Summary
Liberibacter asiaticus is an unculturable parasitic bacterium of the alphaproteobacteria group hosted by both citrus plants and a psyllid insect vector (Diaphorina citri). In the citrus tree, the bacteria thrive only inside the phloem, causing a systemically incurable and deadly plant disease named citrus greening or Huanglongbing. Currently, all commercial citrus cultivars in production are susceptible to L. asiaticus, representing a serious threat to the citrus industry worldwide. The technical inability to isolate and culture L. asiaticus has hindered progress in understanding the biology of this bacterium directly. Consequently, a deep understanding of the biological pathways involved in the regulation of host–pathogen interactions becomes critical to rationally design future and necessary strategies of control. In this work, we used surrogate strains to evaluate the biochemical characteristics and biological significance of CLIBASIA_03135. This gene, highly induced during early stages of plant infection, encodes a 23 kDa protein and was renamed in this work as LotP. This protein belongs to an uncharacterized family of proteins with an overall structure resembling the LON protease N‐terminus. Co‐immunoprecipitation assays allowed us to identify the Liberibacter chaperonin GroEL as the main LotP‐interacting protein. The specific interaction between LotP and GroEL was reconstructed and confirmed using a two‐hybrid system in Escherichia coli. Furthermore, it was demonstrated that LotP has a native molecular weight of 44 kDa, corresponding to a dimer in solution with ATPase activity in vitro. In Liberibacter crescens, LotP is strongly induced in response to conditions with high osmolarity but repressed at high temperatures. Electrophoretic mobility shift assay (EMSA) results suggest that LotP is a member of the LdtR regulon and could play an important role in tolerance to osmotic stress.
Introduction
Free‐living unicellular organisms are constantly exposed to life‐threatening challenges as a direct consequence of sudden, frequent and drastic changes in their environment. This fundamental principle is also true for bacterial species growing in close association with other organisms, which may provide shelter, nutrients or a suitable attachment substrate. Parasitic, commensal or mutualistic bacteria use similar mechanisms to survive the conditions encountered in the host environment. Insect‐vectored, phytopathogenic bacteria use different strategies to adapt their physiology to two organisms with remarkable physiological differences. The fundamental changes can be evaluated by quantifying fluctuations in gene expression and protein synthesis/modification, or by detecting variations in the amount/quality of the synthesized metabolites among others (Okinaka et al., 2002; Bouchart et al., 2007; Oshima et al., 2011; Yan et al., 2013).
Liberibacter asiaticus (from the Rhizobiaceae family) is an unculturable parasitic bacterium hosted by both citrus plants (and a few other species) and a psyllid insect vector (Diaphorina citri). In this work, we used the taxonomic nomenclature L. asiaticus observing the guidelines previously established for the use of the category Candidatus (Stackebrandt et al., 2002). Within the infected plant, this organism is restricted to the phloem, whereas within the insect, L. asiaticus can prosper in several tissues (Ammar et al., 2011). The phloem‐feeding insect works as a propagation vector, spreading the bacteria from infected trees to uninfected ones (Bové, 2006). Once the plant is infected, the bacteria thrive only inside the phloem, causing a systemically incurable and deadly plant disease named citrus greening or Huanglongbing. L. asiaticus infection drastically reduces citrus fruit production and quality, ultimately resulting in plant death in a few years (5–7 years) post‐infection. Currently, all commercial citrus cultivars in production are susceptible to L. asiaticus, representing a serious threat to the citrus industry worldwide (Wang and Trivedi, 2013).
A subset of characteristics distinctive of this pathosystem have made citrus greening a highly complex disease that is very difficult to successfully treat, including a large delay period (6 months to 3 years) from psyllid inoculation to manifestation of the first symptoms of infection in the plant; highly variable, patchy distribution of the bacteria in the roots and canopy (Bové, 2006; Ding et al., 2015; Louzada et al., 2016). These characteristics magnified by the technical inability to isolate and culture L. asiaticus have hindered progress in understanding the biology of this bacterium. Given this complex scenario, an ideal approach to develop a treatment for citrus greening is to impede the biological mechanisms that aid the bacteria in physiologically adapting to the plant host. Consequently, a deep understanding of the biological pathways involved in the regulation of host–pathogen interactions becomes critical to rationally design future strategies of control.
