Abstract
Background & Aims
Patterning of the small intestinal epithelium along its cephalocaudal axis establishes three functionally distinct regions: duodenum, jejunum, and ileum. Efficient nutrient assimilation and growth depend on the proper spatial patterning of specialized digestive and absorptive functions performed by duodenal, jejunal, and ileal enterocytes. When enterocyte function is disrupted by disease or injury, intestinal failure can occur. One approach to alleviate intestinal failure would be to restore lost enterocyte functions. The molecular mechanisms determining regionally defined enterocyte functions, however, are poorly delineated. We previously showed that GATA binding protein 4 (GATA4) is essential to define jejunal enterocytes. The goal of this study was to test the hypothesis that GATA4 is sufficient to confer jejunal identity within the intestinal epithelium.
Methods
To test this hypothesis, we generated a novel Gata4 conditional knock-in mouse line and expressed GATA4 in the ileum, where it is absent.
Results
We found that GATA4-expressing ileum lost ileal identity. The global gene expression profile of GATA4-expressing ileal epithelium aligned more closely with jejunum and duodenum rather than ileum. Focusing on jejunal vs ileal identity, we defined sets of jejunal and ileal genes likely to be regulated directly by GATA4 to suppress ileal identity and promote jejunal identity. Furthermore, our study implicates GATA4 as a transcriptional repressor of fibroblast growth factor 15 (Fgf15), which encodes an enterokine that has been implicated in an increasing number of human diseases.
Conclusions
Overall, this study refines our understanding of an important GATA4-dependent molecular mechanism to pattern the intestinal epithelium along its cephalocaudal axis by elaborating on GATA4’s function as a crucial dominant molecular determinant of jejunal enterocyte identity. Microarray data from this study have been deposited into NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO series accession number GSE75870.
Keywords: Transcriptional Regulation, Jejunal Identity, Enterohepatic Signaling, Fgf15, FXR
Abbreviations used in this paper: bio-ChIP-seq, biotin-mediated chromatin immunoprecipitation with high-throughput sequencing; bp, base pair; cDNA, complementary DNA; cKI, conditional knock-in; cKO, conditional knockout; Cyp7a1, cytochrome P450 family 7 subfamily A member 1; dATP, deoxyadenosine triphosphate; E, embryonic day; EMSA, electrophoretic mobility shift assay; Fabp6, fatty acid binding protein 6; Fgf, fibroblast growth factor; FXR, farnesoid X receptor; lnl, loxP-flanked PGK-Neo-3xSV40 polyadenylation sequence; mRNA, messenger RNA; OSTα/β, organic solute transporter α/β; pA, polyadenylation; PCR, polymerase chain reaction; qRT, quantitative reverse-transcription; SBS, short-bowel syndrome; Slc, solute carrier; TSS, transcription start site; xiFABP, Xenopus I-FABP
Graphical abstract
See editorial on page 297.
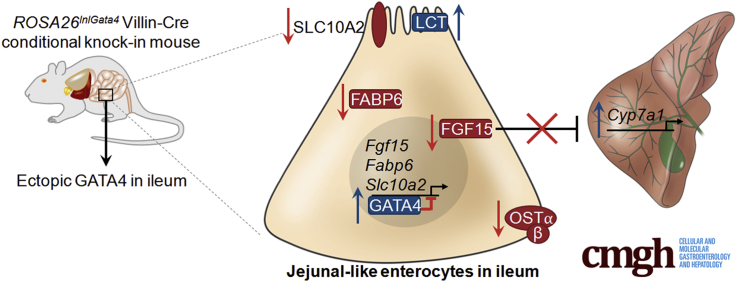
Summary.
GATA binding protein 4 (GATA4) establishes jejunal enterocyte identity and represses ileal enterocyte identity in the intestine, likely through direct activation and repression of expression of key regional-specifying genes. One important GATA4 target is fibroblast growth factor 15, a key regulator of enterohepatic bile acid cycling.
The small intestine is composed of duodenum, jejunum, and ileum. Enterocytes within each perform specialized functions dictated by their position along the cephalocaudal axis to mediate digestion and absorption of nutrients, vitamins, and fluids to sustain growth, hydration, and electrolyte balance. Duodenal enterocytes are specialized to complete the digestive process.1 Enzymes secreted from the pancreas as well as bile synthesized by liver and stored by the gall bladder enter the duodenum and combine with enzymes secreted by the duodenal enterocytes to facilitate digestion. Jejunal enterocytes accomplish the bulk of nutrient uptake by absorbing digestive products, namely lipid–bile acid emulsions, sugars, and oligopeptides/amino acids.1, 2 In addition to absorbing vitamin B12, ileal enterocytes play a critical role in maintaining the enterohepatic circulation of bile acids and in regulating bile acid metabolism.1, 2 Ileal enterocytes absorb bile acids from the intestinal lumen, and bile acids travel via the portal circulation from the intestine to the liver, where they are taken up and re-secreted into bile. Uptake of bile acids by ileal enterocytes activates farnesoid X receptor (FXR)-mediated transactivation of genes encoding proteins required for enterocyte bile acid transport including fatty acid binding protein 6 (Fabp6), solute carrier 51a (Slc51a), Slc51b and the secreted enterokine fibroblast growth factor 15/19 (Fgf15/19).3, 4, 5, 6 Binding of FGF15/19 to its receptors on hepatocytes represses expression of cytochrome P450 family 7 subfamily A member 1 (Cyp7a1), which encodes the rate-limiting enzyme in conversion of cholesterol to bile acids, to control hepatic bile acid synthesis.4, 5 Moreover, bile acids and FGF15/19 may function in the regulation of energy expenditure and lipid and carbohydrate metabolism.7, 8, 9 As such, the importance of bile acid enterohepatic cycling and homeostasis extends beyond intestinal function, and defects in enterohepatic FGF15/19 signaling have been linked to human diseases including cholestatic liver disease, nonalcoholic fatty liver disease, type 2 diabetes, metabolic syndrome, Crohn’s disease, bile acid malabsorption, and bile acid diarrhea.7, 8, 9, 10, 11
Disruption of enterocyte functions caused by Crohn’s disease and other inflammatory bowel diseases, intestinal tumors, trauma, necrotizing enterocolitis, and congenital defects, along with surgical interventions used to treat these disorders, can result in intestinal failure or short-bowel syndrome (SBS).12, 13 High morbidity and mortality are associated with SBS, and the economic and quality-of-life costs for SBS patients are high.12, 14 Better SBS therapies are needed, particularly interventions that restore lost function to remaining intestinal tissue, as well as novel tissue engineering approaches to overcome small-bowel organ shortages for transplant. To make these advances a reality, it will be necessary to understand how duodenal, jejunal, and ileal epithelial identities are patterned along the cephalocaudal axis of the small intestine. Currently, the molecular mechanisms underlying establishment and maintenance of enterocyte populations with regionally defined functions are delineated poorly. Extrinsic cues such as luminal contents and hormones can influence expression of regional-specific enterocyte markers, but tissue transplantation and isograft experiments show that regionalized gene expression programs are intrinsic to the epithelium.15, 16, 17 More recently, using a long-term organoid culture model, Middendorp et al18 showed that adult small intestinal stem cells maintain regional identity and regionally defined gene expression programs in a cell autonomous manner. We propose that the repertoire of transcription factors expressed in duodenal, jejunal, and ileal enterocytes drives patterning by activating and repressing expression of the set of downstream targets defining each region. GATA binding protein 4 (GATA4), a zinc-finger–containing transcription factor with a spatially restricted expression pattern along the cephalocaudal axis of the small intestine, represents one such factor.
GATA4 is expressed in enterocytes of the duodenal and jejunal epithelium but absent from enterocytes of the ileal epithelium.19, 20, 21 Studies of mouse intestinal development show that GATA4 initially is expressed throughout the developing intestinal epithelium and that it becomes excluded from the ileal domain relatively early at embryonic day (E)12.5–13.5.22 GATA4 plays a role in early intestinal development, regulating intestinal epithelial cell proliferation.23 Specifically, GATA4 deletion in the intestinal epithelium via Sonic hedgehog–Cre transiently reduces cellular proliferation in the intestinal epithelium (E10.5–E11.5), resulting in a shorter intestine with decreased epithelial girth. Furthermore, the onset of villus morphogenesis is delayed in the intestine of Gata4 Sonic hedgehog–Cre conditional knockout embryos, perhaps because of reduced epithelial cell proliferation, and villus structure is abnormal. In adult mice, GATA4 is essential for jejunal function.19, 21 Elimination of GATA4 from the intestinal epithelium using Villin-Cre causes a global shift in regional identity within the jejunum.21 In the absence of jejunal GATA4, expression of a wide array of jejunal-specific genes is lost, including expression of genes encoding proteins with important roles in uptake, transport, and processing of cholesterol and lipids.21 Moreover, expression of numerous ileal-specific genes is gained, including expression of genes encoding proteins required for bile acid absorption.21 These sweeping changes in gene expression from a jejunal to an ileal pattern also have important functional consequences, disrupting the normal spatial pattern of the digestive process and causing malabsorption of dietary fat and cholesterol.21 Deletion of Gata4 in the jejunal epithelium of adult mice via an inducible conditional knockout strategy similarly alters jejunal gene expression, shifting it away from a jejunal profile and toward an ileal profile.19 Comparison of jejunal phenotypes between Gata4 and Gata6 Villin-Cre conditional knockout mice further indicates that GATA control of jejunal-ileal epithelial identity is a GATA4-specific function because expression of key jejunal and ileal markers is not altered in GATA6-deficient jejunum.22, 24 Taken together, these studies show that GATA4 is necessary for execution of the enterocyte gene expression program in the jejunum.
The goal of the present study was to test the hypothesis that GATA4 is sufficient to confer jejunal fate within the intestinal epithelium. To test this hypothesis, we used a conditional knock-in approach to generate mice expressing GATA4 in the ileal epithelium, where it normally is absent. If GATA4 is sufficient to drive jejunal identity, the ileal enterocyte gene expression profile should shift from ileal to jejunal. Indeed, we found that the global gene expression profile of GATA4-expressing ileal epithelium differed significantly from control ileal epithelium, aligning more closely with jejunum rather than ileum. We also observed overlap between GATA4-expressing ileum and duodenum, suggesting that ectopic GATA4 expression within the ileum can induce duodenal gene expression, thereby conferring a more proximal-type intestinal identity with both jejunal and duodenal character. Focusing on jejunal vs ileal identity, gene expression changes in the presence or absence of GATA4 and GATA4 chromatin immunoprecipitation experiments strongly suggest that GATA4 patterns regional-specific functions by directly activating expression of genes defining jejunum and by directly repressing expression of genes defining ileum. We further show that enterohepatic signaling was altered in mice expressing GATA4 in the ileum. Taken together, these data extend our understanding of GATA4 as a crucial dominant molecular determinant of enterocyte identity and regulator of enterohepatic signaling. Considering that GATA4 is necessary and sufficient to promote jejunal identity and repress ileal identity, GATA4 represents a logical candidate to consider as a therapeutic target for intestinal failure.
Materials and Methods
Animals
To generate Gata4 conditional knock-in mice (Gt(ROSA)26Sortm1(Gata4)Bat, MGI: 5707906), the coding sequence of mouse Gata4 was amplified by polymerase chain reaction (PCR) using the Expand High Fidelity PCR system (Roche, Madison, WI). The forward primer contained a XhoI restriction site, and the reverse primer contained a SacI restriction site for cloning into the XhoI/SacI sites in the multiple cloning site of the pBigT plasmid25 to create pBigT-Gata4 (Figure 1A). Primer sequences are listed in Table 1. A cassette containing the adenovirus splice acceptor (SA), a loxP-flanked phosphoglycerate kinase (PGK) promoter-neomycin resistance gene (Neo)-3xSV40 polyadenylation (pA) (designated LNL), the Gata4 coding sequence, and the bovine growth hormone polyadenylation (pA) sequence was excised from pBigT-Gata4 with PacI and AscI. This fragment was inserted into the PacI/AscI sites in the plasmid pROSA26PA25 to create the pROSA26PA-Gata4 targeting vector (Figure 1A). Targeting vector, linearized by digestion with Asp718, was introduced into R1 mouse embryonic stem cells by electroporation.26 Embryonic stem cell colonies resistant to 350 μg/mL active G418 (Life Technologies, Grand Island, NY) were genotyped by Southern blot and PCR. Chimeric animals were generated by aggregation of CD-1 morulae with R1 mouse embryonic stem cells containing the modified allele (Gt(ROSA)26Sortm1(Gata4)Bat, designated ROSA26lnlG4), as described previously.27 The modified ROSA26lnlG4 allele was propagated through the germline by breeding chimeras to CD-1 mice, and germline transmission of the modified ROSA26lnlG4 allele was confirmed by Southern blot (Figure 1B). Heterozygous ROSA26lnlG4/+ animals were crossed to generate homozygous ROSA26lnlG4/lnlG4 mice that were crossed with Villin-Cre (Tg(Vil-cre)997Gum) mice28 to generate heterozygous Gata4 conditional knock-in mice (Gata4 cKI) with the genotype ROSA26lnlG4/+Villin-Cre. For conditional knockout of Gata4 in the jejunum, Gata4loxP mice (Gata4tm1.1Sad)29 were crossed with Villin-Cre mice to produce Gata4loxP/loxPVillin-Cre mice. CD-1 mice (Charles River Laboratories, Wilmington, MA) were used as wild-type control. For biotin-mediated chromatin immunoprecipitation (bio-ChIP) studies, the Gata4flbio/flbio(Gata4tm3.1Wtp) and ROSA26BirA(Gt(ROSA)26SorTm1[birA]Mejr) mouse lines were used to generate Gata4flbio/flbio::ROSA26BirA/BirA and ROSA26BirA/BirA mice for bio-ChIP.30, 31 Primers used for PCR genotyping of ear punch DNA are listed in Table 1. Studies with ROSA26lnlG4/+, ROSA26lnlG4/+Villin-Cre, Gata4loxP/loxPVillin-Cre, and CD-1 mice were performed with 6- to 8-week-old male mice. Studies with Gata4flbio/flbio::ROSA26BirA/BirA and ROSA26BirA/BirA were performed with epithelial cells from 4- to 6-month-old male mice. In all studies, duodenum was collected from no more than 8 cm adjacent to the stomach, jejunum was collected 10 cm from the stomach, and ileum was collected from no more than 5 cm adjacent to the cecum. The Medical College of Wisconsin Institutional Animal Care and Use Committee approved all animal procedures.
