 1 cestode), 1 acanthocephalan, and 19 nematodes (Supplementary material S2). Parasite species richness is higher in Alouatta palliata, with 14 taxa reported, followed by A. pigra with 13 taxa, and Ateles geoffroyi with 11 taxa. Alouatta palliata is the most studied primate, since 13 of the 22 available parasitological reports of primates in Mexico address that species, while A. pigra and A. geoffroyi have been the focus of 9 and 8 studies respectively. Most of the parasitological research has been conducted with free-ranging primate populations (73%), while 18% of studies were from host in semi-captivity and 9% in captivity conditions.
3.2. Molecular identification of the eggs and the phylogenetic analysis
We were able to extract and amplify DNA from four of the six different egg morphotypes found in the faeces. The ancylostomatid and the ascarid eggs could not be sequenced because only two eggs for each of these taxa were found in the faeces. We successfully amplified the 28S for all the egg morphotypes. The mitochondrial gene, cox1, was more difficult to amplify, and we were only able to obtain a sequence for Strongyloides sp. and only 3 species of Trypanoxyuris.
3.2.1. Trypanoxyuris eggs
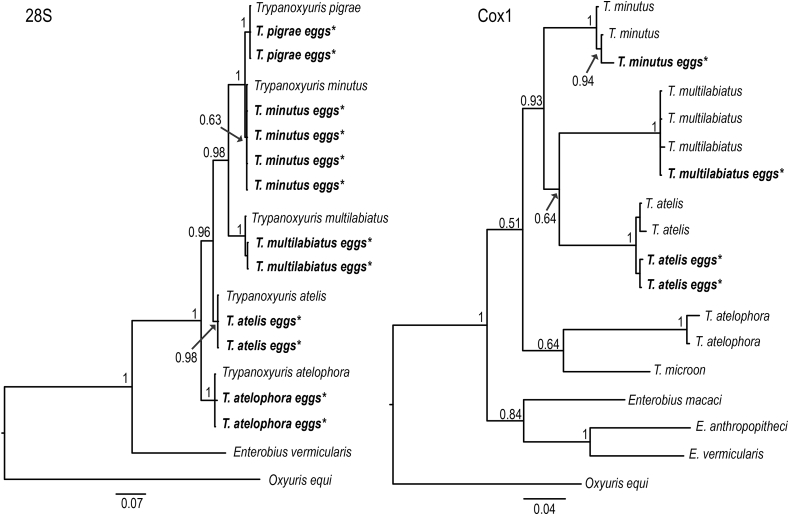
We were able to sequence both the cox1 and 28S genes for Trypanoxyuris eggs obtained from the three species of Mexican primates. We obtained sequences 700 bp long for the 28S gene from the eggs of five Trypanoxyuris species. The final alignment consisted of 19 terminals, including both the sequences from eggs and the sequences from adult individuals obtained from GenBank. This alignment was trimmed to the 700 bp obtained to ensure comparison of the homologous regions. For cox1 gene, we were able to obtain sequences 673 bp long from the eggs of T. minutus, T. atelis and T. multilabiatus. The final alignment was trimmed to 605 bp and consisted of 18 taxa including sequences from Genbank. Phylogenetic analysis on both genes placed each egg with its corresponding pinworm species with high nodal support through posterior probabilities (Fig. 3).
Fig. 3.
Phylogenetic trees based on 28S (left) and cox1 (right) sequences of Trypanoxyuris sp. Sequences obtained from the eggs are bold type and indicated with an *. Numbers at the nodes represent posterior probabilities from Bayesian inference.
3.2.2. Trematodes eggs
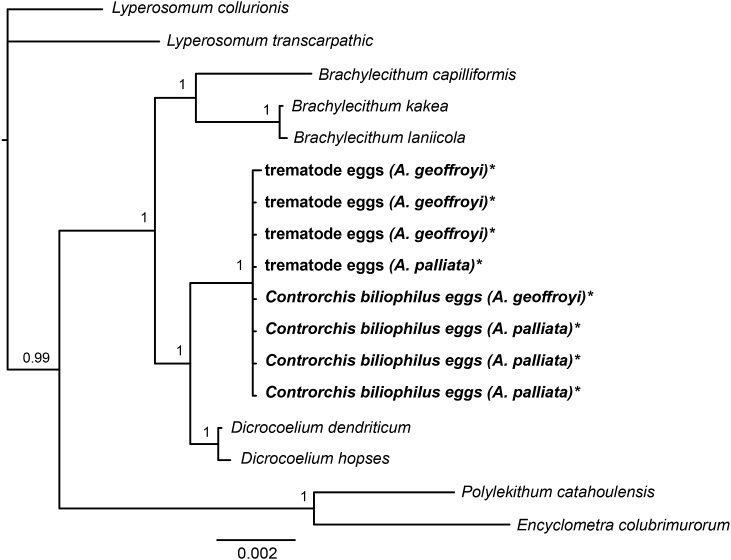
Two different trematode egg morphotypes were found in the faeces of A. palliata and A. geoffroyi: one corresponding to Controrchis biliophilus and the other differing from this in the appearance of the content material inside the egg (see Fig. 2). Sequences of 786 bp for 28S were obtained for a sample of each trematode egg from each of the two host species. The final alignment, including the sequences from GenBank for the family Dicrocoeliidae and other species included in the order Plagiorchiida, consisted of 17 sequences. This alignment was trimmed to the 786 bp obtained to ensure a comparison of homologous regions. The phylogenetic tree shows that all egg sequences belong to the same clade regardless of differences in the egg shape and host species (Fig. 4), indicating that both egg morphotypes correspond to C. biliophilus. These relationships are supported by high posterior probability values. The clade containing the C. biliophilus sequences is placed as a sister taxon of the Dicrocoelium species within the family Dicrocoeliidae.