An important step towards understanding the genetic mechanisms essential to the L. asiaticus–host interaction was published a few years ago (Yan et al., 2013). Interestingly, when the abundance of L. asiaticus mRNA retrieved from infected trees was compared with mRNA obtained from the psyllid, 198 genes showed significant changes in expression. More than 90% of the L. asiaticus genes differentially expressed identified by Yan and coworkers were upregulated in planta. The three genes displaying the largest induction (> sevenfold), unfortunately, encoded for hypothetical or uncharacterized proteins, hindering the discernment of their true biological value. Two of those genes were previously associated with transport systems, whereas CLIBASIA_03135 was a conserved hypothetical protein that has not yet been characterized or associated with any known pathway.
To evaluate the biochemical characteristics and biological significance of CLIBASIA_03135, we have cloned the gene and purified the encoded protein (from now on named LotP). Co‐immunoprecipitation assays using His‐tagged LotP as a bait allowed us to identify the Liberibacter chaperonin GroEL as the main LotP‐interacting protein. The specific interaction between LotP and GroEL was reconstructed and confirmed using a two‐hybrid system in Escherichia coli. Furthermore, it was demonstrated that LotP has a native molecular weight of 44 kDa, corresponding to a dimer in solution with ATPase activity in vitro.
Results
In silico analysis and description
The LotP‐encoding gene, CLIBASIA_03135, is located in a highly conserved cluster in the genome of all sequenced species of the Liberibacter genus. This gene is also conserved in members of the Rhizobiaceae family. In Liberibacter genomes, CLIBASIA_03135 is associated with a small gene, 192 base pairs (bp) long, located directly downstream and separated by a region of only 14 bp (Fig. 1).
Figure 1.
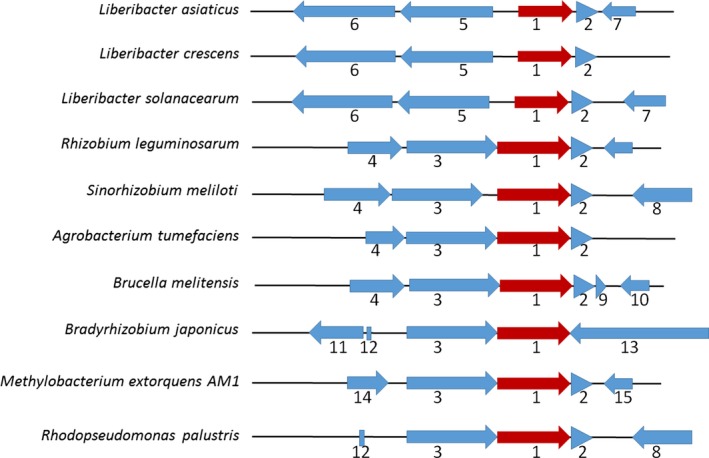
CLIBASIA_03135 locus in L. asiaticus genome. The cluster is highly conserved in Liberibacter species as well as in other Rhizobiaceae members. (1) CLIBASIA_03135 and homologues (red). (2) Hypothetical protein, acid/peroxide resistance. (3) Thioredoxin domain‐containing protein. (4) Aminoacyl‐tRNA editing enzyme. (5) Carboxynorspermidine decarboxylase (EC 4.1.1.96). (6) Carboxynorspermidine synthase (EC 1.5.1.43). (7) Hypothetical protein. (8) Ubiquinone biosynthesis hydroxylase, UbiH/UbiF/VisC/COQ6. (9) Hypothetical protein. (10) 3‐Isopropylmalate dehydrogenase (EC 1.1.1.85). (11) Putative oxidoreductase. (12) tRNA‐Gly. (13) Hypothetical protein. (14) Hypothetical protein. (15) Hypothetical protein.
CLIBASIA_03135 encodes for a hypothetical, uncharacterized, 221‐amino‐acid protein. A BLAST analysis using LotP's linear amino acid sequence as a query indicated a high degree of conservation in homologous proteins encoded in the Rhizobiaceae family, including Liberibacter solanacearum (86% identity), Liberibacter africanus (85% identity), Liberibacter americanus (75% identity), Liberibacter crescens (64% identity), Sinorhizobium meliloti and Agrobacterium tumefaciens (56% identity in both cases). LotP homologues are also present in other taxa like Burkholderia, Mycobacterium and Pseudomonas. LotP's evolutionary history was inferred by using the maximum‐likelihood method based on a JTT matrix‐based model. Our analysis indicated a vast distribution of LotP in the alphaproteobacteria group with scattered presence in members of gammaproteobacteria and betaproteobacteria. Interestingly, 35% of the microorganisms identified in the analysis were plant‐associated bacteria and half of those species are well‐known plant pathogens (Fig. 2).