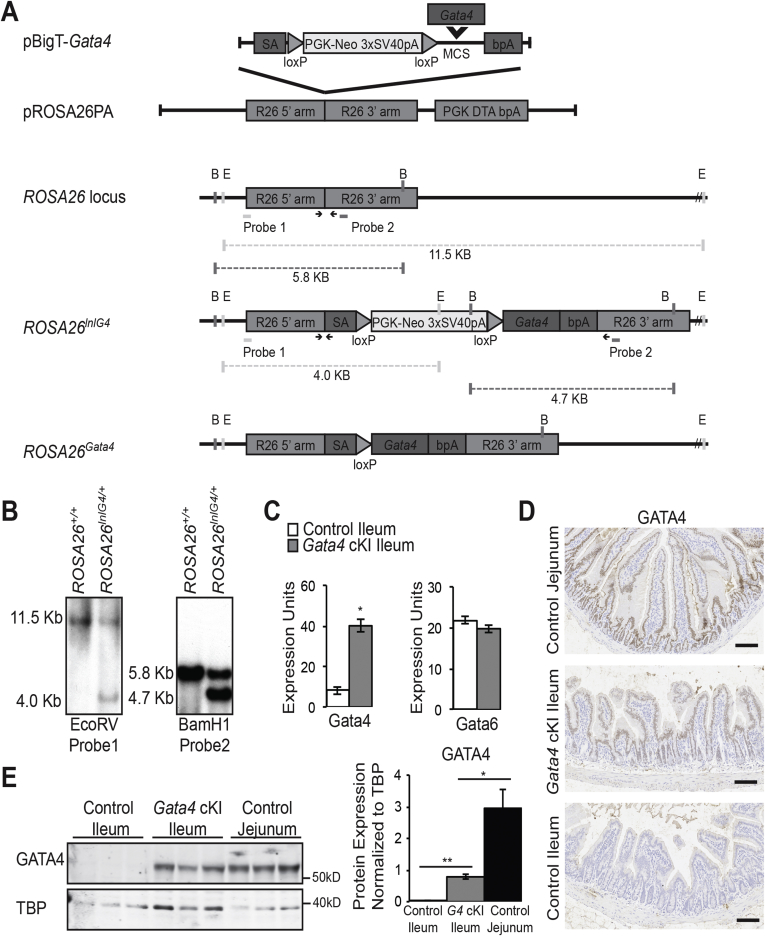
Figure 1.
Gata4 conditional knock-in mice express GATA4 in the ileum. (A) Schematic illustrating the strategy used to generate a conditional Gata4 knock-in mouse line. The coding sequence of the mouse Gata4 gene was amplified by PCR and inserted into XhoI/SacI sites in the multiple cloning site (MCS) of pBig-T to generate pBigT-Gata4. The targeting cassette consisting of an adenoviral splice acceptor (SA), a loxP flanked phosphoglycerate kinase (PGK) promoter-neomycin resistance gene (Neo) and 3×SV40 polyadenylation sequence (pA) sequence (loxP-PGK-Neo-3×SV40pA-loxP, LNL), the Gata4 coding sequence, and a bovine growth hormone polyadenylation (pA) sequence was excised from pBigT-Gata4 with PacI/AscI and inserted into the PacI/AscI sites in pROSA26PA to create pROSA26PA-Gata4. Homologous recombination between pROSA26PA-Gata4 and the endogenous ROSA26 locus in mouse R1 embryonic stem cells yielded the targeted locus Gt(ROSA)26Sortm1(Gata4)Bat, designated ROSA26lnlG4. After Cre recombination to excise the LNL cassette, Gata4 is expressed. BamHI (B) and EcoRV (E) restriction sites used for Southern blot analysis, the position of Southern blot probes, and relevant BamHI and EcoRV restriction digest fragments identified by Southern blot are shown. Arrows mark sites of genotyping primers (Table 1, primers). (B) Southern blot analysis confirmed germline transmission of the ROSA26lnlG4 allele. Representative Southern blot analysis of EcoRV or BamHI digested genomic DNA harvested from a wild-type mouse (ROSA26+/+) or a mouse heterozygous for the modified ROSA26 allele (ROSA26lnlG4/+). We observed the expected fragments representing the wild-type and modified alleles (EcoRV digest, 11.5-kb wild-type allele and 4.0-kb modified allele; BamHI digest, 5.8-kb wild-type allele and 4.7-kb modified allele). (C) qRT-PCR showed that Gata4 mRNA was induced in ileum of ROSA26lnlG4/+Villin-Cre (designated Gata4 cKI) mice compared with ileum of control mice (ROSA26lnlG4/+). Gata6 mRNA remained unchanged in the ileum of Gata4 cKI mice compared with controls (n = ileum of 5 control and 6 Gata4 cKI animals; experiments performed in triplicate). Glyceraldehyde-3-phosphate dehydrogenase was used for normalization. Error bars show SEM. P values were determined by 2-sample Student t test: *P ≤ .05. (D) Immunohistochemistry showed nuclear GATA4 protein (brown staining) in ileal epithelium of Gata4 cKI mice and in the jejunal epithelium of control mice whereas GATA4 protein was absent from ileal epithelium of control mice. Sections from at least 3 control and 3 Gata4 cKI animals were evaluated. Hematoxylin was used to counterstain tissue. Scale bars: 100 μm. (E) Immunoblot analysis of nuclear extracts from jejunal and ileal epithelial cells of control mice and from ileal epithelial cells of Gata4 cKI mice was used to quantify GATA4 protein in ileum of Gata4 cKI mice and to compare GATA4 abundance between control jejunum and GATA4-expressing ileum. The blot shown contains nuclear protein extracts from 3 control and 3 Gata4 cKI animals and is representative of analysis of more than 24 control and Gata4 cKI animals. To quantify protein expression, signal was measured using quantitative infrared immunoblotting (LI-COR) and National Institutes of Health ImageJ software. GATA4 protein levels were normalized to TATA binding protein (TBP) levels. GATA4 expression in ileum of Gata4 cKI mice was 27% the level observed in control jejunum. Molecular weight marker locations are indicated. Error bars show SEM. P values determined by 2-sample Student t test: *P ≤ .05, **P ≤ .001.
Table 1.
Primer and Oligo sSquences for pBigT-Gata4, Southern Probes, Genotyping, RT-PCR, bio-ChIP-PCR, and EMSA
| Gene | Forward | Reverse | Size, bp |
|---|---|---|---|
| pBigT-Gata4 Generation | |||
| Gata4cds | TACTCGAGGGGGCGATGTACCAAAGCCTG | ATGAGCTCTTACGCGGTGATTATGTCCCC | 1348 |
| Southern probes | |||
| pROSA26 5’ probe | CTCTGGCTCCTCAGAGAGCCTC | TCCGGCTGTCTCACAGAACGGC | 137 |
| Genotyping | |||
| ROSA26Gata4 | AAAGTCGCTCTGAGTTGTTAT | GCGAAGAGTTTGTCCTCAACC | 311, transgene |
| AAAGTCGCTCTGAGTTGTTAT | GGAGCGGGAGAAATGGATATG | 603, wt | |
| VillinCre | CAAGCCTGGCTCGACGGCC | CGCGAACATCTTCAGGTTCT | ∼350 |
| Gata4loxP | CCCAGTAAAGAAGTCAGCACAAGGAAAC | AGACTATTGATCCCGGAGTGAACATT | 355, wt |
| 455, loxP | |||
| Gata4 Exon7 | CAGTGCTGTCTGCTCTGAAGCTGT | CCAAGGTGGGCTTCTCTGTAAGAAC | 378, wt |
| 550, flagbio | |||
| BirAJax 14/15 | TTCAGACACTGCGTGACT | GGCTCCAATGACTATTTGC | 500 |
| BirAJax 16/17 | GTGTAACTGTGGACAGAGGAG | GAACTTGATGTGTAGACCAGG | 400, wt |
| RT-PCR | |||
| Aadac | GGCTTAGCCCCAAAACACCA | CAAGAGCCTGAAGGGCAGGA | 241 |
| Abi3bp | ATGAAACATTGGCTTTACCAGCAGA | AAAGCACTAGACAGGGTGGGCTTAG | 124 |
| Acot12 | ACCACAAGATCTCACAGCCCTACTG | ACTCATCAGGTTTTAGCCCACCATT | 188 |
| Asah2 | TGGCCATTTTTGCCATAGCC | ACCACTCTGGGCTCCACGAC | 161 |
| Cd74 | GGCTTGAGACTGGTGTCTGTTTCAT | GAAATGGGGGTTCCCTTTAGATAGC | 115 |
| Dfna5 | TGGGAAACCAGAAATGCTACTTGAG | CACTGGAATTGCCCCAGACATAATA | 150 |
| Dnase1 | TGGCTGTGAACCCTGTGGAA | GTCCTCCAGGCCCCACTTTT | 190 |
| Enpp7 | CATGGTGAAGCAGGTAGACAGGACT | AGTCCAACAGCTCAAACTGGATGTC | 197 |
| Ereg | TGTTTTGACCTCAAAGCTGGGAATA | TTTTAAAACATGTTCACCCGACACC | 151 |
| Gapdh | GATGCCCCCATGTTTGTGAT | GGTCATGAGCCCTTCCACAAT | 150 |
| Nos2 | CTCAGCCCAACAATACAAGATGACC | TGTGGTGAAGAGTGTCATGCAAAAT | 184 |
| Pik3ap | ATGCGACATCCTCATCTTCTACAGC | GTATGTCTGCGTCTTCTGGCTACGA | 109 |
| Prss23 | AGGGCTGGAGAATGGCTAAGTAGTG | GCAAGGACCATCAGTCCTCCTATCT | 144 |
| Rragd | AGTGATCAAAGGTGAACACCACCAT | TGGGAGGCTCAATCTTACATTTCAA | 103 |
| Slc23a1 | TGGATTCCAACCCAGGTGCT | TACCATGCCCATCCCAAAGG | 236 |
| Slc25a48 | CTTTAAGGTGTTTTTCAGGGGCATC | TCCTGATACCTCATTCGCTTCTCAC | 140 |
| Tmigd1 | TTTTGCATCAAGACCATCTCGATTT | TTTCTATTCTGAACCCTTGCCCATT | 180 |
| bio-ChIP-PCR | |||
| Aadac bs | AGCTGAGCTGTGAACAGAAAGGAAC | TTCAATGGCTCGGCTTATGAAGTAG | 101 |
| Abi3bp bs | TGGATTCTTATCTGGGGTGGCTTAT | AAGTCTTCCCAGAGGACTCGTGTTT | 145 |
| Acot12 bs | AGGAGAGCTGTCTGTGCCTCTTTCT | CTCCAACAGATCGTTGATGGGATAG | 183 |
| Asah2_1 bs | TCTCCTGGCCATGTTTGGTCTACTA | AGGGATGGTGAGGGTAAGATTTTCA | 165 |
| Asah2_2 bs | AGGACAGCTGCCTTATCTTATCCTT | AAAACCACCCAAACATTATAGCTG | 109 |
| Alb nc | TACTTAACATAGGGACGAGATGGTA | CATACTAAACGTAGACAAGTTGGCC | 221 |
| Cd36 bs | GCCACACTGTCGTAAGGAGTATTGG | TTTTGCACATTAATCCCTTCGTGAT | 100 |
| Cd74 bs | AGCCTCTAATCTGGAAAAGGGTTCC | GCAGCTCTCTTCCCCTACACACATA | 197 |
| Cdk4 nc46 | CATACAGTGGCTTATTATATTTCC | CTCCACCGCCATGGGGAAACATTC | 259 |
| Dfna5 bs | CCCACTTCCTTGTCAACAATGTCTC | TTTCTTGGCTGATGCTATGGTTTGT | 126 |
| Dll1 nc | TGGCAAGGGAGAGAAGGAAGCA | ACAGCCTTGGTCTGTTCAGTGT | 158 |
| Dnase1 bs | GTGGCCATGAGTCATCCGTCTA | GAGGGAAAGGCTTCCCACAGAT | 146 |
| Enpp7 bs | TAAAAGCCATCCCAGCATCACA | CAAAGTCCCAGGGTCAGACAGG | 108 |
| Ereg bs | CAGGAAAGAGAGTGAAGCGAGTGAG | GAATGAAGTTTCAATGCAGCTCCTG | 192 |
| Fabp1 bs | TATCTCTTTCGGTCTGGTTGCACTC | AGAGAGGCCATTATGTGAAGCTGTG | 123 |
| Fabp6 bs | AGAGTTCTGGGCCAATCTGGTCTAC | TGAGTTACATTAAATGCTCAGCTGCTT | 112 |
| Fgf15 bs | CCAGTTGTCCATATGAATTCTCCA | GGCTGATGGCACTGAGACAGAG | 118 |
| Hprt nc | AGCGCAAGTTGAATCTGC | AGCGACAATCTACCAGAG | 219 |
| Hsd17b6 bs | GAAATCGGAGATTTGAGGTCACTTG | CAGAACTATCCCGCTCCTCTTTCTC | 104 |
| Il33 bs | TCTCTGTTTCTGAGGGGAACCAA | CATCGACCAAGACTGTCAGCATC | 183 |
| Lct bs | CTGGCAGCGCATGATTAAGTTG | TGGGAGGCAGTCTTCATTGTGA | 199 |
| Mep1a bs | TCCCTTTGGGTAGCCTGAATGA | TCGTGGTGACTCCCAACCATAA | 148 |
| Prss23 nc | AGATGATTGATCGGGGCTTTCA | GGTAGAGGCAGCAGGCAACAAG | 106 |
| Prss23 bs | ATGGCCAAGGCAACTTAGAGAAGAG | TTTGTGCTTGGGATTTAAGGTCTGA | 122 |
| Rnf180 bs | CTGGAGCTCAACTGTACCACAGGAT | AAGCAATCCCTGCAGGAAAATAGAC | 173 |
| Slc10a2_1 bs38 | GAATTAAGCCCAACCACACAGT | TTTTAACCCCGTCTTTCCTCTACT | 150 |
| Slc10a2_2 bs38 | GTACAATGATGGGCTGAATGGG | CTCAGATACTTTAACCCACAGGCA | 201 |
| Slc10a2 nc | GGTCTGTTAAACAAAGGGCAAACAA | AGCTCAGTGTTCACCCAACCAGATA | 133 |
| Slc23a1 bs | TCCTGAAGAGCAGCAATGGTGA | GGGCCAGAGTCTACAGGGTGAA | 143 |
| Slc25a48 bs | GATTCATTCCTGGAGGAGGAAAATG | GTTGGGGTGTCATCTTGCTAACCTT | 141 |
| Sqrdl bs | CAGCTGCTTCTGGTAATGAGTGACA | CACAGGACCAATCTTGTTTTCCATC | 140 |
| Suox bs | ATCAGGTAGGTTTTCCCAAGCCAGT | TGGCTTTCTCTTTCCTCCAATACTGT | 122 |
| Tmigd1 bs | TTAATAAGCCTCTCAGGTGGCTGTG | GCAGTGCTAGATCCTTCCCTTTGAT | 135 |
| Ugt2a3 bs | AACTCTCTTGAGCTCCGTGTCTTTG | GTTTCGCAAAGATTGAGGGTTTGTT | 104 |
| Ugt2a3 nc | GAATCAGAGAGGCAAAATGAAGTCC | AAATCATGTTGCATTCCATTGATATTT | 143 |
| Gene | ID | Gene | ID |
|---|---|---|---|
| TaqMan | |||
| Akr1b8 | Mm00484314_m1 | Lactase | Mm01285112_m1 |
| Cck | Mm00446170_m1 | Mep1a | Mm00484970_m1 |
| Cd36 | Mm01135198_m1 | Pdx1 | Mm00435565_m1 |
| Cyp7a1 | Mm00484150_m1 | Rnf180 | Mm01304501_m1 |
| Fabp1 | Mm00444340_m1 | Slc10a2 | Mm00488258_m1 |
| Fabp6 | Mm00434316_m1 | Sqrdl | Mm00502443_m1 |
| Fgf15 | Mm00433278_m1 | Sst | Mm00436671_m1 |
| Gapdh | Mm99999915_g1 | Suox | Mm00620388_g1 |
| Gata4 | Mm00484689_m1 | Trpm6 | Mm00463112_m1 |
| Gata6 | Mm00802636_m1 | Ugt2a3 | Mm00472170_m1 |
| Hsd17b6 | Mm00457343_m1 | ||
| EMSA Probes | |||
| Lct | CATTGTGATAAGATAAAGCTGT | Lct Mutant | CATTGTCGGTGCGGTAAGCTGT |
| Cd36 | TTAGATAAGATTATAGTATGTT | Cd36 Mutant | TTACGGTACGGTTCCGCATGTT |
| Fgf15 | GAACTGTAGATAAACACAGG | Fgf15 Mutant | GAACTGGCCGGTCACACAGG |
| Fgf15_2 | AGGCTCTGATAACAGGCTAA | Fgf15_2 Mutant | CGGCTCTCGGTGCAGGGCCA |
| Slc10a2 | AACTGATGATAAGTTGGCTGCTTATCAAGC | Slc10a2 Mutant | AACTCGGCGGTGCTTGGCTGCCACCGAAGC |
| xiFABP | GGAGATCCCTGTACAGATATGGGGAGAC | ||
bs, binding site; nc, negative control.