Fig. 4.
Phylogenetic tree based on 28S sequences of Controrchis biliophilus. Sequences obtained from the eggs are bold type and indicated with an *. Numbers at the nodes represent posterior probabilities from Bayesian inference. Host species are indicated within parenthesis.
3.2.3. Strongyloides eggs
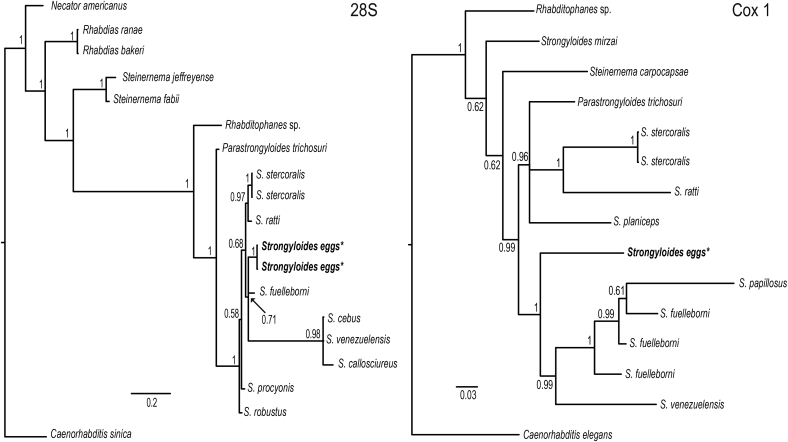
We were able to sequence both the cox1 (1079 bp) and 28S (686 pb) genes for Strongyloides eggs obtained from Ateles geoffroyi. Final alignment of cox1 included 9 species of Strongyloides from different host species; it was trimmed to 721 bp because some of the GenBank sequences were shorter. For 28S, the alignment consisted of 19 terminals including 10 species of Strongyloides from different host species. This alignment was trimmed to 686 bp to ensure a comparison of homologous regions; nevertheless, missing data (“?”) was allowed to expand the number of taxa compared, specifically to include S. cebus, which in conjunction with S. venezuelensis had 52% and 55% of missing data, respectively.
Phylogenetic analyses were carried on separately for each molecular marker to assess its utility in species identification. Molecular analysis confirmed that these eggs belonged to the genus Strongyloides; however, its position within each phylogenetic tree varied between genes. For the 28S the two sequences obtained were identical to each other. The tree shows that our sequences are nested in an unresolved clade along with S. fuelleborni and a group containing 3 species, i.e., S. cebus, S. venezuelensis and S. callosciureus (Fig. 5). Genetic divergence between our samples and the other 4 species included in the clade varied from 4.7 to 5.1%. The cox1 tree shows that the only sample we were able to sequence for this marker for Strongyloides is nested as a sister species of a clade formed by S. papillosus, S. fuelleborni and S. venezuelensis (Fig. 5), with a sequence divergence of 18.9%, 14.4% and 16.4%, respectively. Needless to say, not all the same species of the family Strongyloididae are represented in both trees because sequences of both molecular markers are not yet available.
Fig. 5.
Phylogenetic trees based on 28S (left) and cox1 (right) sequences of Strongyloides eggs. Sequences obtained from the eggs are bold type and indicated with an *. Numbers at the nodes represent posterior probabilities from Bayesian inference.
4. Discussion
Though primates are a relatively well studied group in Mexico (Estrada and Mandujano, 2003), only a few studies have focused on assessing their parasite diversity, and only free-ranging primate populations of six regions have been previously surveyed (Stroner and Gonzalez-Di Pierro, 2006, Trejo-Macias et al., 2007, Vitazkova and Wade, 2007, Cristobal-Azkarate et al., 2010). The information presented here increases the number of localities where parasites of these primates have been studied, contributing to a more complete parasitological evaluation across the distribution range of these primates in Mexico. The parasites found in the present study correspond with the previously reported taxa for the three Mexican primates; however, despite the more extensive sampling, the parasite species richness found was lower. Eggs of Raillietina sp., Strongylidae and Necator sp. have been previously found in the faeces of Mexican primates (Supplementary material S2), and species such as Ascaris lumbricoides, Calodium hepaticum, Dipetalonema gracile and Parabronema bonnei, have been reported from necropsies of A. palliata and A. geoffroyi (Caballero, 1948, Caballero and Grocott, 1952, Villanueva-Jimenez, 1988); however, we did not find eggs matching these descriptions in any of the revised samples.