Figure 2.
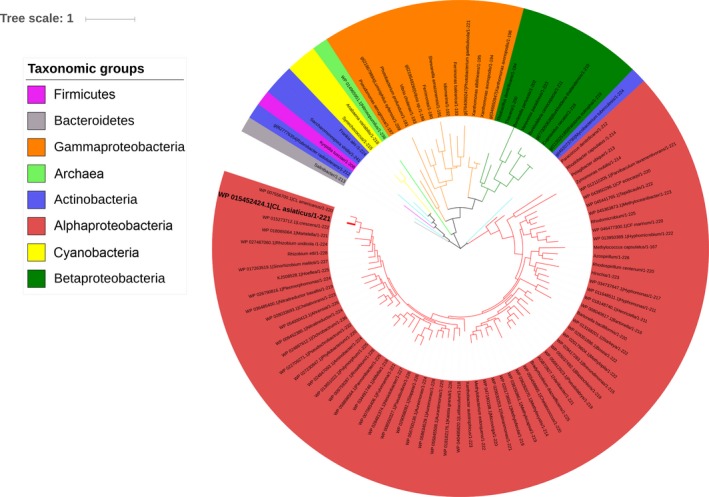
Phylogenetic analysis of LotP. The evolutionary history of LotP was inferred by using the maximum‐likelihood method based on the JTT matrix‐based model. The phylogenetic tree with the highest log likelihood (−15397.4801) is shown. Initial trees for the heuristic search were obtained automatically by applying neighbor‐joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model. The topology with superior log‐likelihood value was selected, the branch lengths measured in the number of substitutions per site. The analysis involved 96 amino acid sequences. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA7.
A structural‐based search, using Phyre2 and SWISS‐MODEL servers, identified the N‐terminal domain of Bacillus subtilis ATP‐dependent protease La1 (LON Protease) as the protein with the highest structural similarities. A predicted LotP model was obtained using this La1 domain (PDB: 3M65) that comprises the first 209 amino acids of the LON protein. This peptide possesses minimal sequence identity with LotP (identity = 18%) but a comparable tertiary structure. The structural model constructed, with 90% of sequence coverage, consists of two subdomains: one globular domain formed by the first 120 amino acids of the N‐terminal region and a distant α‐helical domain shaped by a four‐helix bundle (Fig. 3). These two regions are connected by a flexible loop formed by six amino acids. Consequently, and based only on structural similarities with this small domain, the product of the CLIBASIA_03135 gene was previously annotated in protein databases as an LA1 aminopeptidase, also known as a LON protease. However, the total length of the amino acid sequence of the LON protease is four times longer than LotP and two critical LON protease domains are absent in LotP, the central ATPase motif (200–240 amino acids) and the C‐terminal protease domain (240–300 amino acids). Collectively, the in silico evidence retrieved suggests that LotP belongs to a different, as of yet uncharacterized protein family.
Figure 3.
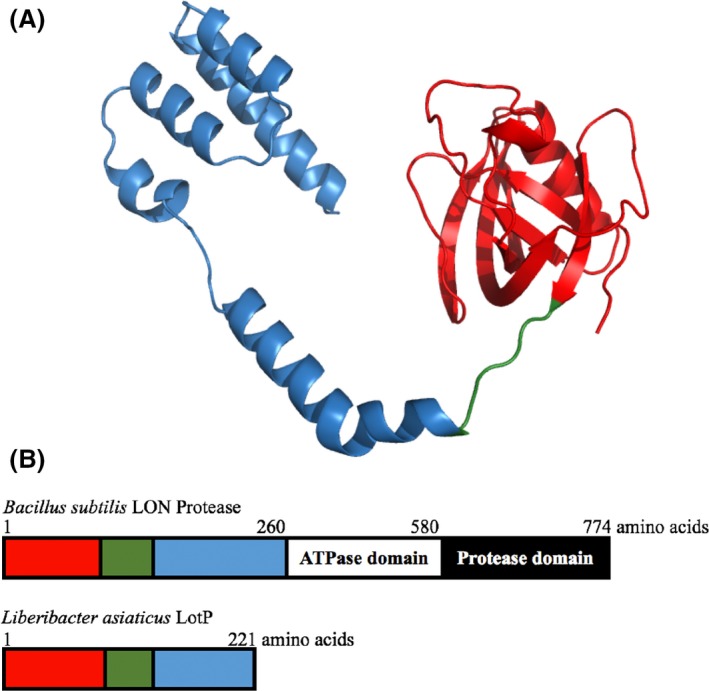
In silico structural analysis of LotP.