Southern Blot
Genomic DNA was isolated from mouse tails by digesting tissue in lysis buffer (4 mol/L urea, 10 mmol/L 1,2-Cyclohexylenedinitrilotetraacetic acid, 0.5% sarkosyl, 0.1 mol/L Tris HCl pH 8.0, 0.2 mol/L NaCl, and 2 μg proteinase K) at 55°C overnight followed by DNA precipitation. Southern blot was performed using standard conditions with EcoRV or BamHI digested DNA and probes illustrated in Figure 1A. Probe 1 was generated by PCR using primers listed in Table 1 and the plasmid pROSA26-5’ as the template.25, 32 Probe 2 was generated by isolating a 381–base pair (bp) product from HindIII digestion of the plasmid pROSA26PA.25
Epithelial Cell Isolation
Epithelial cells (crypt and villus) were isolated from duodenum, jejunum, and ileum following a protocol modified from Guo et al33 in 2009. Briefly, intestine was everted on a glass rod and placed in a conical tube containing a balanced salt solution (BSS) buffer with EDTA (1.5 mmol/L KCl, 96 mmol/L NaCl, 27 mmol/L sodium citrate, 8 mmol/L KH2PO4, 5.6 mmol/L Na2HPO4, and 15 mmol/L EDTA) with 200 μmol/L phenylmethylsulfonyl fluoride. After 25 minutes of vortexing at 4°C to release epithelial cells from villi and crypts, remaining mesenchymal and muscle tissues were removed. Epithelial cell isolates, containing both crypt and villus cells, were pelleted by centrifuging at 2000 rpm for 10 minutes at 4°C. Cells were washed twice with 1× phosphate-buffered saline containing 200 μmol/L phenylmethylsulfonyl fluoride and used for RNA or protein extraction.
Reverse-Transcription PCR
Total RNA was isolated from duodenal, jejunal, and ileal epithelial cells and from liver (RNeasy; Qiagen, Valencia, CA). Complementary DNA (cDNA) was generated from DNase-treated RNA as previously described.34, 35 Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and RT-PCR using [α-32P] deoxyadenosine triphosphate (dATP) were performed as previously described.22, 35, 36 For qRT-PCR and radioactive RT-PCR, glyceraldehyde 3 phosphate dehydrogense (Gapdh) expression was used for normalization. A Storm820 Phosphor Imager (Amersham Biosciences, Pittsburgh, PA) was used to quantitate band intensities from radioactive RT-PCR. Each gene was assayed in at least 3 independent experiments. For TaqMan (Thermo Scientific, Rockford, IL) qRT-PCR of gene expression in duodenum and jejunal epithelium, cDNA from 3 control ROSA26lnlG4/+ and 3 Gata4 cKI ROSA26lnlG4/+Villin-Cre mice were used. For TaqMan qRT-PCR of gene expression in ileal epithelium, cDNA from 5 control ROSA26lnlG4/+ and 6 Gata4 cKI ROSA26lnlG4/+Villin-Cre mice were used. For RT-PCR with [α-32P] dATP to determine gene expression in ileal epithelium, cDNA from 3 control ROSA26lnlG4/+ and 3 Gata4 cKI ROSA26lnlG4/+Villin-Cre mice were used. For all RT-PCR of gene expression in jejunal epithelium (TaqMan qRT-PCR and RT-PCR with [α-32P] dATP), cDNA from 3 control Gata4loxP/loxP and 3 mutant Gata4loxP/loxPVillin-Cre mice were used. Statistical analysis was performed using StatPlus Software (AnalystSoft Inc, Walnut, CA). Error bars represent the SEM. P values were determined by 2-sample Student t test. Table 1 contains primer sequences and TaqMan assay identifiers.
Histochemistry, Immunohistochemistry, and Immunofluorescence
Duodenal, jejunal, and ileal tissue (0.5 cm) was fixed in 4% paraformaldehyde overnight, processed, and paraffin-embedded. H&E staining was performed per standard procedures. Citric acid antigen retrieval was performed for immunohistochemistry. Staining was visualized using R.T.U. Vectastain Elite ABC reagent (Vector Labs, Burlingame, CA) and a Metal Enhanced DAB substrate kit (Thermo Scientific, Rockford, IL). For immunofluorescence, fresh-frozen sections were fixed with 4% paraformaldehyde before antibody staining. Sections from at least 3 control (ROSA26lnlG4/+) and 3 Gata4 cKI animals (ROSA26lnlG4/+ Villin-Cre) were analyzed. Histochemistry and immunohistochemistry slides were scanned using a NanoZoomer slide scanner (Hamamatsu, Bridgewater, NJ), and images from scanned slides were captured using NDP.view 2 software (Hamamatsu). Fluorescent images were captured on a Nikon Eclipse 80i microscope (Melville, NY), using a Nikon digital smart DS-QiMc camera. Table 2 shows antibody details.
Table 2.
Antibodies
| Antibody | Dilution | Manufacturer | Catalog number |
|---|---|---|---|
| Immunohistochemistry and immunofluorescence | |||
| GATA4 C-20 (goat polyclonal) | 1:250 | Santa Cruz Biotechnology, Santa Cruz, CA | sc-1237 |
| ASBT/SLC10A2 (rabbit polyclonal) | 1:4000 | Gift from Paul Dawson | |
| LCT T-14 (goat polyclonal) | 1:50 | Santa Cruz Biotechnology | sc-240614 |
| ECAD (mouse polyclonal) | 1:4000 | BD Transduction Laboratories | 610181 |
| Biotinylated rabbit anti-goat (heavy and light chains) IgG | 15 μl/mL | Vector Labs | BA-5000 |
| Biotinylated goat anti-rabbit (heavy and light chains) IgG | 15 μl/mL | Vector Labs | BA-4000 |
| Immunoblotting | |||
| GATA4 C-20 (goat polyclonal) | 1:1000 | Santa Cruz Biotechnology | sc-1237 |
| TATA binding protein | 1:1000 | Abcam, Cambridge, MA | ab818 |
| ASBT/SLC10A2 (rabbit polyclonal) | 1:1500 | Gift from Paul Dawson | |
| OSTα | 1:1000 | Gift from Paul Dawson | |
| OSTβ | 1:1000 | Gift from Paul Dawson | |
| LCT T-14 (goat polyclonal) | 1:1000 | Santa Cruz Biotechnology | sc-240614 |
| IRDye 800CW donkey anti-goat IgG (heavy and light chains) | 1:15,000 | LI-COR Biosciences | 926-32214 |
| IRDye 680RD donkey anti-mouse IgG (heavy and light chains) | 1:15,000 | LI-COR Biosciences | 926-68072 |
| IRDye 800CW donkey anti-rabbit IgG (heavy and light chains) | 1:15,000 | LI-COR Biosciences | 926-32213 |
Immunoblotting
Nuclear and cytoplasmic protein extracts were prepared from duodenal, jejunal, and ileal epithelial isolates from control ROSA26lnlG4/+ and Gata4 cKI (ROSA26lnlG4/+Villin-Cre) mice using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) and HALT Protease and Phosphatase Inhibitor cocktails without EDTA (Thermo Scientific). Whole-cell lysates were prepared from jejunal and ileal epithelial isolates from control ROSA26lnlG4/+ and Gata4 cKI (ROSA26lnlG4/+Villin-Cre) mice using a 1× NP40 lysis buffer (0.5% NP40, 50 mmol/L Tris pH 8, 10% glycerol, 0.1 mmol/L EDTA pH 8, and 250 mmol/L NaCl) containing HALT Protease and Phosphatase Inhibitor cocktails without EDTA (Thermo Scientific, Rockford, IL). Nuclear proteins (5 μg), cytoplasmic proteins (25 μg), or whole-cell lysates (10 μg) were separated using Nu-PAGE Bis-Tris 4%–12% gradient gels (Invitrogen, Carlsbad, CA), and transferred to an Immobilon-FL polyvinylidene difluoride membrane (Millipore, Darmstadt, Germany). Immunoblots were visualized using an Odyssey Infrared Imaging System (LI-COR, Lincoln, NE) and quantitated using National Institutes of Health ImageJ software (Bethesda, MD). Table 2 shows antibody details.
Oligonucleotide Array Analysis
Total RNA was extracted from duodenal, jejunal, or ileal epithelial cells (RNeasy; Qiagen) harvested from wild-type CD-1 control (n = 3 each duodenum, jejunum, and ileum), ROSA26lnlG4/+ control (n = 3 ileum), Gata4 cKI (ROSA26lnlG4/+Villin-Cre, n = 3 ileum), and Gata4 conditional knockout (cKO) (Gata4loxP/loxPVillin-Cre, n = 3 jejunum) animals. The standard manufacturer’s procedure for GeneChip Mouse Gene 1.0 ST Arrays (Affymetrix, Santa Clara, CA) was followed. Arrays were scanned using a 7G Affymetrix GeneChip Scanner (Affymetrix). Data were analyzed using Partek Genomics Suite software version 6.6 (Partek, Inc, St. Louis, MO). A threshold of at least 2.0-fold change with an unadjusted P value of .05 or less was used to identify transcripts of interest for all comparisons. To generate gene sets with regionally enriched expression, we performed comparisons between duodenum and jejunum, jejunum and ileum, and duodenum and ileum. By using these 3 comparisons, duodenal-enriched transcripts were defined as those with 2.0-fold (P ≤ .05) enrichment over both the jejunum and ileum; jejunal-enriched transcripts were defined as those with 2.0-fold (P ≤ .05) enrichment over both duodenum and ileum; and ileal-enriched transcripts were defined as those with 2.0-fold (P ≤ .05) enrichment over both duodenum and jejunum. In addition, jejunal/duodenal-enriched transcripts were those with equivalent expression between jejunum and duodenum but with 2.0-fold (P ≤ .05) enrichment over ileum. Lists of genes with enriched expression in duodenum, jejunum/duodenum, jejunum, or ileum (Supplementary Table 1) and CEL files from Affymetrix oligonucleotide analysis of ileal epithelium of control and Gata4 cKI mice were used for Gene Set Enrichment Analysis, and normalized enrichment scores were calculated using default parameters.37 Microarray data from this study have been deposited into NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO series accession number GSE75870.
Bio-ChIP Sequencing Re-analysis
Previously published and publicly available alignment files for the input and GATA4–bio-ChIP with high-throughput sequencing (GATA4–bio-ChIP-seq) (GSE6895738) were used for peak calling with model-based analysis of ChIP sequencing v1.0 using a P value of 10-5.39 The remaining parameters were set to the default. The nearest RefSeq gene for each peak within 100 kb was annotated using CisGenome.40
Bio-ChIP
Epithelial cell extracts were obtained from the jejunum of Gata4flbio/flbio::ROSA26BirA/BirA and ROSA26BirA/BirA (n = 6 per genotype).30, 31 Cells were fixed in 1% formaldehyde for 10 minutes with rocking followed by 5 minutes quenching in 125 mmol/L glycine. Cell pellets were frozen before sonication using a Misonix Sonicator 3000 (Misonix, Inc, Farmingdale, NY). Bio-ChIP was performed per previously published protocols with modifications described herein.36, 41 Pull-downs were performed at 4°C using magnetic streptavidin beads (M-280 Dynabeads; Life Technologies) after preclearing chromatin with protein A magnetic beads (Dynabeads). Bead-chromatin complexes were washed for 5 minutes twice with 2% sodium dodecyl sulfate buffer, twice with a high-salt buffer, once with lithium chloride buffer, and twice with Tris-EDTA buffer before chromatin elution in sodium dodecyl sulfate–ChIP elution buffer overnight at 70°C. Eluted chromatin was treated with proteinase K and ribonuclease A followed by phenol-chloroform extraction and ethanol precipitation. GATA4 occupancy was detected by PCR with primers within the GATA4 binding peaks and [α-32P] dATP. PCR products were separated by 4% polyacrylamide gels in 0.5 × Tris-borate-EDTA buffer and visualized by autoradiography using a Storm820 Phosphor Imager (Amersham Biosciences). Band intensity was measured in ImageQuant 5.2 (Molecular Dynamics, Sunnyvale, CA). Percentage enrichment was calculated by dividing the band intensity for the ChIP sample by 10× the band intensity for the input sample to account for the 1:10 dilution of input sample used in the PCR reaction. A 2-sample Student t test was used to compare means.
Electrophoretic mobility Shift Assay
A double-stranded oligonucleotide probe containing an experimentally validated consensus GATA4-binding site from the Xenopus intestinal fatty acid binding protein (xiFABP) gene42, 43 was radiolabeled using Klenow fragment and [α-32P] dATP. Electrophoretic mobility shift assay (EMSA) reactions contained 1× shift buffer (4% Ficoll, 20 mmol/L HEPES pH 7.9, 0.1 mmol/L ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, 0.5 mmol/L dithiothreitol), 2 μg nonspecific DNA poly(dIdC), approximately 1 ng of radiolabeled xiFABP GATA4 probe, and approximately 1 μg nuclear extract from 293T cells expressing exogenous mouse GATA4 from a plasmid-containing GATA4 (pcDNA-GATA4 plasmid, gift from Stephen Duncan). Each competition EMSA reaction contained a 200-fold molar excess of a specific cold competitor double-strand oligonucleotide probe. Two distinct probes were designed for the Fgf15 gene because there were 2 consensus GATA4 binding sites within approximately 100 bp of each other within the peak of interest. Each reaction was incubated for 20 minutes at room temperature. A 4% polyacrylamide gel in 0.5 × Tris-borate-EDTA buffer, pre-electrophoresed at 300 V for approximately 2 hours at 4°C, was loaded with 5 µl of the reaction and run at 300 V for 1.5 hours at 4°C. After drying the gel, bands were visualized by autoradiography. Probe sequences are provided in Table 1.