Given that the majority of parasitological studies on Mexican primates are based on egg identification, we cannot discard the possibility that some of those reported parasite taxa are a result of sample contamination or even a possible misidentification. Another limiting factor in accomplishing proper taxonomic identification is that egg samples are not deposited in parasite collections, and thus the identification cannot be independently verified and relies on the photographs provided by the authors. For example, Enterobius sp. have been reported for the three Mexican primates (García-Serrano, 1995, Rodríguez-Velázquez, 1996, Stroner and Gonzalez-Di Pierro, 2006; Supplementary material S2); nevertheless, co-evolutionary studies have shown that pinworms of the genus Enterobius only parasitize Old World primates and that Trypanoxyuris is the genus of pinworms found in New World monkeys (Brooks and Glen, 1982, Hugot et al., 1996, Hugot, 1999, Hugot, 1998). Oxyurid eggs are very similar among members of the Enterobiinae subfamily, making it possible to mistake species. Moreover, molecular studies on pinworm diversity in Mexican primates have shown that five Trypanoxyuris species are found in these hosts (Solórzano-García et al., 2015, Solórzano-García et al., 2016). For these reasons, we believe that the records of Enterobius previously mentioned in the literature are in fact Trypanoxyuris.
Similarly, ancylostomatid eggs have been reported for A. palliata only in one location (Cristobal-Azkarate et al., 2010). These nematodes have not been reported as parasites of A. palliata outside Mexico, and have only been reported as parasites of A. caraya (Stuart et al., 1998). According to two photographs of the ancylostomatid eggs presented by Cristobal-Azkarate et al. (2010), these eggs lack the characteristic features of ancylostomatid eggs, such as a thin, smooth and colourless shells containing embryonic blastomeres (Rai et al., 1996) and instead resemble the eggs of Parabronema bonnei and a trematode, respectively.
Two other nematodes, Necator sp. and Trichostrongylus sp., have been reported as parasites of spider monkeys in captivity in Mexico (González Hernández, 2004, Villa-Espinoza, 2011; Supplementary material S2) and Colombia (Castañeda et al., 2010). Since these parasites have not been reported in a free-ranging population, their presence could be the result of enclosure conditions, close contact with humans and other animals in captivity, and the health status of other animals in the zoos; thus, these parasites do not necessarily belong to the natural parasitic fauna of this primate.
The results presented here show that the application of molecular and phylogenetic methods could help overcome the limitations imposed by traditional non-invasive sampling. As suggested by Criscione et al. (2005), one of the three key uses of molecular markers is to link morphologically indistinguishable life stages to adult stages of known species. However, the utility of this approach relies in the availability of molecular information from previous parasitological studies, as exemplified by the three types of parasite eggs that we were able to sequence. Trypanoxyuris is undoubtedly the taxa with the most available information. Molecular data from the adults of the five pinworm species parasitizing Mexican primates have been published (Solórzano-García et al., 2015, Solórzano-García et al., 2016), which made egg identification via DNA analysis straightforward. This nematode genus contains 21 species that parasitize primates across the neotropics. The identification of the different Trypanoxyuris species can be easily obtained by sequencing samples from different host species and areas, increasing the extant genetic library.
Controrchis biliophilus is the only reported trematode species in Mexican primates (Supplementary material S2). The eggs are characterized by its brown colour, a thick shell and the presence of two readily visible eyespot remnants (Jiménez-Quiros and Brenes, 1957). Even though no molecular information is available for the trematode C. biliophilus, adult worms are held in the Colección Nacional de Helmintos (CNHE), Instituto de Biología, Universidad Nacional Autónoma de México, which allowed us to confirm the identity of these eggs. Samples of C. biliophilus were collected from a troop of howler monkeys translocated to Agaltepec Island in Catemaco, Veracruz (Villanueva-Jimenez, 1988). We obtained sequences of C. biliophilus eggs from the same locality, enabling a greater confidence in the identification of the parasite. Trematode eggs with a slightly different morphology, specifically lacking the two eyespot remnants, has been previously reported by Trejo-Macias et al. (2007), and this morphology was also observed in the present study for samples from A. palliata and A. geoffroyi. The molecular and phylogenetic analysis showed no differences in the 28S DNA sequences between C. biliophilus and the trematode egg with a slightly different morphology, confirming that this particular egg morphotype also corresponds with C. biliophilus.
Finally, Strongyloides sp. has been reported in Mexican primates, but species determination has not been established because the eggs of Strongyloides lack morphological features that allow for discrimination among a wide diversity of species. There are over 40 species of Strongyloides that parasitize vertebrates (Dorris et al., 2002). Strongyloides stercoralis and S. fuelleborni have been found in primates (Gillespie and Chapman, 2006, Chapman et al., 2009, Dupain et al., 2009), and S. cebus has been mentioned so far as the only species that naturally infects Neotropical primates (Mati et al., 2013). The phylogenetic analysis presented here confirmed that the eggs belonged to Strongyloides; however, their phylogenetic associations with S. fuelleborni and with S. cebus have not been resolved. Furthermore, the genetic divergence between the eggs found and the species of Strongyloides for which sequences are available suggests that these might represent a new species, although this cannot be established at the moment due to the lack of additional DNA sequences from Strongyloides eggs occurring in Mexican primates. Unfortunately, we were not able to find any larvae or adults in the faeces that would allow us to take this inquiry any further.