A. Ribbon representation of predicted LotP model. The model was constructed based on the structural similarities with B. subtilis N‐terminus LON protease (PDB: 3M65). The globular domain is depicted in red; a flexible loop in green connects this domain with the four‐helix bundle coloured in blue.
B. Schematic representation of the domains present in LotP and in B. subtilis LON protease. LotP conserved scaffolds are represented with the same colours used in the predicted model described above. B. subtilis LON protease N‐terminal domain is 40 residues longer, the primary sequence not being conserved. LON protease is composed of two additional domains, the ATPase and protease in the C‐terminus of the protein.
Identification of LotP protein interaction partners
The LON protease is a key component of the cellular protein control system (Bezawork‐Geleta et al., 2015; Cho et al., 2015). This system is responsible for maintaining cell protein quality (proper folding maintenance and optimal biological function). The N‐terminal module of the LON protease is involved in recognition and binding of the substrates to be hydrolysed (Roudiak and Shrader, 1998; Chir et al., 2009; Li et al., 2010; Wohlever et al., 2014). Consequently, because of the high similarities in protein folding, we hypothesize that the biological role of LotP may depend upon interaction with other intracellular proteins. With the aim of testing our hypothesis, CLIBASIA_03135 was cloned, His‐tagged, and the recombinant His6X‐LotP protein was purified by affinity chromatography as previously described (Pagliai et al., 2014). The purified protein was dialysed in the presence of MgCl2, which was critical to maintain the protein in solution (see Experimental Procedures section).
The purified protein showed an apparent molecular weight (MW) of 23 kDa in denaturing SDS‐PAGE (Fig. 4A). The native molecular mass was 44 kDa as determined by size‐exclusion chromatography using a Superose 12 resin, while similar results were obtained in the presence of ATP (Fig. S3). These results indicate that LotP is a dimer in solution and, in contrast to LON protease, its dimerization is not affected by ATP.
Figure 4.
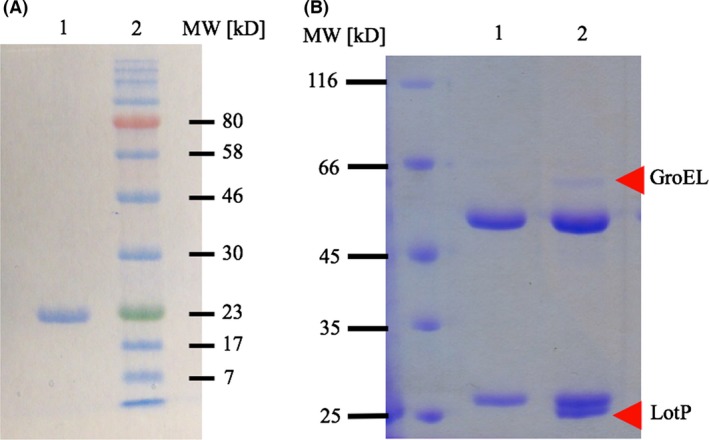
Identification of LotP interaction partners.
A. Lane 1, SDS‐PAGE, His6X‐LotP purified by affinity chromatography. Lane 2, Color Plus pre‐stained MWM (New England BioLabs).
B. SDS‐PAGE showing the results of the immunoprecipitation assay Lane 1: control, L. crescens cell‐free extract was mixed with monoclonal anti‐His6X antibody in the absence of LotP. The light and heavy chains of the monoclonal antibody are visible. Lane 2: the immunoprecipitation was carried out with the same monoclonal antibody using His6X‐LotP as a bait and L. crescens cell‐free extract. LotP ~23‐kDa band and L. crescens GroEL (~60 kDa) are indicated with arrows. In this gel, EZ‐run was used as molecular weight marker (Fisher Scientific).