Results
GATA4 Is Expressed Ectopically in the Ileal Epithelium of ROSA26lnlGata4/+Villin-Cre Mice
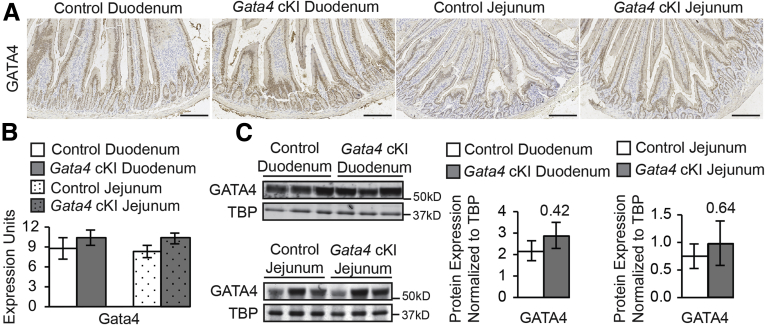
We used a conditional knock-in approach to generate mice that ectopically expressed GATA4 in the ileal epithelium (Figure 1A). We targeted the Gata4 conditional knock-in allele to the ROSA26 locus because ROSA26 promoter activity is ubiquitous and its disruption results in no adverse phenotype.32, 44, 45 The targeted allele consisted of the mouse Gata4 coding sequence downstream of a loxP flanked neomyocin resistance gene expression cassette containing a triple polyadenylation (3×pA) signal sequence (LNL cassette). The 3×pA sequence blocks transcriptional read-through to prevent GATA4 expression in the absence of Cre (Figure 1A). GATA4 expression, therefore, was activated by Cre-mediated excision of the LNL cassette (Figure 1A). Southern blot analysis confirmed germline transmission of the conditional knock-in allele (Gt(ROSA)26Sortm1(Gata4)Bat, designated hereafter as ROSA26lnlG4) (Figure 1B). ROSA26lnlG4 mice were crossed with heterozygous Villin-Cre mice to activate GATA4 expression within the ileal epithelium. Villin-Cre is expressed in all epithelial cells of the small intestine beginning at E13.5.22, 28 To verify GATA4 expression in ileum of ROSA26lnlG4/+Villin-Cre mice (designated Gata4 cKI), we analyzed Gata4 messenger RNA (mRNA) levels by qRT-PCR and GATA4 protein levels by immunohistochemistry and immunoblot. Gata4 mRNA was increased significantly in ileum of Gata4 cKI mice compared with that of controls (Figure 1C). Gata6 transcript levels were equivalent between ileum from control and Gata4 cKI mice, showing that GATA4 induction in the ileum did not affect Gata6 expression (Figure 1C). Immunohistochemistry staining of ileal tissue sections and immunoblot analysis of ileal epithelial cell nuclear protein fractions showed nuclear GATA4 protein within the ileal epithelium of Gata4 cKI mice and its absence within nuclei of control ileal epithelial cells (Figure 1D and E). GATA4 protein levels also were assessed in jejunal samples from control mice to compare protein levels between normal jejunum and GATA4-expressing ileum. Quantification of immunoblots showed the level of GATA4 in Gata4 cKI ileum to be approximately 27% of control jejunal level (Figure 1E). In contrast to ileum, we observed no significant increase in GATA4 mRNA or protein in the epithelium of the duodenum or jejunum from Gata4 cKI mice (Figure 2). This may reflect the strength of the ROSA26 promoter, or it may indicate that there are post-transcriptional and/or post-translational mechanisms in place in the duodenum and jejunum to regulate the total level of GATA4.
Figure 2.
Duodenal and jejunal epithelial cells in Gata4 cKI mice express normal levels of GATA4. (A) Immunohistochemistry showed nuclear GATA4 protein (brown staining) in duodenal and jejunal epithelium of Gata4 cKI mice at similar staining intensity compared with controls. Sections from at least 3 control and 3 Gata4 cKI animals were evaluated. Hematoxylin was used to counterstain tissue. Scale bars: 100 μm. (B) qRT-PCR showed that Gata4 mRNA was unchanged in epithelial cells of the duodenum and jejunum of ROSA26lnlG4/+Villin-Cre (designated Gata4 cKI) mice compared with control mice (ROSA26lnlG4/+) (n = 3 per genotype; experiments performed in triplicate). Glyceraldehyde-3-phosphate dehydrogenase was used for normalization. Error bars show SEM. P values were determined by 2-sample Student t test. (C) Immunoblot analysis of nuclear extracts from duodenal and jejunal epithelial cells of control and Gata4 cKI mice was used to quantify GATA4 protein (n = 3 per genotype). To quantify protein expression, signal was measured using quantitative infrared immunoblotting (LI-COR) and National Institutes of Health ImageJ software. GATA4 protein levels were normalized to TATA binding protein (TBP) levels. GATA4 expression was unchanged in duodenum and jejunum of Gata4 cKI animals compared with control. Molecular weight marker locations are indicated. Error bars show SEM. P values were determined by 2-sample Student t test.
GATA4-Expressing Ileum Loses Ileal Identity
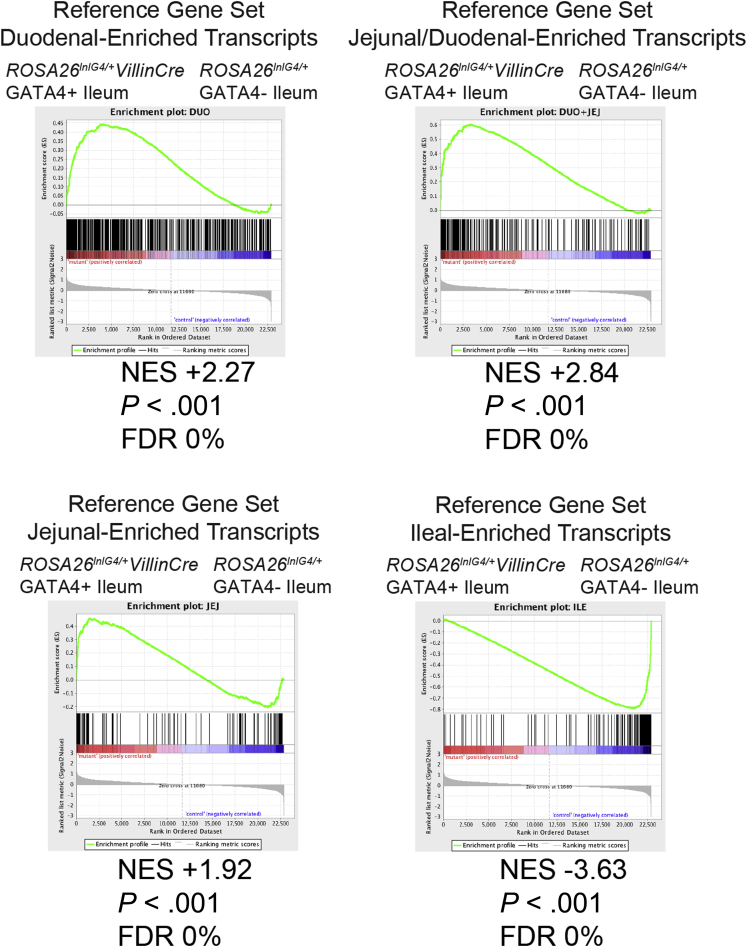
To determine the extent to which GATA4 expression in the ileum altered cellular identity, we defined gene sets with regionally enriched expression along the cephalocaudal axis of the normal intestine and compared these sets of regionally enriched transcripts with the gene expression profile of GATA4-expressing ileal epithelium. By using Affymetrix oligonucleotide array analysis, we compared gene expression among epithelial cell populations from the duodenum, jejunum, and ileum. We included duodenal epithelial cells in our analysis to comprehensively characterize regional gene expression along the cephalocaudal intestinal axis. To identify genes with regionally enriched expression in the intestine, we measured differential gene expression between regions (duodenum vs jejunum, jejunum vs ileum, and duodenum vs ileum) using a 2-fold change threshold and an unadjusted P value of .05 or less (Figure 3A and Supplementary Table 1). We performed gene set enrichment analysis using jejunal-enriched (130), jejunal/duodenal-enriched (288), duodenal-enriched (846), and ileal-enriched (212) gene sets (Figure 3A) and Affymetrix microarray data profiling gene expression of ileal epithelial cells from Gata4 cKI and control mice. Gene set enrichment analysis offers an unbiased computational method to determine whether a defined gene set (jejunal-, jejunal/duodenal-, duodenal-, or ileal-enriched gene sets) shows significant enrichment in 1 of 2 biological states (ileal epithelium of Gata4 cKI vs control mice).37 We found that the global gene expression profile of Gata4 cKI ileal epithelium differed significantly from that of control ileal epithelium, aligning more closely with jejunum and duodenum rather than ileum (Figure 4). We conclude that GATA4-expressing ileum loses ileal identity and that ectopic GATA4 expression within the ileum shifts the ileal transcriptome toward a more proximal identity characteristic of jejunum and duodenum.
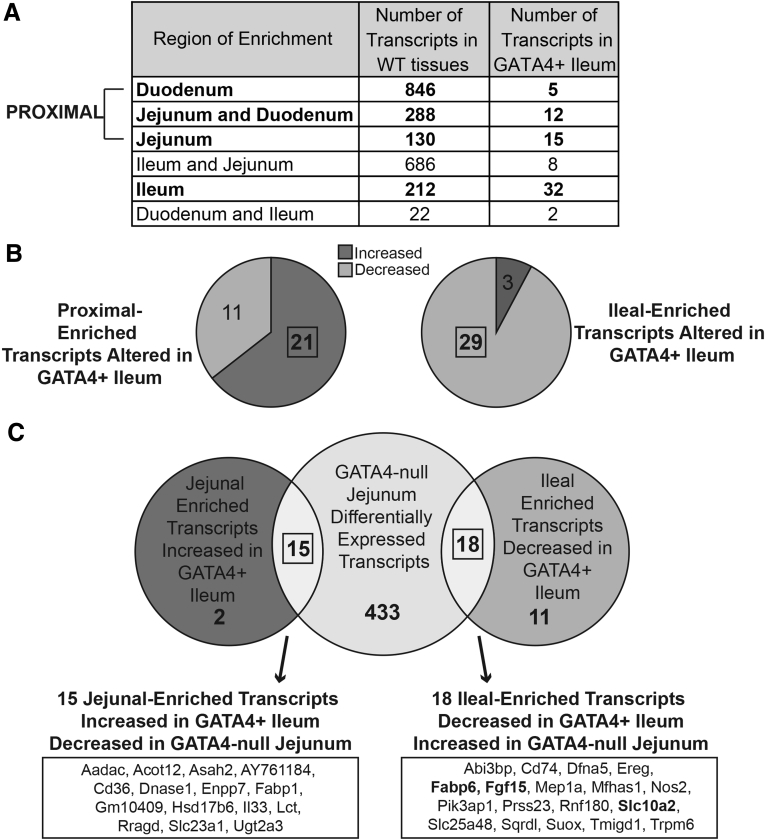
Figure 3.
Identification of high-confidence GATA4 targets that define jejunal vs ileal enterocyte identity. (A) We used Affymetrix oligonucleotide array analysis with RNA from wild-type (WT) CD-1 duodenal, jejunal, and ileal epithelial cells (n = 3) to identify sets of regionally enriched transcripts and with RNA from ileal epithelial cells of control (Rosa26lnlG4/+) and Gata4 cKI (Rosa26lnlG4/+Villin-Cre) mice (n = 3 per genotype) to identify transcripts altered in GATA4-expressing (GATA+) ileum compared with control ileum. A threshold of at least a 2-fold change and an unadjusted P value ≤ .05 was used for comparisons. We determined overlap between transcripts differentially expressed in GATA4+ ileum (136 transcripts) and those identified as either duodenal-enriched, jejunal/duodenal-enriched, jejunal-enriched, or ileal-enriched (bolded text). We found that 47% of transcripts with altered expression in GATA4+ ileum overlapped with proximal-enriched (duodenal, jejunal/duodenal, and jejunal sets, indicated by bracket) or ileal-enriched transcript sets (64 of 136, 32 proximal-enriched, 32 ileal-enriched). (B) Of the 32 proximal-enriched transcripts identified, the expression of 21 was increased in GATA4+ ileum. Of the 32 ileal-enriched transcripts identified, the expression of 29 was decreased in GATA4+ ileum. (C) Focusing on the jejunal-enriched set (17 transcripts, including 15 jejunal-enriched and 12 jejunal/duodenal-enriched) and the ileal-enriched set (29 transcripts), we reasoned that high-confidence GATA4 direct targets would show converse expression changes in the presence or absence of GATA. Therefore, to identify transcripts differentially expressed in GATA4-deficient jejunum, we used Affymetrix oligonucleotide array analysis with RNA from jejunal epithelial cells of Gata4 cKO (Gata4loxP/loxPVillin-Cre) mice (n = 3) and WT CD-1 mice (n = 3, same arrays as in panel A). A threshold of at least a 2-fold change and an unadjusted P value ≤ .05 was used. We determined overlap between the 46 jejunal- or ileal-enriched transcripts and those transcripts differentially expressed in GATA4-deficient jejunum (466 transcripts). We found that 15 of 17 jejunal-enriched transcripts with increased expression in GATA4+ ileum and 18 of 29 ileal-enriched transcripts with decreased expression in GATA4+ ileum overlapped with those altered in GATA4-deficient jejunum and that all 33 overlapping transcripts showed a converse expression pattern in GATA4-deficient jejunum compared with GATA4+ ileum (ie, expression of all 15 jejunal-enriched transcripts was decreased in GATA4-deficient jejunum and expression of all 18 ileal-enriched transcripts was increased in GATA4-deficient jejunum). Specific jejunal- and ileal-enriched transcripts are listed in the box. Those bolded are important in the enterohepatic circulation pathway.
Figure 4.
GATA4-expressing ileum loses ileal identity. Transcripts identified as having enriched expression in the epithelium of duodenum, jejunum/duodenum, jejunum, or ileum of wild-type animals (Figure 3A and Supplementary Table 1) were used as reference gene sets in gene set enrichment analysis. CEL files for microarrays performed with RNA isolated from ileal epithelial cells of control (Rosa26lnlG4/+) and Gata4 cKI (Rosa26lnlG4/+Villin-Cre) mice (n = 3 per genotype) were tested for enrichment of gene sets by gene set enrichment analysis. The top panels and bottom left panel show gene set enrichment analysis using transcripts enriched in duodenal (top left), jejunal/duodenal (top right), or jejunal (bottom left) epithelium as the reference gene set; the bottom left panel shows gene set enrichment analysis using transcripts enriched in ileal epithelium as the reference gene set. Proximal transcripts were highly enriched (normalized enrichment score [NES] = +2.27, +2.84, and +1.92) in GATA4-expressing (GATA4+) ileum, whereas ileal transcripts were enriched in control ileum (NES = -3.63). All were statistically significant by P value and false-discovery rate (FDR).