Another important parasite is Ascaris lumbricoides. Adults of this species were found in a necropsy of an A. palliata specimen that died from natural causes in Los Tuxtlas, Mexico (see García-Prieto et al., 2012). This record adds to those made by several authors based on eggs found in the faeces, described as Ascaris sp., in the three Mexican primates (González Hernández, 2004, Cristobal-Azkarate et al., 2010, Trejo-Macias, 2010). Ascaris lumbricoides is a human parasite commonly found in other primates such as chimpanzees and gorillas (Lilly et al., 2002, Dupain et al., 2009). It has also been reported in several howler monkey species (Stuart et al., 1990, Stuart et al., 1998) and other Neotropical primates (Michaud et al., 2003) but is unusual, and in most cases these primates were in close contact with humans. In the present study, we found two eggs of an ascarid in samples from A. geoffroyi. Unfortunately, we were not able to extract DNA from these eggs, but we believe that a more intensive sampling, where a large number of eggs could be gathered, would provide the molecular data needed to clarify the taxonomic identity of the ascarid and to elucidate its zoonotic potential.
The presence of parasites such as Strongyloides fuelleborni and Ascaris lumbricoides, both common human parasites and with the capability of causing severe illness (Crompton, 2001, Olsen et al., 2009), is of major concern for the conservation of free-ranging primate populations. A precise confirmation that these parasite species are occurring in Neotropical primates remains essential to determine possible transmission routes and the potential effects that habitat fragmentation and the increase of human encroachment into wildlife territory could have on the spread of these parasites. For example, by applying molecular techniques, Gasser et al. (2009) were able to show that Oesophagostomum bifurcum in humans was genetically distinct from those harboured by non-human primates, concluding that non-human primates were not reservoir hosts for human oesophagostomiasis and that the genetic variants had different transmission patterns. Moreover, molecular analysis have been conducted to diagnose, asses host specificity, and explore the zoonotic potential of parasitic protozoans in howler monkeys (Alouatta spp.) (Soares et al., 2011, Helenbrook et al., 2015, Martínez et al., 2016, Villanueva-Garcia et al., 2017).
There is no doubt that more research is needed to properly characterize the parasites of Neotropical primates. The combination of molecular techniques with non-invasive sampling methods has proven to be effective for a better understanding of parasite diversity, transmission modes, and evolutionary history. Even though parasite eggs found in faeces are vast sources of information, the isolation of eggs along with DNA extraction and amplification are highly laborious tasks. A wide variety of methods have been described to obtain DNA from parasite eggs found in faeces (Štefanić et al., 2004, Harmon et al., 2006, Trachsel et al., 2007, Demeler et al., 2013, Federer et al., 2016). Nevertheless the standardization of the molecular diagnostic procedures still remains a critical issue in order for such techniques to be widely applied in the parasitological study of endangered species. As new molecular methods emerge, such as new generation sequencing and meta-genomic analysis, and the costs for their application become more accessible, the surveillance of the parasitic fauna of endangered species through non-invasive sampling will be easily accomplished and more accurate (Srivathsan et al., 2016). Nevertheless, this surveillance will rely on current efforts to molecularly typify the parasites found in these hosts.
The principal aim of this study was to present molecular data from the parasite eggs found in the faeces that could serve as a basis for future parasitological assessments on Mexican but also on New World primates. The results presented here support the contention that ribosomal genes are more suitable than mitochondrial DNA for species diagnosis (Blouin, 2002). Since the divergence levels found for the amplified region of the 28S gene (within the same parasite species) was really low, we suggest this as an efficient and appropriate tool for parasite species diagnosis. A more accurate parasite diagnosis would enable us to understand the ecological and evolutionary background of parasite-host associations, possibilities for cross-transmissions and their implications for primate conservation. Likewise, the proper identification of parasites when managing primate populations in captivity or for conservation purposes is essential. This is particularly important when moving individuals among zoos around the world, or when they are subjected to reintroduction and translocation programmes (Nunn and Altizer, 2006), to avoid disease outbreaks by the introduction of novel parasites that could threat the resident populations, including non-primates.