Cell‐free extract of L. crescens, the closest culturable relative to L. asiaticus, was used as the sample to perform immunoprecipitation assays. Pure His6X‐LotP was used as a bait and the interaction complex was recovered using magnetic beads coated with anti‐His‐tag antibodies. The complexes obtained were analysed via SDS‐PAGE (Fig. 4B). Several minor protein bands (~5) were observed as a result of the co‐immunoprecipitation but only one was prominent, with an apparent monomeric MW of 60 kDa. The samples were analysed by mass spectrometry, and the results obtained are summarized in Table 1. Collectively, the results suggest that LotP interacts with proteins involved in the prokaryotic cellular stress response, with the chaperone GroEL being a prominent target. Equivalent assays were carried out using S. meliloti (Fig. S2) and E. coli cell‐free extracts and comparable results were obtained.
Table 1.
LotP Immunoprecipitation assays: protein identified with MS/MS
| Protein identified | Accession Number (gi) | Molecular weight | Total unique peptide count |
|---|---|---|---|
| Hsp40 chaperone | 940652580 | 41 kDa | 5 |
| Serine endoprotease | 940652534 | 49 kDa | 2 |
| ClpX | 254039948 | 47 kDa | 5 |
| GroEL | 254040526 | 58 kDa | 34 |
Furthermore, the protein–protein interaction between LotP and GroEL was assessed by cloning the genes encoding the interacting proteins in a two‐hybrid system (Vallet‐Gely et al., 2005). The assay was carried out using E. coli as previously described (Wrench et al., 2013). The gene encoding LotP was fused to the Zif protein and the CLIBASIA_03720 gene (encoding GroEL) was fused to the ω subunit of the RNAP; the reciprocal fusions were also constructed and included in the assay (see Table 2 for strain descriptions). In this system, the target proteins fused to the Zif protein and ω subunit of the RNAP must physically interact in order to stimulate transcription of the lacZ gene, encoding the β‐galactosidase, in the E. coli reporter strain. This physical interaction is then quantitatively measured by monitoring the resulting amount of β‐galactosidase activity produced in each strain. Physical interaction between the target proteins results in increased β‐galactosidase activity compared with control strains. The strains were grown in MOPS minimal media, the chimerical proteins induced with IPTG, and the β‐galactosidase activity was determined. The strains FL03 and FL05 carrying both recombinant plasmids demonstrated a significantly higher β‐galactosidase (1988 ± 49 and 1249 ± 16 AU, respectively) activity compared with strains carrying empty plasmids (941 AU). The protein–protein interaction was detected in a range of temperature (30–42°C). The values described in Table 3 belong to the enzymatic activities obtained at 37°C. The β‐galactosidase activity at 25°C was significantly lower (170 AU), probably due to the slow growth of E. coli. These results, the co‐immunoprecipitation and higher β‐galactosidase activity, are in support of the physical interaction between these two L. asiaticus proteins. The results are summarized in Table 3.
Table 2.
Strains and plasmids used in this study
| Strains | Relevant genotype | References |
|---|---|---|
| E. coli DH5α | ϕ80 dlacZΔM15Δ(lacZYA‐argF)U169 recA1 endA1 hsdR17 (rK− mK+) supE44 thi‐1gyrA relA1 | Laboratory stock |
| E. coli BL21 (DE3) | F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5‐T7 gene 1 ind1 sam7 nin5]) | Stratagene |
| E. coli JM109 | endA1, recA1, gyrA96, thi‐1, hsdR17 (rK− mK+), relA1, supE44, Δ(lac‐proAB), [F’ traD36,proAB, laqIqZΔM15] | New England Biolabs |