Based on the phenotype of GATA4-deficient jejunum, which gained ileal character at the expense of jejunal character,21 we predicted that transcripts characteristic of proximal intestine (jejunal, jejunal/duodenal, duodenal) would be induced and that transcripts characteristic of distal intestine (ileum) would be reduced in the presence of GATA4 in the ileum. Of the 136 transcripts identified as differentially expressed by at least 2-fold (P ≤ .05) between control and GATA4-expressing ileum, 47% (64 of 136) were defined as genes with proximal-enriched (32 transcripts) or distal-enriched (32 transcripts) expression (Figure 3A and Supplementary Table 2). Among these 64 genes, expression of 78% (50 of 64) behaved as predicted, 66% (21 of 32) of the identified jejunal-, jejunal/duodenal-, and duodenal-enriched transcripts increased in ileum of Gata4 cKI mice compared with controls (Figure 3B an Table 3), and 91% (29 of 32) of the identified ileal-enriched transcripts decreased in ileum of Gata4 cKI mice compared with controls (Figure 3B and Table 3). Within the set of proximal-enriched transcripts up-regulated in GATA4-expressing ileum, 81% (17 of 21) were among the top 25% most duodenal or jejunal-enriched transcripts (Table 3). Within the set of ileal-enriched transcripts down-regulated in GATA4-expressing ileum, 45% (13 of 29) were among the top 25% most ileal-enriched transcripts (Table 3). Because the remaining 62 genes identified as differentially expressed between controls and Gata4 cKI mice were expressed equivalently along the intestinal cephalocaudal axis in wild-type animals, these were not included in subsequent analyses. Of course, it is possible that GATA4 could contribute to regulation of these genes for a purpose other than defining regional specificity.
Table 3.
Duodenal- and Jejunal-Enriched Transcripts With Increased Expression and Ileal-Enriched Transcripts With Decreased Expression in GATA4+ Ileum
| Duodenal/jejunal transcripts increased in GATA4+ ileum | ||
|---|---|---|
| Gene symbol | cKI fold change | Jejunal fold enrichment |
| Fabp1a | 6.03 | 292.04 |
| Lctb | 6.93 | 140.53 |
| Cd36a | 2.87 | 45.39 |
| Dnase1b | 2.93 | 38.10 |
| Enpp7b | 2.98 | 33.69 |
| Hsd17b6a | 3.25 | 27.78 |
| Slc23a1b | 2.79 | 25.06 |
| Aadaca | 3.74 | 24.56 |
| Ugt2a3a | 3.37 | 23.52 |
| Acot12a | 3.28 | 18.37 |
| Gata4a | 2.61 | 15.24 |
| Asah2b | 3.00 | 14.75 |
| Gstm3a | 2.75 | 8.55 |
| Il33b | 2.79 | 7.95 |
| Rragdb | 2.21 | 7.89 |
| AY761184a | 2.18 | 2.78 |
| Gm10409/Gm3020/Gm3194b | 2.28 | 2.34 |
| Duodenal fold enrichment | ||
|---|---|---|
| Cd59a | 2.19 | 7.49 |
| Slc16a12 | 2.11 | 6.22 |
| Lmcd1 | 2.07 | 5.30 |
| Akr1b8 | 2.47 | 3.32 |
| Ileal transcripts decreased in GATA4+ ileum | ||
|---|---|---|
| Gene symbol | cKI fold change | Ileal fold enrichment |
| Fabp6 | -3.35 | -310.51 |
| Slc10a2 | -3.26 | -112.94 |
| Tmigd1 | -3.51 | -26.15 |
| Suox | -3.53 | -24.13 |
| Nos2 | -2.14 | -13.47 |
| Slc25a48 | -3.63 | -11.92 |
| Scd1 | -3.84 | -6.30 |
| Sqrdl | -2.32 | -5.87 |
| Dfna5 | -2.05 | -4.54 |
| Bex1 | -2.03 | -4.41 |
| Prss23 | -2.45 | -4.39 |
| Mep1a | -2.05 | -4.33 |
| Ly6a | -2.65 | -4.18 |
| Mfhas1 | -2.69 | -3.99 |
| Pik3ap1 | -2.17 | -3.90 |
| Rnf180 | -2.77 | -3.72 |
| Abi3bp | -4.13 | -3.67 |
| Trpm6 | -2.53 | -3.64 |
| Ereg | -2.01 | -3.52 |
| Fgf15 | -5.56 | -3.42 |
| Acta2 | -2.76 | -3.10 |
| Ebf1 | -2.08 | -2.58 |
| Marcksl1 | -2.07 | -2.30 |
| Cd74 | -2.02 | -2.30 |
| Cd53 | -2.62 | -2.20 |
| Mapk4 | -2.04 | -2.15 |
| Dcn | -2.34 | -2.15 |
| Nlrc5 | -2.15 | -2.09 |
| Ighv1-63 | -2.52 | -2.07 |
Jejunal/duodenal-enriched transcripts (equivalently expressed between duodenum and jejunum).
Jejunal-enriched transcripts.
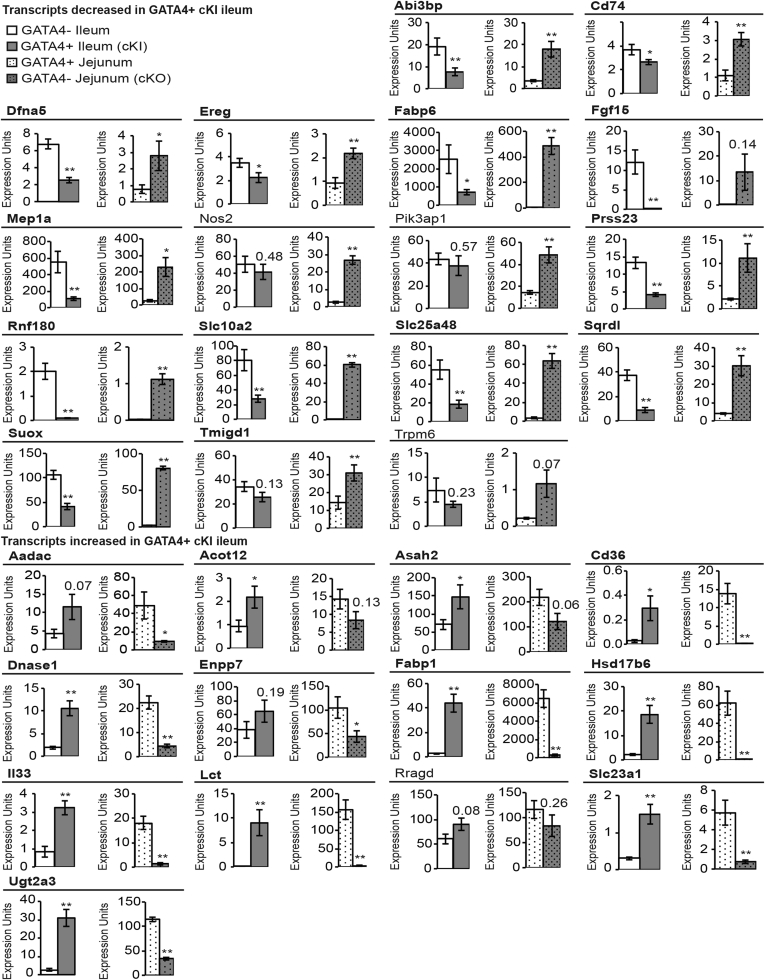
Our next goal was to identify among these genes those most likely to be regulated directly by GATA4 to establish regional specificity along the cephalocaudal intestinal axis. Because we previously identified a functional defect in jejunum lacking GATA4,21 we chose to focus the remainder of our analysis on GATA4’s regulation of jejunal vs ileal gene expression. Therefore, we focused subsequent analyses on the ileal-enriched genes with repressed expression in GATA4-expressing ileum (29 genes) (Table 3) and the jejunal-enriched genes with induced expression in GATA4-expressing ileum (17 genes) (Table 3). For this analysis, jejunal-enriched transcripts were defined as those enriched specifically in the jejunum compared with the duodenum and ileum (8 jejunal-enriched genes) (Table 3) and those equivalently enriched in both the duodenum and jejunum over the ileum (9 jejunal/duodenal-enriched genes) (Table 3).
To identify high-confidence GATA4 targets among these 46 genes, we filtered our data further to identify those with converse expression changes in the presence or absence of GATA4 in the jejunum (ie, increased in Gata4 cKO jejunal epithelium/decreased in Gata4 cKI ileal epithelium or decreased in Gata4 cKO jejunal epithelium/increased in Gata4 cKI ileal epithelium). We determined gene expression changes in jejunal epithelium lacking GATA4 using Affymetrix oligonucleotide array analysis to compare jejunal epithelium of Gata4 cKO (Gata4loxP/loxPVillin-Cre) mice with that of CD-1 control mice (Figure 3C and Supplementary Table 3). Of the 17 jejunal-enriched transcripts up-regulated in Gata4 cKI ileum, we identified 15 as also down-regulated in the GATA4-null jejunum (Figure 3C). Of the 29 ileal-enriched transcripts down-regulated in Gata4 cKI ileum, we found 18 as also up-regulated in the GATA4-null jejunum (Figure 3C). In sum, these analyses identified 33 genes as putative high-confidence GATA4 direct targets.
GATA4 Activates and Represses Gene Expression Within the Intestine to Distinguish Jejunal and Ileal Epithelial Regional Identities
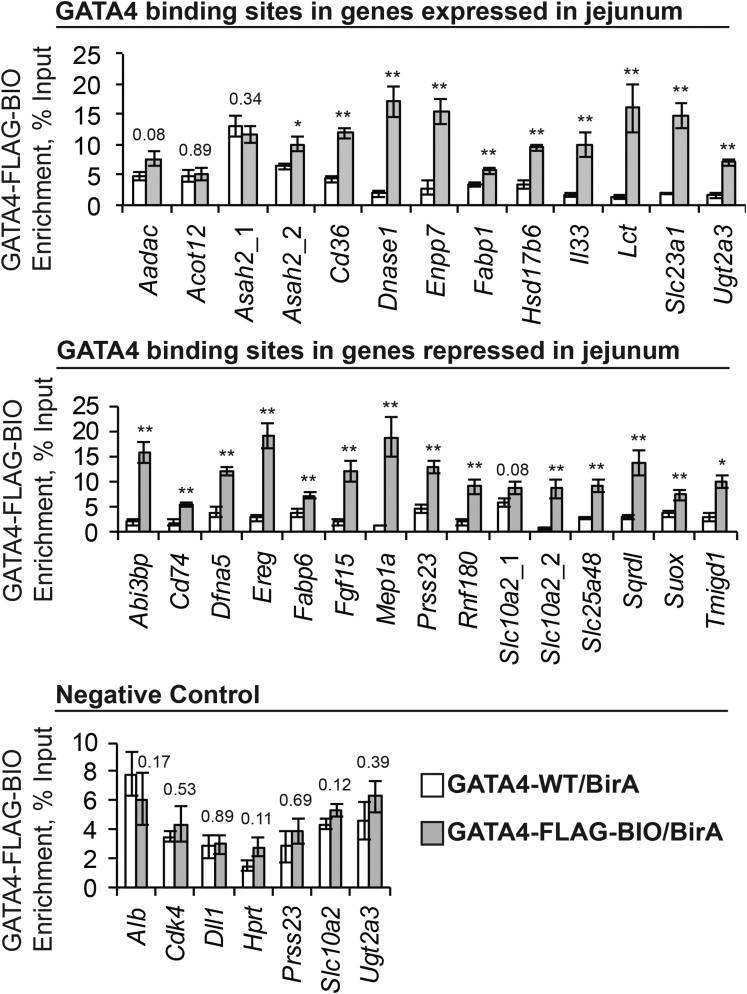
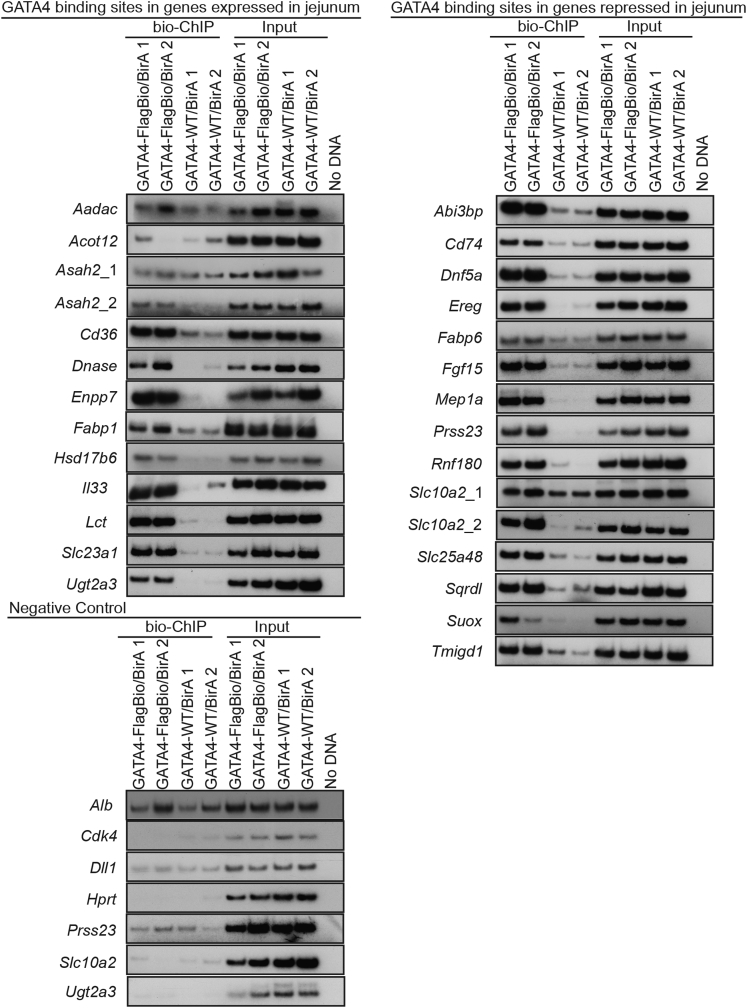
To determine whether GATA4 has the capacity to directly regulate expression of the 33 genes with altered expression in both GATA4-expressing ileum and GATA4-deficient jejunum, we obtained publically available GATA4 bio-ChIP-seq data38 and identified GATA4 chromatin occupancy using model-based analysis of ChIP-seq for peak calling with a 2-sided and increased P value threshold to reduce identification of false-positive peaks.39 Peaks within 100 kb upstream or downstream of the nearest transcription start site (TSS) were annotated (Supplementary Table 4), and GATA4 binding peaks were identified in 30 of the 33 genes. We subsequently eliminated any genes that failed RT-PCR validation for differential expression between ileum of Gata4 cKI and control or between jejunum of Gata4 cKO and control (Figure 5), reducing the set to 26 potential high-confidence GATA4-regulated targets (Table 4). We performed bio-ChIP–PCR using sonicated chromatin from isolated jejunal epithelial cells of Gata4flbio/flbio::ROSA26BirA/BirA mice and ROSA26BirA/BirA mice30, 31, 41 (Figure 6). Biotinylation of endogenous GATA4 protein in Gata4flbio/flbio::ROSA26BirA/BirA mice allows precipitation of GATA4-chromatin complexes with streptavidin.30, 31, 41 We performed 3 independent streptavidin pull-downs, each including chromatin harvested from 2 Gata4flbio/flbio::ROSA26BirA/BirA animals and 2 ROSA26BirA/BirA animals (n = 6 animals of each genotype assayed by bio-ChIP–PCR). We identified the GATA4 binding peak closest to each gene’s TSS and verified that each contained at least 1 consensus GATA4 binding site (A/T-GATA-A/G). Primers flanking the tallest point of the bio-ChIP-seq peak were used for PCR. For negative controls, we designed primers to assay GATA4 occupancy at regions in intestinal-expressed genes that lack GATA4 bio-ChIP-seq peaks (Dll1, Hprt, Prss23, Slc10a2, and Ugt2a3) and to assay GATA4 occupancy at sequences in genes identified as GATA4 targets in other tissues but that are either equivalently expressed in ileum of control and Gata4 cKI mice (Cdk4)46 or absent in ileum of control and Gata4 cKI mice (Alb).47, 48 GATA4 enrichment at putative binding sites in jejunal chromatin from Gata4flbio/flbio::ROSA26BirA/BirA animals compared with that of controls was measured and expressed as a percentage of input (Figure 6, Figure 7 shows representative PCR autoradiographs for each site assayed). We observed GATA4 binding at 9 of 12 regions identified as encoding jejunal-enriched transcripts up-regulated in GATA4-expressing ileum and at 13 of 14 regions identified within genes encoding ileal-enriched transcripts down-regulated in GATA4-expressing ileum (Figure 6). For the 4 genes lacking statistically significant GATA4 enrichment at the peaks residing closest to the gene’s TSS, we examined GATA4 bio-ChIP-seq data to identify the next GATA4 binding peak containing at least 1 consensus GATA binding site. We identified such peaks in 2 of the 4 genes (Slc10a2, peak ∼68 kb downstream of TSS within an intron; Asah2, peak ∼26 kb upstream of the TSS) and assayed GATA4 occupancy at these regions by bio-ChIP-PCR. We observed GATA4 binding to both regions (Figure 6). Taken together, we found that GATA4 binds to sites in all genes identified as encoding ileal-enriched transcripts down-regulated in GATA4-expressing ileum and that GATA4 binds to sites in 10 of 12 genes (83%) identified as encoding jejunal-enriched transcripts up-regulated by expression of GATA4 in the ileum (Figure 7). As expected, GATA4 was not enriched at any negative control sequence queried (0 of 7) (Figure 6).