Acknowledgements
We would like to thank Lino Mendoza, Ruben Mateo and Pablo Gutierrez for their support during field work. We also thank Hacienda de la Luz, Tabasco; the Arqueological Park of Comalcalco, Tabasco, Parque Museo La Venta, Tabasco, Jardín Botánico Dr. Alfredo Barrera Marín, Puerto Morelos, Quintana Roo, and all the NPAs that kindly granted permission for collecting samples. We thank David Hernández Mena for his help during field work and data analysis. We are grateful with Berenit Mendoza Garfias for her technical support taking the SEM pictures, and with Laura M. Márques for sequencing services. This study was partially funded by the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT-UNAM IN204514) to GPPL. This paper is part of the fulfillments to accomplish the PhD degree of BSG within the Posgrado en Ciencias Biológicas of UNAM.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ijppaw.2017.04.001.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- Ash L.R., Orihel T.C. fifth ed. American Society of Clinical Pathology Press; Chicago, Illinois, USA: 2007. Atlas of Human Parasitology. [Google Scholar]
- Blouin M. Molecular prospecting for cryptic species of nematodes: mitochondrial DNA versus internal transcribed spacer. Int. J. Parasitol. 2002;32:527–531. doi: 10.1016/s0020-7519(01)00357-5. [DOI] [PubMed] [Google Scholar]
- Brooks D.R., Glen D.R. Pinworms and primates: a case study in coevolution. Proc. Helminth. Soc. Wash. 1982;49:76–85. [Google Scholar]
- Brooks D.R., McLennan D. Smithsonian Institution Press; Washington D.C: 1993. Parascript: Parasites and the Language of Evolution. [Google Scholar]
- Caballero E. Estudios helmintológicos de la región oncocercosa de México y de la República de Guatemala. Nemátoda, 4ta parte. An. Inst. Bio. 1948;19:137–151. [Google Scholar]
- Caballero E., Grocott R.G. Nota sobre la presencia de Capillaria hepatica en un mono araña (Ateles geoffroyi vellerosus) de Mexico. An. Inst. Bio. 1952;23:211–215. [Google Scholar]
- Castañeda F.E., Rubiano J.O., Cruz L.J., Rodríguez L.C. Prevalencia de helmintos intestinales en primates neotropicales cautivos alojados en la ciudad de Ibagué. Rev. Colomb. Cienc. Anim. 2010;3:33–40. [Google Scholar]
- Chapman C.A., Speirs M.L., Gillespie T.R., Holland T., Austad K.M. Life on the edge: gastrointestinal parasite from the forest edge and interior primate groups. Am. J. Primatol. 2009;68:397–409. doi: 10.1002/ajp.20233. [DOI] [PubMed] [Google Scholar]
- Criscione C.D., Poulin R., Blouin M.S. Molecular ecology of parasites: elucidating ecological and microevolutionary processes. Mol. Ecol. 2005;14:2247–2257. doi: 10.1111/j.1365-294X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- Cristobal-Azkarate J., Hervier B., Vegas-Carrillo S., Osorio-Sarabia D., Rodriguez-Luna E., Vea J.J. Parasitic infections of three mexican howler monkey groups (Alouatta palliata mexicana) exposed to differing degrees of habitat disturbance in fragmented forests. Primates. 2010;51:231–239. doi: 10.1007/s10329-010-0193-7. [DOI] [PubMed] [Google Scholar]
- Crompton D.W.T. Ascaris and ascariasis. Adv. Parasitol. 2001;14:285–375. doi: 10.1016/s0065-308x(01)48008-0. http://dx.doi.org/10.1016/S0065-308X(01)48008-0 [DOI] [PubMed] [Google Scholar]
- Demeler J., Ramünke S., Wolken S., Ianiello D., Rinaldi L., Gahutu J.B., Cringoli G., von Samson-Himmelstjerna G., Krücken J. Discrimination of gastrointestinal nematode eggs from crude fecal egg preparations by inhibitor-resistant conventional and real-time PCR. PLoS One. 2013;8:e61285. doi: 10.1371/journal.pone.0061285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorris M., Viney M., Blaxter M. Molecular phylogenetic analysis of the genus Strongyloides and related nematodes1. Int. J. Parasitol. 2002;32:1507–1517. doi: 10.1016/s0020-7519(02)00156-x. [DOI] [PubMed] [Google Scholar]
- Duarte-Quiroga A., Estrada A. Primates as pets in Mexico City: an assessment of the species involved, source of origin, and general aspects of treatment. Am. J. Primatol. 2003;61:53–60. doi: 10.1002/ajp.10108. [DOI] [PubMed] [Google Scholar]
- Dupain J., Nell C., Petrzelkova K.J., Garcia P., Modrý D., Ponce-Gordo F. Gastrointestinal parasites of bonobos in the lomako forest, democratic republic of Congo. In: Huffman M.A., Chapman C., editors. Primate Parasite Ecology. The Dynamics and Study of Host-parasite Relationships. Cambridge University Press; Cambridge: 2009. pp. 297–310. [Google Scholar]