| E. coli KDZif1ΔZ | araD (gpt‐lac)5, rpsL (Strr), ΔspoS3::cat (Camr) [F’ lacIq (Z321(‐61) lacZYA*) Kanr] | (Vallet‐Gely et al., 2005) |
| E. coli BW25113 | F−, DE(araD‐araB)567, lacZ4787(del)::rrnB‐3, LAM−, rph‐1, DE(rhaD‐rhaB) 568, hsdR514 | Keio collection |
| E. coli JW4103 | F−, Δ(araD‐araB)567, ΔlacZ4787(::rrnB‐3), λ −, rph‐1, Δ(rhaD‐rhaB)568, ΔgroL768::kan, hsdR514 | Keio collection |
| L. crescens BT‐1 | Standard strain (Wild Type). | (Leonard et al., 2012) |
| S. meliloti 1021 | expR102::ISRm2011‐1expR.Smr | (Galibert et al., 2001) |
| FL01 | KDZif1ΔZ pACTR‐Zif, pBRGP‐ω. Tetr, Ampr. Kanr | This work |
| FL02 | KDZif1ΔZ pACTR‐CLIBASIA_03135‐Zif, pBRGP‐ω. Tetr, Ampr. Kanr | This work |
| FL03 | KDZif1ΔZ pACTR‐Zif, pBRGP‐CLIBASIA_03135‐ω. Tetr, Ampr. Kanr | This work |
| FL04 | KDZif1ΔZ pACTR‐CLIBASIA_03720‐Zif, pBRGP‐CLIBASIA_03135‐ω. Tetr, Ampr. Kanr | This work |
| FL05 | KDZif1ΔZ pACTR‐CLIBASIA_03135‐Zif, pBRGP‐CLIBASIA_03720‐ω. Tetr, Ampr. Kanr | This work |
| FL06 | KDZif1ΔZ pACTR‐CLIBASIA_03720‐Zif, pBRGP‐ω. Tetr, Ampr. Kanr | This work |
| FL07 | KDZif1ΔZ pACTR‐Zif, pBRGP‐CLIBASIA_03720‐ω. Tetr, Ampr. Kanr | This work |
| Plasmids | Characteristics | Reference |
| p15TV‐L | Expression vector Ampr. | (Pagliai et al., 2010) |
| pBRGP‐ω | Translational fusion vector | (Vallet‐Gely et al., 2005) |
| pACTR‐AP‐Zif | Translational fusion vector. Tetr | (Vallet‐Gely et al., 2005) |
| pBAD24 | Expression vector, arabinose inducible promoter, Ampr | (Guzman et al., 1995) |
| pFL02 | pACTR‐CLIBASIA_03135‐Zif. Tetr | This work |
| pFL03 | pBRGP‐CLIBASIA_03135‐ω. Ampr | This work |
| pFL04 | pBRGP‐CLIBASIA_03720‐ω. Ampr | This work |
| pFL05 | pACTR‐CLIBASIA_03720‐Zif. Tetr | This work |
| pFL06 | pBAD24‐CLIBASIA_03135. Ampr | This work |
Table 3.
Analysis of LotP interactions with GroEL
| Strains | Plasmids and fused proteins | β‐Galactosidase activity | |
|---|---|---|---|
| pBRPG‐ω | pACTR‐AP‐Zif | ||
| FL01 | – | – | 941 ± 44 |
| FL02 | – | LotP | 811 ± 12 |
| FL03 | LotP | GroEL | 1988 ± 49* |
| FL04 | – | GroEL | 866 ± 57 |
| FL05 | GroEL | LotP | 1249 ± 16* |
| FL06 | GroEL | – | 1030 ± 17 |
LotP = CLIBASIA_03135 gene; GroEL = CLIBASIA_03720 gene. The mean ± SD is shown. ANOVA test indicated statistically significant variation in mean β‐galactosidase activity across strains (F 5,18 = 232.7; P‐value = 1.07e‐15). Strains followed by an asterisk (*) are significantly different from all other strains according to Tukey's HSD test (α = 0.05). The experiments were performed in triplicate.
LotP displays phosphatase activity in solution
In silico analyses were not conclusive in predicting any catalytic activity associated with this protein. Laboratory assays were performed to evaluate the two activities classically associated with the LON protease, namely protease and phosphatase activity. No proteolytic activity was detected, but LotP showed a weak activity towards p‐NPP (phosphatase model substrate) (Proudfoot et al., 2008). In a subsequent enzymatic assay, LotP's activity towards several nucleosides was tested in parallel. Malachite green was used to detect the Pi released by the enzyme action (Crowe et al., 2010). The results demonstrated that LotP preferentially used ATP as an enzyme substrate (Fig. 5A). The ATPase activity was consistent in a wide range of pH (5.5–9), and the optimal temperature for in vitro catalysis was 50°C. The enzyme demonstrates classical hyperbolic saturation kinetics with an apparent V max = 10.2 μMoles min−1 and a K m = 133.98 μM (Fig. 5B). LotP alone, or used in combination with purified GroEL, showed no chaperone activity in vitro (data not shown).