Figure 5.
Validation of converse gene expression patterns for jejunal- or ileal-enriched transcripts in GATA4-expressing (GATA4+) ileum and GATA4-deficient (GATA4-) jejunum. RT-PCR was used to determine transcript abundance for the 30 jejunal- and ileal-enriched transcripts, identified as having GATA4 binding peaks by bio-ChIP-seq, in ileal epithelial cells from Gata4 cKI (ROSA26lnlG4/+Villin-Cre) and control (ROSA26lnlG4/+) mice and in jejunal epithelial cells from Gata4 cKO (Gata4loxP/loxPVillin-Cre) and control (WT CD-1) mice. The 26 genes confirmed to have converse gene expression patterns in GATA4+ ileum and GATA4- jejunum are shown in bold. Glyceraldehyde-3-phosphate dehydrogenase was used for normalization. Expression of each gene was assayed in at least 3 independent experiments using cDNA from n = 3–6 for control, Gata4 cKI, and Gata4 cKO animals. Error bars represent SEM. *P ≤ .05. **P ≤ .01.
Table 4.
High-Confidence GATA4 Regulated Targets
| Jejunal-enriched transcripts | ||
|---|---|---|
| Gene symbol | cKI Ileum fold change | cKO jejunum fold change |
| Lct | 6.93 | -19.17 |
| Fabp1 | 6.03 | -10.84 |
| Aadac | 3.74 | -2.76 |
| Ugt2a3 | 3.37 | -2.89 |
| Acot12 | 3.28 | -3.75 |
| Hsd17b6 | 3.25 | -18.32 |
| Asah2 | 3.00 | -3.31 |
| Enpp7 | 2.98 | -6.80 |
| Dnase1 | 2.93 | -17.57 |
| Cd36 | 2.87 | -29.24 |
| Slc23a1 | 2.79 | -15.73 |
| Il33 | 2.79 | -5.02 |
| Ileal-enriched transcripts | ||
|---|---|---|
| Gene symbol | cKI Ileum fold change | cKO jejunum fold change |
| Fgf15 | -5.56 | 8.48 |
| Abi3bp | -4.13 | 10.40 |
| Slc25a48 | -3.63 | 7.38 |
| Suox | -3.53 | 22.73 |
| Tmigd1 | -3.51 | 23.35 |
| Fabp6 | -3.35 | 308.59 |
| Slc10a2 | -3.26 | 80.81 |
| Rnf180 | -2.77 | 3.32 |
| Prss23 | -2.45 | 4.77 |
| Sqrdl | -2.32 | 4.83 |
| Dfna5 | -2.05 | 2.79 |
| Mep1a | -2.05 | 4.68 |
| Cd74 | -2.02 | 2.07 |
| Ereg | -2.01 | 2.72 |
Figure 6.
GATA4 occupies sites in jejunal- and ileal-enriched genes, suggesting GATA4 directly regulates expression of jejunal- and ileal-enriched genes in the jejunum to define jejunal enterocyte identity. Bio-ChIP–PCR showed GATA4 enrichment at GATA4 binding sites within genes expressed in jejunum (top panel) and within genes repressed in jejunum (middle panel). No GATA4 enrichment was observed at sites lacking GATA4 bio-ChIP-seq binding sites (Dll1, Hprt, Prss23, Slc10a2, and Ugt2a3) or in genes identified as GATA4 targets in other tissues but that are either equivalently expressed in ileum of control and Gata4 cKI mice (Cdk4) or absent in ileum of control and Gata4 cKI mice (Alb) (bottom panel). Audioradiographic band intensity was measured using a Storm820 Phosphor Imager and ImageQuant software. Representative autoradiographs for each site assayed are shown in Figure 7. Enrichment per sample was normalized to input (n = 6 Gata4flbio/flbio::ROSA26BirA/BirA mice, designated GATA4-FlagBio/BirA, and 6 ROSA26BirA/BirA mice, designated GATA4-WT/BirA). Error bars show SEM. P values were determined by 2-sample Student t test: *P ≤ .05, **P ≤ .005. P values > .05 are listed on graphs. GATA4 occupancy at the binding sites in the Slc10a2 gene (Slc10a2_1 and Slc10a2_2) was analyzed previously by qPCR.38
Figure 7.
Representative autoradiographs of bio-ChIP–PCR. Bio-ChIP–PCR was used to evaluate GATA4 occupancy at predicted binding sites in the 26 high-confidence direct targets we identified and in 7 negative controls (Alb, Cdk4, Dll1, Hprt, Prss23, Slc10a2, and Ugt2a3). GATA4 occupied chromatin was isolated by performing streptavidin pull-down with chromatin from jejunal epithelial cells of GATA4-FlagBio/BirA or GATA4-WT/BirA mice. As representative data, PCR with chromatin from 2 mice per genotype is shown here. Input PCR confirmed that equivalent chromatin amounts were used in pull-downs. In all, 6 mice per genotype were assayed by bio-ChIP–PCR.
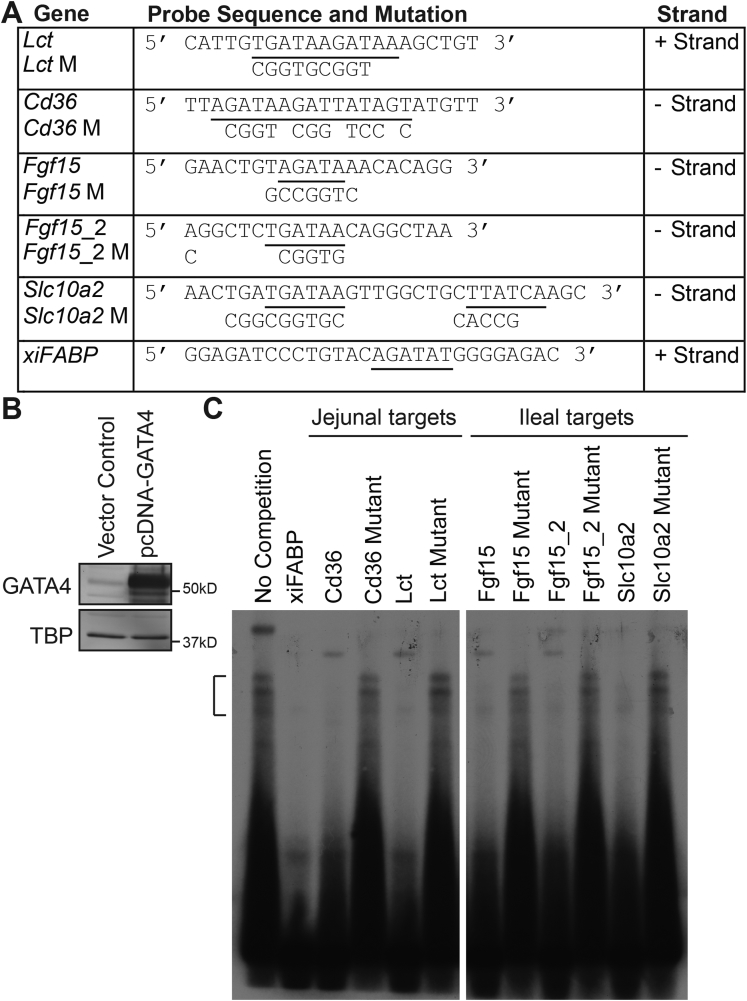
As proof in principle that our methodology identified bona fide GATA4 binding sequences, we selected 2 genes with jejunal-enriched expression (Lct and Cd36) and 2 with ileal-enriched (Fgf15 and Slc10a2) expression and used an EMSA to test GATA4 binding to its binding sites within these genes (Figure 8). Nonradiolabeled probes containing gene-specific GATA4 consensus sequence(s) previously queried by bio-ChIP-PCR (Figure 8A) were tested for their ability to compete with a well-characterized GATA4 binding site from the Xenopus intestinal fatty acid binding protein (xiFABP) gene42, 43 for binding to exogenous GATA4 protein (Figure 8B). We also introduced mutations into each GATA4 consensus sequence (Figure 8A) and tested the ability of each mutant to compete for GATA4 binding by EMSA. The unlabeled xiFABP sequence as well as every unlabeled wild-type GATA4 consensus binding site from the targets tested inhibited GATA4 binding to the radiolabeled xiFABP probe (Figure 8C). In contrast, all mutated GATA4 consensus sequences failed to compete with or minimally competed with the radiolabeled xiFABP probe for GATA4 binding (Figure 8C). Taken together, GATA4 binding data (EMSA and ChIP) and our observation of converse changes in gene expression in ileum expressing GATA4 and jejunum lacking GATA4 strongly suggest that GATA4 regulates regional gene expression within the intestine by directly binding to genes encoding jejunal transcripts to activate transcription and to genes encoding ileal transcripts to repress transcription. Of course, to definitively prove direct regulation of these genes by GATA4, it would be necessary to mutate the GATA4 binding sites within these targets and show that such mutations abolish GATA4-dependent regulation of target gene expression within mouse jejunal epithelium.
Figure 8.
GATA4 binds to GATA consensus binding sites in jejunal-enriched genes and in ileal-enriched genes regulated by GATA4 expression in the intestine. (A) Table shows double-stranded oligonucleotide EMSA probes (5’ to 3’). Underlined sequences identify the location of GATA4 consensus binding sites within probes. Nucleotides mutated to eliminate GATA4 consensus binding sites in mutant probes are indicated. Two distinct probes were designed for the Fgf15 gene because there were 2 consensus GATA4 binding sites within approximately 100 bp of each other within the peak of interest. (B) Immunoblot analysis of nuclear extracts from 293T cells transfected with either control empty plasmid or GATA4 containing plasmid (pcDNA-GATA4) shows overexpression of GATA4 in pcDNA-GATA4–transfected cells. GATA4 protein levels were normalized to TATA binding protein (TBP) levels. Molecular weight marker locations are indicated. (C) Representative EMSA competition assay is shown. Nuclear extract from pcDNA-GATA4–transfected 293T cells was incubated with radiolabeled xiFABP probe in the absence (lane 1) or presence (lanes 2–11) of 200-fold molar excess of unlabeled competitors. A bracket indicates the location of GATA4-bound xiFABP probe. Data are representative of 3 independent assays.
Expression of GATA4 Jejunal Targets Is Not Increased in Jejunum of Gata4 cKI Mice
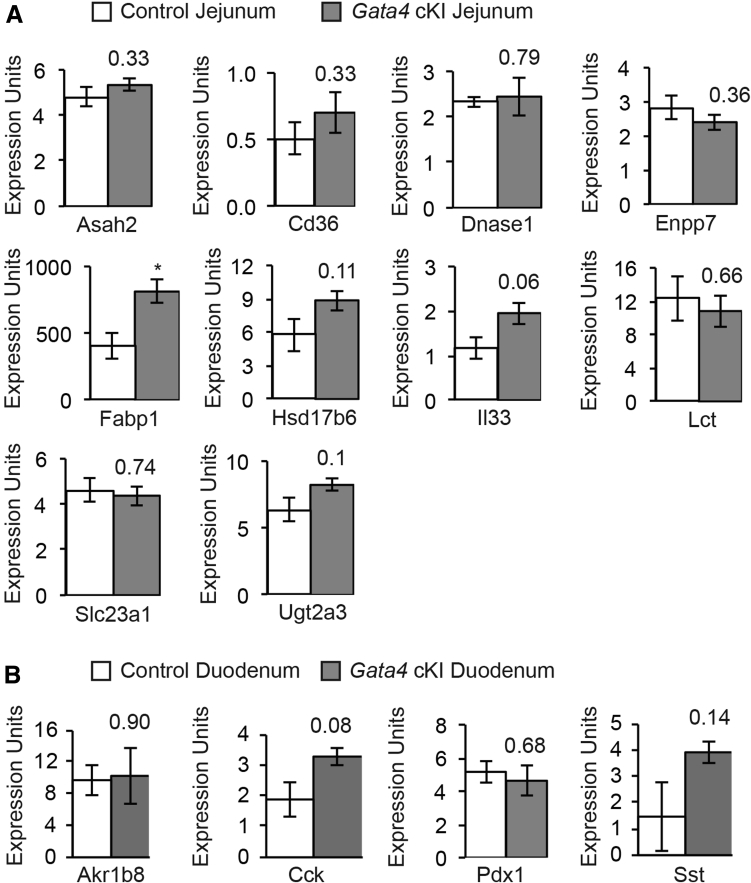
We compared the expression of the 10 jejunal-enriched targets occupied by GATA4 in jejunal epithelium from control and Gata4 cKI mice by qRT-PCR. We found only Fabp1 transcript to be increased significantly between these groups (Figure 9A). Although interleukin 33 (Il33) transcript was increased, the P value for this change was just outside the significance cut-off value of P < .05. We also used qRT-PCR to compare expression of 4 transcripts identified by our analysis as duodenal-enriched (Akr1b8, Cck, Pdx1, and Sst) (Supplementary Table 1) in duodenal epithelium from control and Gata4 cKI mice. We observed no difference in these markers between control and Gata4 cKI mice (Figure 9B). Although Cck and Sst transcript levels trended toward slightly higher levels in the duodenum of Gata4 cKI mice compared with controls, the P value for these was outside of the significance cut-off value of P < .05. These results were not unexpected because we showed earlier that GATA4 protein was not increased significantly in the duodenum or jejunum of Gata4 cKI mice compared with controls (Figure 2).