- Estrada A., Mandujano S. Investigaciones con Alouatta y Ateles en México. Neotrop. Primates. 2003;11:145–154. [Google Scholar]
- Federer K., Armua-Fernandez M.T., Gori F., Hoby S., Wenker C., Deplazes P. Detection of taeniid (Taenia spp., Echinococcus spp.) eggs contaminating vegetables and fruits sold in European markets and the risk for metacestode infections in captive primates. Int. J. Parasitol. Parasites Wildl. 2016;5:249–253. doi: 10.1016/j.ijppaw.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- García-Prieto L., Falcón-Ordaz J., Guzmán-Cornejo C. Helminth parasites of wild Mexican mammals: list of species, hosts and geographical distribution. Zootaxa. 2012;3290:1–92. [Google Scholar]
- García-Serrano O.G. Universidad Nacional Autónoma de México; 1995. Identificación de nemátodos en monos aulladores (Alouatta palliata) cautivos en el estado de Veracruz. [Google Scholar]
- García-Varela M., Nadler S.A. Phylogenetic relationships of Palaeacanthocephala (Acanthocephala) inferred from SSU and LSU rDNA gene sequences. J. Parasitol. 2005;91:1401–1409. doi: 10.1645/GE-523R.1. [DOI] [PubMed] [Google Scholar]
- Gasser R.B. Molecular tools - advances, opportunities and prospects. Vet. Parasitol. 2006;136:69–89. doi: 10.1016/j.vetpar.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Gasser R.B., De Gruijter J.M., Polderman A.M. The utility if molecular methods for elucidating primate-pathogen relationships-the Oesophagostomun bifurcum example. In: Huffman M.A., Chapman C.A., editors. Primate Parasite Ecology. The Dynamics and Study of Host-parasite Relationships. Cambridge University Press; Cambridge: 2009. pp. 47–62. [Google Scholar]
- Gillespie T.R., Chapman C.A. Prediction of parasite infection dynamics in primate metapopulations based on attributes of forest fragmentation. Conserv. Biol. 2006;20:441–448. doi: 10.1111/j.1523-1739.2006.00290.x. [DOI] [PubMed] [Google Scholar]
- Gómez A., Nichols E. Neglected wild life: parasitic biodiversity as a conservation target. Int. J. Parasitol. Parasites Wildl. 2013;2:222–227. doi: 10.1016/j.ijppaw.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Hernández M. Universidad Veracruzana; 2004. Prevalencia de helmintiasis gastrointestinales en monos araña (Ateles geoffroyi) del Parque zoológico botánico Miguel Ángel de Quevedo en Veracruz, Veracruz, México. [Google Scholar]
- Greiner E.C., McIntosh A. Collection methods and diagnostic procedures for primate parasitology. In: Huffman M.A., Chapman C.A., editors. Primates Parasite Ecology. The Dynamics and Study of Host-parasite Relationships. Cambridge University Press; 2009. pp. 3–27. [Google Scholar]
- Harmon A.F., Zarlenga D.S., Hildreth M.B. Improved methods for isolating DNA from Ostertagia ostertagi eggs in cattle feces. Vet. Parasitol. 2006;135:297–302. doi: 10.1016/j.vetpar.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Hasegawa H., Chapman C.A., Huffman M.A. Useful diagnostic references and images of protozoans, helminths, and nematodes commonly found in wild primates. In: Huffman M.A., Chapman C., editors. Primate Parasite Ecology. The Dynamics and Study of Host-parasite Relationships. Cambridge University Press; Cambridge: 2009. pp. 507–513. [Google Scholar]
- Helenbrook W.D., Shields W.M., Whipps C.M. Characterization of Blastocystis species infection in humans and mantled howler monkeys, Alouatta palliata aequatorialis, living in close proximity to one another. Parasitol. Res. 2015;114:2517–2525. doi: 10.1007/s00436-015-4451-x. [DOI] [PubMed] [Google Scholar]
- Hugot J. Primates and their pinworm parasites: the Cameron Hypothesis revisited. Syst. Biol. 1999;48:523–546. doi: 10.1080/106351599260120. [DOI] [PubMed] [Google Scholar]
- Hugot J. Phylogeny of Neotropical monkeys: the interplay of morphological, molecular, and parasitological data. Mol. Phylogenet. Evol. 1998;9:408–413. doi: 10.1006/mpev.1998.0497. [DOI] [PubMed] [Google Scholar]
- Hugot J.P., Gardner S.L., Morand S. The Enterobiinae Subfam. Nov. (Nematoda, Oxyurida) pinworm parasites of primates and rodents. Int. J. Parasitol. 1996;26:147–159. doi: 10.1016/0020-7519(95)00108-5. [DOI] [PubMed] [Google Scholar]
- Jiménez-Quiros O., Brenes R. Helmintos de la República de Costa Rica: V. Sobre la validez del género Controrchis Price, 1928 (Trematoda, Dicrocoeliidae) y descripción de Controrchis caballeroi n.sp. Rev. Biol. Trop. 1957;5:103–121. [Google Scholar]
- Lafferty K.D. Environmental parasitology: what can parasites tell us about human impacts on the environment? Parasitol. Today. 1997;13:251–255. doi: 10.1016/s0169-4758(97)01072-7. [DOI] [PubMed] [Google Scholar]