Figure 5.
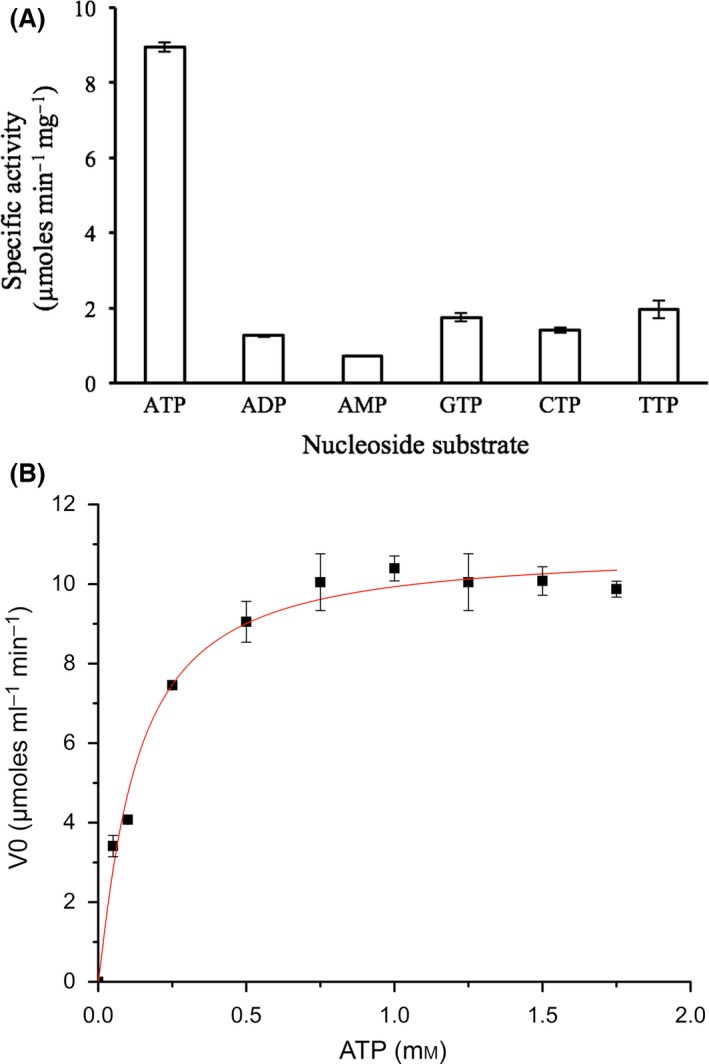
LotP phosphatase activity determinations.
A. Phosphatase activity profile using several nucleosides as enzyme substrate. Typical enzyme reaction contains 1 mM of each substrate and 2–10 μg ml−1 of enzyme.
B. Steady‐state kinetic characterization of ATPase activity. Initial rates of the reactions were determined at fixed concentrations of ATP, and the Pi released was quantified by using Malachite green reagent as previously described (Crowe et al., 2010).
LotP is able to interact with E. coli proteins affecting cell viability
The main biological role of bacterial GroEL is related to the maintenance of correct protein folding during stress (Georgopoulos, 2006; Hayer‐Hartl et al., 2016). Specifically, this protein protects cell integrity by refolding denatured proteins, mostly those proteins affected by heat denaturation (Cho et al., 2015). With the aim of assessing the biological effects of LotP expression on cell growth during temperature stress, LotP was cloned in the pBAD24 expression vector (Guzman et al., 1995). The protein was expressed in the E. coli wild‐type strain, BW25113, and the isogenic JW4103, a mutant with impaired ability to respond to heat shock due to lower levels of GroEL production. The growth of the wild‐type strain expressing LotP at 42°C was less than the growth observed at 37°C. In both conditions, the strains expressing LotP grew slower since the beginning of the assay (Fig. 6A). This kind of effect is typical of strains with flawed adaptation to important environmental challenges, such as thermal stress in this case. The effect of temperature was drastic when LotP was expressed in the mutant strain. E. coli JW4103 expressing LotP was unable to grow at 42°C after 24 h of incubation (Fig. 6B). To explain these results, a new immunoprecipitation assay was performed, this time using E. coli cell‐free extract obtained from both strains (BW25113 and JW4103). Mass spectrometry analysis of the proteins recovered demonstrates that LotP is also able to interact with E. coli GroEL. Surprisingly, seven other proteins, including DnaJ and ClpX, co‐precipitate with LotP when the JW4103 cell‐free extract was used. Therefore, we hypothesize that LotP interacts with GroEL in E. coli. However, in the absence of GroEL, other proteins essential for survival during temperature challenges can interact with LotP, affecting the overall cell viability (Fig. S1).