Figure 9.
Expression of jejunal and duodenal transcripts is unchanged in Gata4 cKI animals. (A) qRT-PCR was used to determine transcript abundance of the 10 jejunal-enriched transcripts, identified as having enriched GATA4 binding by bio-ChIP–PCR (Figure 7) in jejunal epithelial cells from control and Gata4 cKI mice. (B) qRT-PCR was used to determine transcript abundance of 4 duodenal transcripts in duodenal epithelial cells from control and Gata4 cKI mice. (A and B) Glyceraldehyde-3-phosphate dehydrogenase was used for normalization. Expression of each gene was assayed in at least 3 independent experiments (n = 3 per genotype). Error bars represent SEM. P values were determined by 2-sample Student t test.
Enterohepatic Circulation Is Altered in Gata4 cKI Mice
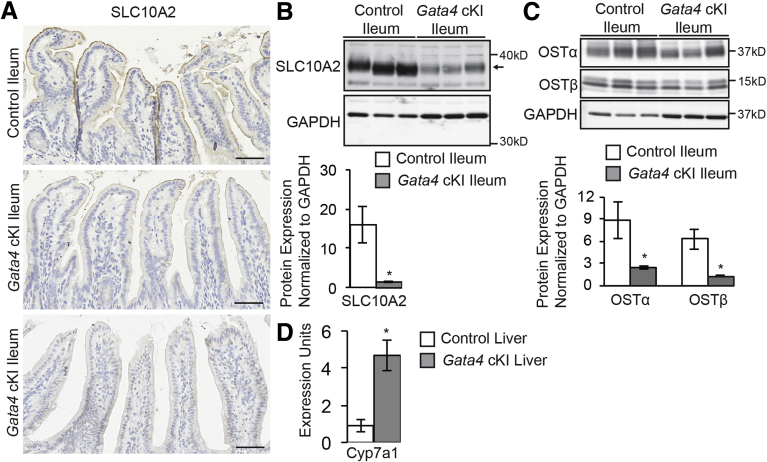
Among the set of GATA4 targets repressed in GATA4-expressing ileum were those involved in regulating an important enterohepatic circulation pathway. Uptake of luminal bile acids by ileal enterocytes occurs via the bile acid transporter SLC10A2.49 Within enterocytes, bile acids bind to the intracellular bile acid binding protein FABP6 and ultimately are transported from the enterocyte into the circulation by the basolateral heterodimeric organic solute transporter (OST), which consists of OSTα and OSTβ proteins (encoded by the Slc51a and Slc51b genes, respectively).6, 50 Bile acids also act as ligands for the nuclear receptor FXR.6 Bile acid–bound FXR promotes Fgf15 expression, and secretion of FGF15 into the portal venous circulation results in FGF receptor 4 activation on hepatocytes.4, 5 This activation inhibits expression of Cyp7a1, the gene encoding the rate-limiting enzyme in bile acid synthesis.4 Genes encoding proteins involved in this pathway that were decreased significantly in GATA4-expressing ileum included Slc10a2, Fabp6, and Fgf15 (Table 3 and Figure 3). Moreover, by relaxing the fold-change criterion in analyzing transcripts altered in GATA4-expressing ileum, we found expression of 3 additional transcripts encoding ileal-enriched bile acid uptake pathway proteins to be decreased: Nr1h4 (Fxr), -1.4-fold, P ≤ .05; Slc51a (Ostα), -1.7-fold, P ≤ .01; and Slc51b (Ostβ), -1.6-fold, P ≤ .05. To determine if changes in mRNA abundance correlated with changes in protein abundance, we performed immunohistochemistry to compare SLC10A2 levels in ileum between Gata4 cKI mice and control mice. Overall, we observed decreased SLC10A2 protein at the epithelial brush border of ileum from Gata4 cKI mice compared with control mice (n = 7 control, n = 14 Gata4 cKI) (Figure 10A). In some regions, SLC10A2 was undetectable (Figure 10A, bottom panel); in other regions, SLC10A2 levels were greatly reduced (Figure 10A, middle panel). We used immunoblot to quantify total SLC10A2, OSTα, and OSTβ proteins in GATA4-expressing ileal enterocytes and control ileal enterocytes and found that ileum of Gata4 cKI mice contained only 9% of SLC10A2 protein typical of control tissue (Figure 10B) and only 26% and 19% of OSTα and OSTβ proteins typical of control tissue, respectively (Figure 10C). Because we observed lower levels of transcripts and proteins encoded by genes required for bile acid uptake and enterohepatic signaling, we examined the expression of hepatic Cyp7a1. We found Cyp7a1 mRNA expression to be increased significantly in livers of Gata4 cKI mice compared with that of controls (Figure 10D). These findings are consistent with decreased Cyp7a1 transcript observed in livers of Gata4 cKO animals.21 Therefore, we conclude that enterohepatic signaling was altered in mice expressing GATA4 in the ileum.
Figure 10.
Enterohepatic signaling is altered in animals expressing GATA4 in the ileum. (A) Immunohistochemistry for SLC10A2 (brown stain) shows SLC10A2 protein lining the brush border of control ileum (n = 7, upper panel). In contrast, SLC10A2 staining was faint to nearly absent along the brush border of ileum from Gata4 cKI mice (n = 14, 7 of 14 faint SLC10A2 staining, middle panel, and 7 of 14 low to no SLC10A2 staining, lower panel). Immunohistochemistry from 2 independent Gata4 cKI mice is shown as representative of the 2 types of SLC10A2 staining observed. (B) Immunoblot using whole-cell extracts from ileal epithelium of control and Gata4 cKI mice was used to measure expression of SLC10A2 protein. To quantify protein expression, signal was measured using quantitative infrared immunoblotting (LI-COR) and National Institutes of Health ImageJ software. SLC10A2 protein levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. SLC10A2 expression in ileum of Gata4 cKI mice (ROSA26lnlG4/+Villin-Cre) was 9% of the level observed in control ileum (ROSA26lnlG4/+; n = 3 animals per genotype). Arrowhead indicates the SLC10A2 band measured. Molecular weight marker locations are indicated. Error bars show SEM. P values were determined by 2-sample Student t test: *P ≤ .05. (C) Immunoblot using whole-cell extracts from ileal epithelium of control and Gata4 cKI mice was used to measure expression of OSTα and OSTβ proteins. To quantify protein expression, signal was measured using quantitative infrared immunoblotting (LI-COR) and National Institutes of Health ImageJ software. OSTα and OSTβ protein levels were normalized to GAPDH levels. Expression of OSTα and OSTβ proteins in ileum of Gata4 cKI mice (ROSA26lnlG4/+Villin-Cre) was 26% and 19% of the level observed in control ileum (ROSA26lnlG4/+), respectively (n = 3 animals per genotype). Molecular weight marker locations are indicated. Error bars show SEM. P values were determined by 2-sample Student t test: *P ≤ .05. (D) qRT-PCR shows increased Cyp7a1 expression in liver from Gata4 cKI mice (ROSA26lnlG4/+Villin-Cre) compared with control mice (ROSA26lnlG4/+; n = 3 animals per genotype). Glyceraldehyde-3-phosphate dehydrogenase was used for normalization. Error bars represent SEM. *P ≤ .05.
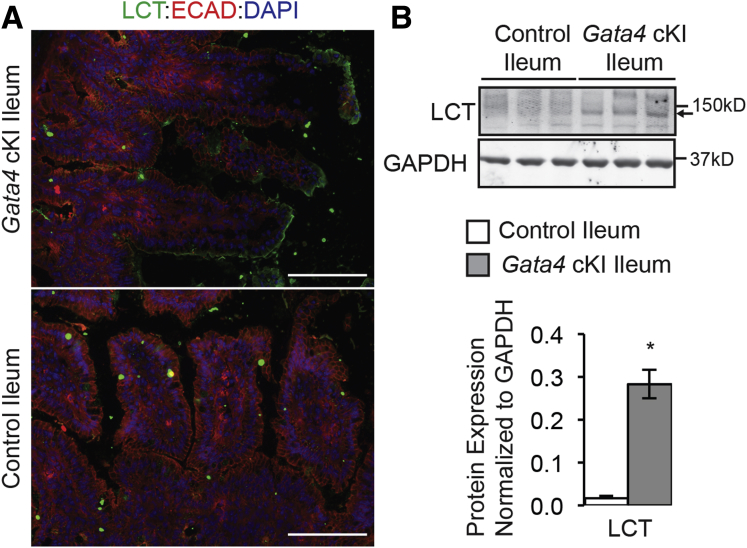
Lactase Protein Is Induced in the Ileum of Gata4 cKI Mice
The finding that changes in ileal-specific gene transcription affected ileal-specific protein expression led us to examine the status of jejunal protein expression in GATA4-expressing ileum. Because lactase (LCT), a well-documented marker of jejunal function, was identified as the top jejunal target induced by expression of GATA4 in the ileum (Table 3), we chose to examine LCT protein expression in ileum of control and Gata4 cKI mice. We used immunostaining and immunoblot analyses to examine LCT protein expression in ileum from control and Gata4 cKI mice. We observed brush-border expression of LCT protein only in GATA4-expressing ileum (Figure 11A). Immunoblot analysis further showed induction of LCT protein in ileal epithelial cells of Gata4 cKI mice and not in cells of control mice (Figure 11B). These data show that GATA4-expressing ileal cells can express jejunal proteins and suggest that these cells would be capable of accomplishing jejunal functions.
Figure 11.
Lactase protein is expressed in Gata4 cKI ileum. (A) Immunofluorescent staining showed Lactase protein (LCT, green) at the brush border in ileal epithelium of Gata4 cKI mice, whereas LCT protein was absent from ileal epithelium of control mice. Sections from at least 3 control and 3 Gata4 cKI animals were evaluated. E-cadherin (ECAD, red) was used to stain epithelial cell membranes. 4′,6-diamidino-2-phenylindole (DAPI) (blue) stained nuclei. Scale bars: 100 μm. (B) Immunoblot analysis of protein extracts from ileal epithelial cells of control and Gata4 cKI mice was used to quantify LCT protein (n = 3). To quantify protein expression, signal was measured using quantitative infrared immunoblotting (LI-COR) and National Institutes of Health ImageJ software. Arrowhead indicates the LCT band measured. LCT protein levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. Molecular weight marker locations are indicated. Error bars show SEM. P values were determined by 2-sample Student t test: *P ≤ .05.
Discussion
Together, our data show that GATA4 is sufficient to confer jejunal identity and to repress ileal identity in the small intestinal epithelium. These findings refine our understanding of GATA4’s critical function in patterning the intestinal epithelium along its cephalocaudal axis, initially uncovered through studies with Gata4 cKO mice.19, 21, 24 Moreover, we identified a novel mechanism through which GATA4 may directly repress Fgf15 expression to influence enterohepatic signaling. Although the level of GATA4 protein expressed in Gata4 cKI ileal epithelium was only 27% of that expressed in jejunal epithelium, it nevertheless was sufficient to alter the global ileal gene expression profile such that it aligned more closely with jejunum and duodenum rather than ileum. Of the genes with expression changed by addition of GATA4 to the ileum, roughly half were genes with proximal-enriched (jejunal and duodenal) or ileal-enriched expression patterns. Moreover, gene expression not only was affected positively by the presence of GATA4 but also was affected negatively, implying that GATA4 normally patterns the jejunal-ileal boundary by activating and repressing transcription.
Our approach to identify and to validate GATA4 transcriptional targets among those genes encoding jejunal-enriched or ileal-enriched transcripts was stringent. The facts that we confirmed GATA4 occupancy by ChIP in all but 2 of the 26 genes (92%) we identified and that we showed GATA4 binding to its consensus sites within a subset of these genes by EMSA show the validity of our approach to identify GATA4 targets involved in regulating jejunal vs ileal identity. Because we limited our analysis of GATA4 binding sites to those satisfying specific and stringent criteria, we cannot exclude the possibility that additional important GATA4 targets involved in determining jejunal vs ileal identity exist and were omitted by our analysis. The number of genes encoding jejunal- or ileal-enriched transcripts regulated by GATA4 to define the jejunal-ileal boundary, therefore, likely was under-represented by this analysis. Nevertheless, we conclude that GATA4 patterns the intestinal jejunal-ileal boundary by activating and repressing transcription of key regional-defining transcripts within the jejunum.
An important consequence of patterning discrete jejunal and ileal domains is the restriction of bile acid absorption to the ileum. We conclude that GATA4 controls spatial patterning of bile acid absorption along the cephalocaudal axis of the small intestine by repressing expression of key genes encoding proteins essential for bile acid homeostasis. Our work supports a model in which GATA4 prevents inappropriate proximal absorption of bile acids by directly suppressing Slc10a2, Fabp6, and Fgf15 expression; exclusion of GATA4 from the terminal ileum thereby provides a permissive environment for transcription of these same genes and, therefore, for bile acid uptake. Moreover, GATA4 may directly repress additional genes encoding important regulators of bile acid homeostasis that were not identified by our study. Our strategy required 2-fold changes in gene expression in both GATA4-deficient jejunum and GATA4-expressing ileum. The level of GATA4 reconstituted in mutant ileum, however, was not equivalent to that normally expressed in jejunum. Therefore, it is possible that the amount of GATA4 present in ileum of Gata4 cKI mice was insufficient to reduce expression of some targets by the 2-fold threshold. In fact, by relaxing the fold-change criterion in analyzing transcripts altered in GATA4-expressing ileum, we identified 3 additional key bile acid pathway genes—Nr1h4, Slc51a, and Slc51b— with decreased expression in GATA4-expressing ileum. Moreover, expression of Nr1h4, Slc51a, and Slc51b was increased in GATA4-deficient jejunum (Supplementary Table 3 and Battle et al21), and each contains GATA4 binding peaks (Supplementary Table 4), suggesting these genes as additional possible direct GATA4 targets. These data suggest that GATA4 coordinates the repression of expression of multiple genes required for bile acid absorption, thereby excluding this function from the jejunum, which is important for efficient lipid uptake by the jejunum.