- Lilly A.A., Mehlman P.T., Doran D. Intestinal parasites in gorillas, chimpanzees, and humans at Mondika research site, Dzanga-Ndoki National Park, Central African Republic. Int. J. Primatol. 2002;23:555–573. [Google Scholar]
- Martínez M.F., Kowalewski M.M., Salomón O.D., Schijman A.G. Molecular characterization of trypanosomatid infections in wild howler monkeys (Alouatta caraya) in northeastern Argentina. Int. J. Parasitol. Parasites Wildl. 2016;5:198–206. doi: 10.1016/j.ijppaw.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mati V.L.T., Ferreira Junior F.C., Pinto H.A., de Melo A.L. Strongyloides cebus (Nematoda: Strongyloididae) in Lagothrix cana (Primates: atelidae) from the Brazilian Amazon: aspects of clinical presentation, anatomopathology, treatment, and parasitic biology. J. Parasitol. 2013;99:1009–1018. doi: 10.1645/13-288.1. [DOI] [PubMed] [Google Scholar]
- Michaud C., Tantalean M., Ique C., Montoya E., Gozalo A. A survey for helminth parasites in feral New World non-human primate populations and its comparison with parasitological data from man in the region. J. Med. Primatol. 2003;32:341–345. doi: 10.1046/j.1600-0684.2003.00037.x. [DOI] [PubMed] [Google Scholar]
- Miller M., Pfeiffer W., Schwartz T. Proceedings of the Gateway Computing Environments Workshop. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; pp. 1–8. New Orleans. [Google Scholar]
- Monis P.T., Andrews R.H., Saint C.P. Molecular biology techniques in parasite ecology. Int. J. Parasitol. 2002;32:551–562. doi: 10.1016/s0020-7519(01)00352-6. [DOI] [PubMed] [Google Scholar]
- Nakano T., Okamoto M., Ikeda Y., Hasegawa H. Mitochondrial cytochrome c oxidase subunit 1 gene and nuclear rDNA regions of Enterobius vermicularis parasitic in captive chimpanzees with special reference to its relationship with pinworms in humans. Parasitol. Res. 2006;100:51–57. doi: 10.1007/s00436-006-0238-4. [DOI] [PubMed] [Google Scholar]
- Nunn C.L., Altizer S. Oxford University Press; 2006. Infectious Diseases in Primates. Behavior, Ecology and Evolution. [Google Scholar]
- Nylander J.A.A. 2004. MrModeltest v.2. [Google Scholar]
- Olsen A., van Lieshout L., Marti H., Polderman T., Polman K., Steinmann P., Stothard R., Thybo S., Verweij J.J., Magnussen P. Strongyloidiasis - the most neglected of the neglected tropical diseases? Trans. R. Soc. Trop. Med. Hyg. 2009;103:967–972. doi: 10.1016/j.trstmh.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Overstreet R.M. Parasitological data as monitors of environmental health. Parassitologia. 1997;39:169–175. [PubMed] [Google Scholar]
- Pérez-Ponce de León G. Los parásitos de peces como bioindicadores de la salud de los ecosistemas. In: González Zuarth A., Vallarino A., Pérez Jiménez J.C., Low Pfeng A.M., editors. Bioindicadores: Guardianes de Nuestro Futuro Ambiental. 2014. pp. 253–270. El Colegio de la Frontera Sur (Ecosur) e Instituto Nacional de Ecología y Cambio Climatico (INEEC) [Google Scholar]
- Poulin R. The functional importance of parasites in animal communities: many roles at many levels? Int. J. Parasitol. 1999;29:903–914. doi: 10.1016/s0020-7519(99)00045-4. [DOI] [PubMed] [Google Scholar]
- Rai S.K., Uga S., Kataoka N., Matsumura T. Kyokuseisya; Kobe, Japan: 1996. Atlas of Medical Parasitology. [Google Scholar]
- Rodríguez-Luna E., Solórzano-García B., Shedden-González A., Rangel-Negrin A., Dias P.A.D., Cristobal-Azkarate J., Cortes-Ortiz L., Dunn J.C., Domingo-Balcells C., Sánchez S., Vea-Baró J., Conerjo J. Universidad Veracruzana; 2009. Taller de conservación, análisis y manejo planificado para los primates mexicanos, 2006. (CBSG/IUCN) [Google Scholar]
- Rodríguez-Velázquez G. Universidad Nacional Autónoma de México; 1996. Presencia de nematodos gastroentericos en monos araña (Ateles geoffroyi) en cautiverio en Pipiapan (Catemaco, Veracruz) mediante examenes coproparasitoscopicos. [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Samuel W.M., Pybus M.J., Kocan A., editors. Parasitic Diseases of Wild Mammals. second. ed. Iowa State University Press; Iowa, USA: 2001. [Google Scholar]
- SEMARNAT . Diario Oficial de La Federación. 2010. NormaOficial Mexicana NOM-059-ECOL-2010, protección ambiental-especies nativas de México de flora y fauna silvestres-Categorías de riesgo y especificaciones para su inclusión, exclusión o cambio-Lista de especies en riesgo. Jueves 30 de diciembre de 2010, México. [Google Scholar]
- Soares R.M., de Souza S.L.P., Silveira L.H., Funada M.R., Richtzenhain L.J., Gennari S.M. Genotyping of potentially zoonotic Giardia duodenalis from exotic and wild animals kept in captivity in Brazil. Vet. Parasitol. 2011;180:344–348. doi: 10.1016/j.vetpar.2011.03.049. [DOI] [PubMed] [Google Scholar]
- Solórzano-García B., Nadler S.A., Pérez-Ponce de León G. Pinworm diversity in free-ranging howler monkeys (Alouatta spp.) in Mexico: morphological and molecular evidence for two new Trypanoxyuris species (Nematoda: Oxyuridae) Parasitol. Int. 2016;65:401–411. doi: 10.1016/j.parint.2016.05.016. [DOI] [PubMed] [Google Scholar]
- Solórzano-García B., Nadler S.A., Pérez Ponce de León G. Trypanoxyuris atelis and T. atelophora (Nematoda: Oxyuridae) in wild spider monkeys (Ateles geoffroyi) in tropical rain forest in Mexico: morphological and molecular evidence. Parasitol. Int. 2015;64:229–235. doi: 10.1016/j.parint.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Srivathsan A., Ang A., Vogler A.P., Meier R. Fecal metagenomics for the simultaneous assessment of diet, parasites, and population genetics of an understudied primate. Front. Zool. 2016;13:17. doi: 10.1186/s12983-016-0150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Štefanić S., Shaikenov B.S., Deplazes P., Dinkel A., Torgerson P.R., Mathis A. Polymerase chain reaction for detection of patent infections of Echinococcus granulosus (“sheep strain”) in naturally infected dogs. Parasitol. Res. 2004;92:347–351. doi: 10.1007/s00436-003-1043-y. [DOI] [PubMed] [Google Scholar]
- Stroner K.E., Gonzalez-Di Pierro A.M. Intestinal parasitic infections in A. pigra in tropical rainforest in Lacandona, Chiapas, Mexico: implications for behavioral ecology and conservation. In: Estrada A., Garber P.A., Pavelka M., Luecke L., editors. New Perspectives in the Study of Mesoamerican Primates: Distribution, Ecology, Behavior and Conservation. Springer; 2006. pp. 215–240. [Google Scholar]
- Stuart M., Pendergast V., Rumfelt S., Pierberg S., Greenspan L., Glander K., Clarke M. Parasites of wild howlers (Alouatta spp.) Int. J. Primatol. 1998;19:493–512. [Google Scholar]
- Stuart M.D., Greenspan L.L., Glander K.E., Clarke M.R. A coprological survey of parasites of wild mantled howling monkeys, Alouatta palliata palliata. J. Wildl. Dis. 1990;26:547–549. doi: 10.7589/0090-3558-26.4.547. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.A., Coop R.L., Wall R.L. fourth ed. Wiley-Blackwell; 2015. Veterinary Parasitology. [Google Scholar]
- Trachsel D., Deplazes P., Mathis A. Identification of taeniid eggs in the faeces from carnivores based on multiplex PCR using targets in mitochondrial DNA. Parasitology. 2007;134:911–920. doi: 10.1017/S0031182007002235. [DOI] [PubMed] [Google Scholar]
- Trejo-Macias G. Universidad Nacional Autónoma de México; 2010. Estacionalidad y fragmentacion de selva: efectos sobre los parasitos gastrointestinales en poblaciones silvestres de monos aulladores (Alouatta palliata y A. pigra) del sureste de México. [Google Scholar]
- Trejo-Macias G., Estrada A., Mosqueda-Cabrera M.A. Survey of helminth parasites in populations of Alouatta palliata mexicana and A. pigra in continuous and fragmentes habitat in southern Mexico. Int. J. Primatol. 2007;28:931–945. [Google Scholar]
- Villa-Espinoza L. Universidad Michoacana de San Nicolás de Hidalgo; 2011. Identificación de endoparásitos en monos araña (Ateles geoffroyi) del parque zoológico “Benito Juárez” de Morelia, Michoacán. [Google Scholar]
- Villanueva-Garcia C., Gordillo-Chavez E.J., Lopez-Escamilla E., Rendon-Franco E., Muñoz-Garcia C., Gama L., Martinez-Flores W., Gonzalez-Rodriguez N., Romero-Valdovinos M., Diaz-Lopez H., Galian J., Villalobos G., Maravilla P., Martinez-Hernandez F. Clarifying the cryptic host specificity of Blastocystis spp. isolates from Alouatta palliata and A. pigra howler monkeys. PLoS One. 2017:1–16. doi: 10.1371/journal.pone.0169637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva-Jimenez E.A. Universidad Veracruzana; 1988. Identificación de helmintos del tracto digestivo del mono aullador (Alouatta palliata) en poblaciones silvestres. [Google Scholar]
- Vitazkova S.K., Wade S.E. Effects of ecology on gastrointestinal parasites of Alouatta pigra. Int. J. Primatol. 2007;28:1327–1343. [Google Scholar]
- Zajac A.M., Conboy G.A. seventh ed. Blackwell Publisher; 2006. Veterinary Clinical Parasitology. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.