By contributing to spatial regulation of bile acid homeostasis, GATA4 influences FGF15-mediated enterohepatic signaling. FGF15 functions as a hormone with a primary target being the liver where it controls hepatic expression of Cyp7a1, the gene encoding the rate-limiting enzyme for synthesis of bile acids from cholesterol.3, 4, 6, 51, 52, 53 Supporting a role for GATA4 in modulating FGF15-mediated enterohepatic signaling, we found that hepatic Cyp7a1 levels were increased 4-fold in Gata4 cKI mice, which express little to no intestinal Fgf15. This agrees with our previous finding that Gata4 intestinal cKO mice, which overexpress Fgf15 in the jejunum, have decreased hepatic Cyp7a1 transcript.21 GATA4 likely influences Fgf15 levels through direct and indirect mechanisms. GATA4 repression of Slc10a2 expression would suppress cellular bile acid uptake and downstream bile acid activation of FXR-mediated Fgf15 transcription.4, 5, 52, 53 Moreover, as described earlier, GATA4 also may repress transcription of Fxr itself, and in this way could reinforce silencing of Fgf15 and other FXR transcriptional targets such as Fabp6, Slc51a, and Slc51b in the jejunum.50, 54 Importantly, we show that GATA4 likely directly represses Fgf15 expression by binding to a consensus GATA4 regulatory element in intron 2 of the Fgf15 gene. These sites are approximately 140-bp upstream of a consensus FXR binding site shown to be important for Fgf15 expression.4 The proximity of GATA4 and FXR binding sites within the Fgf15 gene suggests that GATA4 potentially could modulate FXR binding to Fgf15. Because defects in enterohepatic FGF15/19 signaling have been linked to human diseases including cholestatic liver disease, nonalcoholic fatty liver disease, type 2 diabetes, metabolic syndrome, Crohn’s disease, bile acid malabsorption, and bile acid diarrhea,7, 8, 9, 11 it is important to understand how Fgf15 is regulated, and our study contributes a novel mechanism through which GATA4 represses Fgf15 transcription within the proximal intestine to maintain appropriate FGF15 levels.
Our finding that lactase protein was expressed in the brush border of ileal enterocytes in Gata4 cKI mice implies that in addition to changing the ileal transcriptome, GATA4 expression in the ileum has the capacity to confer jejunal function to the ileum. Directly showing enhanced jejunal function in Gata4 cKI mice, however, would be virtually impossible given that the endogenous jejunal domain is fully intact and fully functional in Gata4 cKI mice. The jejunum is very efficient in performing its digestive and absorptive tasks. For example, we previously have shown that wild-type mice absorb approximately 97% of dietary fat.21 Given the saturated level of jejunal function in mice with an intact jejunum, it would be quite difficult to experimentally detect an enhancement of jejunal function. It is likely that detection of a measurable enhancement of jejunal function in the ileum of Gata4 cKI mice would require disruption of endogenous jejunal function, which is beyond the scope of this study.
In summary, we show that GATA4 is sufficient to establish jejunal enterocyte identity and to repress ileal enterocyte identity in the intestinal epithelium. Not only are key jejunal- and ileal-enriched transcript levels changed in GATA4-expressing ileum, but levels of jejunal- and ileal-enriched proteins also are altered in GATA4-expressing ileum. Changes in liver gene expression in Gata4 cKI mice, downstream of bile acid and FGF15 signaling, show that there are important physiological consequences resulting from the addition of GATA4 to the ileum. This study hones our understanding of a key GATA4-dependent molecular mechanism that patterns the intestinal epithelium along its cephalocaudal axis by elaborating on GATA4’s critical function as an essential dominant molecular determinant of jejunal enterocyte identity. Future studies to determine how GATA4 discriminates between targets that it activates or represses will be important. Genetic evidence suggests that FOG1 is essential for GATA4-mediated repression of at least Slc10a2 and Fabp6.24 GATA4-dependent inhibition of histone H3 lysine 27 acetylation also has been implicated as a mechanism through which GATA4 represses transcription.38 Finally, as a factor that is both necessary and sufficient to program jejunal enterocyte identity, GATA4 represents a logical candidate to consider in developing therapies to treat intestinal failure, particularly for those designed to restore lost functions to remaining intestinal tissue or for tissue-engineering approaches to overcome small-bowel organ shortages for transplant.
Acknowledgments
The authors thank Drs Stephen Krasinski and Boaz Aronson (Division of Gastroenterology and Nutrition, Department of Medicine, Children’s Hospital Boston, and Harvard Medical School, Boston, MA) for providing Gata4flbio/flbio::ROSA26BirA/BirA and ROSA26BirA/BirA mouse lines, and Dr Frank Costantini (Columbia University, New York, NY) for providing pBigT and pROSA26PA plasmids. The authors thank Jixuan Li and Dr Roman Stavniichuk (Department of Cell Biology, Neurobiology, and Anatomy, Medical College of Wisconsin, Milwaukee, WI) for expert technical assistance and Dr Stephen Duncan (Department of Regenerative Medicine and Cell Biology, Medical University of South Carolina, Charleston, SC) for providing the pcDNA-GATA4 plasmid and for helpful discussions and input into the project.
Cayla A. Thompson contributed to the experimental design, performed most of the experiments, analyzed data, and prepared the manuscript draft; Kevin Wojta generated the Gata4 conditional knock-in mouse line; Kirthi Pulakanti and Sridhar Rao analyzed the bioChIP-seq data and contributed to manuscript preparation; Paul Dawson contributed to the experimental design and reviewed the manuscript; and Michele A. Battle conceived and coordinated the project, contributed to the experimental design and data analysis, prepared figures, and prepared the final version of the manuscript. All authors read and approved the final manuscript.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the US National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (DK087873 to M.A.B. and DK047987 to P.D.), Advancing a Healthier Wisconsin (M.A.B.), Hyundai Hope on Wheels Award (S.R.), and the Midwest Athletes against Childhood Cancer fund (S.R.).
Supplementary Material
References
- 1.Binder H.J., Reuben A. Nutrient digestion and absorption. In: Boron W.F., Boulpaep E.L., editors. Medical physiology: a cellular and molecular approach. Updated ed. Elsevier Saunders; Philadelphia: 2005. pp. 947–974. [Google Scholar]
- 2.Davis R.A., Attie A.D. Deletion of the ileal basolateral bile acid transporter identifies the cellular sentinels that regulate the bile acid pool. Proc Natl Acad Sci U S A. 2008;105:4965–4966. doi: 10.1073/pnas.0801194105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landrier J.F., Eloranta J.J., Vavricka S.R. The nuclear receptor for bile acids, FXR, transactivates human organic solute transporter-alpha and -beta genes. Am J Physiol Gastrointest Liver Physiol. 2006;290:G476–G485. doi: 10.1152/ajpgi.00430.2005. [DOI] [PubMed] [Google Scholar]
- 4.Inagaki T., Choi M., Moschetta A. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Song K.H., Li T., Owsley E. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009;49:297–305. doi: 10.1002/hep.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grober J., Zaghini I., Fujii H. Identification of a bile acid-responsive element in the human ileal bile acid-binding protein gene. Involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J Biol Chem. 1999;274:29749–29754. doi: 10.1074/jbc.274.42.29749. [DOI] [PubMed] [Google Scholar]
- 7.Jahn D., Rau M., Hermanns H.M. Mechanisms of enterohepatic fibroblast growth factor 15/19 signaling in health and disease. Cytokine Growth Factor Rev. 2015;26:625–635. doi: 10.1016/j.cytogfr.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 8.Kliewer S.A., Mangelsdorf D.J. Bile acids as hormones: the FXR-FGF15/19 pathway. Dig Dis. 2015;33:327–331. doi: 10.1159/000371670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen B.M., Mangelsdorf D.J., Kliewer S.A. Tissue-specific actions of the metabolic hormones FGF15/19 and FGF21. Trends Endocrinol Metab. 2015;26:22–29. doi: 10.1016/j.tem.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrebee C.B., Dawson P.A. Metabolic effects of intestinal absorption and enterohepatic cycling of bile acids. Acta Pharm Sin B. 2015;5:129–134. doi: 10.1016/j.apsb.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keely S.J., Walters J.R.F. The farnesoid X receptor: good for BAD. Cell Mol Gastroenterol Hepatol. 2016;2:725–732. doi: 10.1016/j.jcmgh.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez I.M., Kang K.H., Jaksic T. Neonatal short bowel syndrome. Semin Fetal Neonatal Med. 2011;16:157–163. doi: 10.1016/j.siny.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Helmrath M.A., Deonarine K. Clinical small intestine. In: Gumucio D.L., Samuelson L.C., Spence J.R., editors. Translational gastroenterology: organogenesis to disease. Hoboken, NJ; John Wiley & Sons: 2014. p. 99. [Google Scholar]
- 14.Wales P.W., de Silva N., Kim J. Neonatal short bowel syndrome: population-based estimates of incidence and mortality rates. J Pediatr Surg. 2004;39:690–695. doi: 10.1016/j.jpedsurg.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 15.Sanderson I.R. Dietary regulation of genes expressed in the developing intestinal epithelium. Am J Clin Nutr. 1998;68:999–1005. doi: 10.1093/ajcn/68.5.999. [DOI] [PubMed] [Google Scholar]
- 16.Spence J.R., Lauf R., Shroyer N.F. Vertebrate intestinal endoderm development. Dev Dyn. 2011;240:501–520. doi: 10.1002/dvdy.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Brink G.R. Hedgehog signaling in development and homeostasis of the gastrointestinal tract. Physiol Rev. 2007;87:1343–1375. doi: 10.1152/physrev.00054.2006. [DOI] [PubMed] [Google Scholar]
- 18.Middendorp S., Schneeberger K., Wiegerinck C.L. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells. 2014;32:1083–1091. doi: 10.1002/stem.1655. [DOI] [PubMed] [Google Scholar]
- 19.Bosse T., Piaseckyj C.M., Burghard E. Gata4 is essential for the maintenance of jejunal-ileal identities in the adult mouse small intestine. Mol Cell Biol. 2006;26:9060–9070. doi: 10.1128/MCB.00124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang R., Olds L.C., Sibley E. Spatio-temporal patterns of intestine-specific transcription factor expression during postnatal mouse gut development. Gene Expr Patterns. 2006;6:426–432. doi: 10.1016/j.modgep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Battle M.A., Bondow B.J., Iverson M.A. GATA4 is essential for jejunal function in mice. Gastroenterology. 2008;135:1676–1686.e1. doi: 10.1053/j.gastro.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker E.M., Thompson C.A., Kohlnhofer B.M. Characterization of the developing small intestine in the absence of either GATA4 or GATA6. BMC Res Notes. 2014;7:902. doi: 10.1186/1756-0500-7-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohlnhofer B.M., Thompson C.A., Walker E.M. GATA4 regulates epithelial cell proliferation to control intestinal growth and development in mice. Cell Mol Gastroenterol Hepatol. 2016;2:189–209. doi: 10.1016/j.jcmgh.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beuling E., Bosse T., aan de Kerk D.J. GATA4 mediates gene repression in the mature mouse small intestine through interactions with friend of GATA (FOG) cofactors. Dev Biol. 2008;322:179–189. doi: 10.1016/j.ydbio.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srinivas S., Watanabe T., Lin C.S. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy A., Rossant J., Nagy R. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood S.A., Allen N.D., Rossant J. Non-injection methods for the production of embryonic stem cell-embryo chimaeras. Nature. 1993;365:87–89. doi: 10.1038/365087a0. [DOI] [PubMed] [Google Scholar]
- 28.Madison B.B., Dunbar L., Qiao X.T. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 29.Watt A.J., Battle M.A., Li J. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Driegen S., Ferreira R., van Zon A. A generic tool for biotinylation of tagged proteins in transgenic mice. Transgenic Res. 2005;14:477–482. doi: 10.1007/s11248-005-7220-2. [DOI] [PubMed] [Google Scholar]
- 31.He A., Shen X., Ma Q. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26:37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 33.Guo J., Longshore S., Nair R. Retinoblastoma protein (pRb), but not p107 or p130, is required for maintenance of enterocyte quiescence and differentiation in small intestine. J Biol Chem. 2009;284:134–140. doi: 10.1074/jbc.M806133200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bondow B.J., Faber M.L., Wojta K.J. E-cadherin is required for intestinal morphogenesis in the mouse. Dev Biol. 2012;371:1–12. doi: 10.1016/j.ydbio.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duncan S.A., Nagy A., Chan W. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of Hnf-4(-/-) embryos. Development. 1997;124:279–287. doi: 10.1242/dev.124.2.279. [DOI] [PubMed] [Google Scholar]
- 36.Walker E.M., Thompson C.A., Battle M.A. GATA4 and GATA6 regulate intestinal epithelial cytodifferentiation during development. Dev Biol. 2014;392:283–294. doi: 10.1016/j.ydbio.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A., Tamayo P., Mootha V.K. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aronson B.E., Rabello Aronson S., Berkhout R.P. GATA4 represses an ileal program of gene expression in the proximal small intestine by inhibiting the acetylation of histone H3, lysine 27. Biochim Biophys Acta. 2014;1839:1273–1282. doi: 10.1016/j.bbagrm.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Liu T., Meyer C.A. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji H., Jiang H., Ma W. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He A., Pu W.T. Genome-wide location analysis by pull down of in vivo biotinylated transcription factors. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb2120s92. Chapter 21: Unit 21.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao X., Sedgwick T., Shi Y.B. Distinct functions are implicated for the GATA-4, -5, and -6 transcription factors in the regulation of intestine epithelial cell differentiation. Mol Cell Biol. 1998;18:2901–2911. doi: 10.1128/mcb.18.5.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Wering H.M., Bosse T., Musters A. Complex regulation of the lactase-phlorizin hydrolase promoter by GATA-4. Am J Physiol Gastrointest Liver Physiol. 2004;287:G899–G909. doi: 10.1152/ajpgi.00150.2004. [DOI] [PubMed] [Google Scholar]
- 44.Friedrich G., Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 45.Zambrowicz B.P., Imamoto A., Fiering S. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci U S A. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rojas A., Kong S.W., Agarwal P. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol. 2008;28:5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bossard P., Zaret K.S. Repressive and restrictive mesodermal interactions with gut endoderm: possible relation to Meckel’s diverticulum. Development. 2000;127:4915–4923. doi: 10.1242/dev.127.22.4915. [DOI] [PubMed] [Google Scholar]
- 48.Zheng R., Rebolledo-Jaramillo B., Zong Y. Function of GATA factors in the adult mouse liver. PLoS One. 2013;8:e83723. doi: 10.1371/journal.pone.0083723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hagenbuch B., Dawson P. The sodium bile salt cotransport family SLC10. Pflugers Arch. 2004;447:566–570. doi: 10.1007/s00424-003-1130-z. [DOI] [PubMed] [Google Scholar]
- 50.Dawson P.A., Hubbert M., Haywood J. The heteromeric organic solute transporter alpha-beta, Ostalpha-Ostbeta, is an ileal basolateral bile acid transporter. J Biol Chem. 2005;280:6960–6968. doi: 10.1074/jbc.M412752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee F.Y., Lee H., Hubbert M.L. FXR, a multipurpose nuclear receptor. Trends Biochem Sci. 2006;31:572–580. doi: 10.1016/j.tibs.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 52.Kim I., Ahn S.H., Inagaki T. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res. 2007;48:2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- 53.Jung D., Inagaki T., Gerard R.D. FXR agonists and FGF15 reduce fecal bile acid excretion in a mouse model of bile acid malabsorption. J Lipid Res. 2007;48:2693–2700. doi: 10.1194/jlr.M700351-JLR200. [DOI] [PubMed] [Google Scholar]
- 54.Landrier J.F., Grober J., Zaghini I. Regulation of the ileal bile acid-binding protein gene: an approach to determine its physiological function(s) Mol Cell Biochem. 2002;239:149–155. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.