Figure 6.
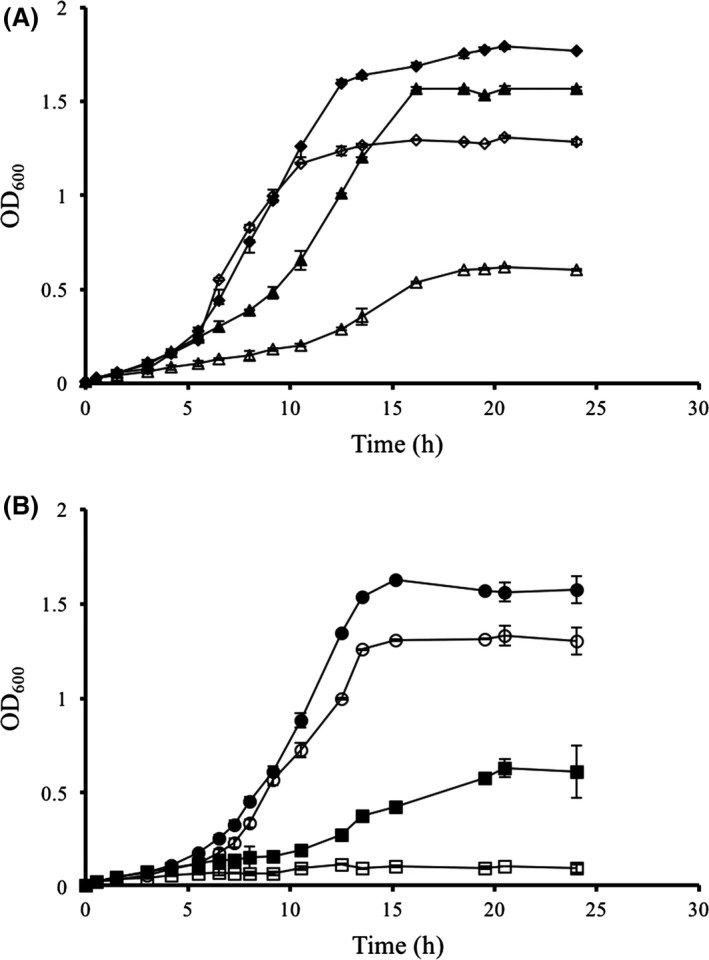
Effect of temperature on the growth of E. coli strains expressing LotP from an inducible expression vector. The gene CLIBASIA_03135 was cloned in pBAD24. The cells were grown in MOPS minimal medium with glycerol as carbon source. LotP expression was induced with 10 mM l‐arabinose starting at the beginning of the assay. The assays were carried out at 37°C (closed symbol) and 42°C (open symbols).
A. BW25113 (WT): pBAD empty plasmid (diamonds, ♦ – ♢); pBAD‐LotP (triangles, ▲ – ▵); B. JW4103 (ΔgroEL): pBAD empty plasmid (circles, ● – ○); pBAD‐LotP (squares, ■ – □). Plotted values represent the mean ± SD of three biological replicates.
LotP transcription is repressed at high temperatures in L. crescens
Liberibacter asiaticus cannot be cultured in a laboratory setting, but the closely related L. crescens can be used as surrogate strain (Pagliai et al., 2014; Lai et al., 2016). In general, Liberibacter species are temperature sensitive, with an optimal growth temperature of 25°C (Fagen et al., 2014). Consequently, heat stress was induced by culturing the bacteria at 30 and 32°C (Fig. 7A). The highest temperature evaluated (32°C) was the maximal temperature tolerated by L. crescens in the culture conditions used. A comparable growth kinetic was obtained for all temperatures of incubation assayed. The rate of transcription for lotP was drastically affected by temperature; fivefold repression was observed when the temperature was higher than 30°C (Fig. 7B). The results obtained in L. crescens are in agreement with the results described in E. coli. Altogether, the results suggest that the biological role of LotP is not directly related to the resistance of this bacterium to high temperatures.
Figure 7.
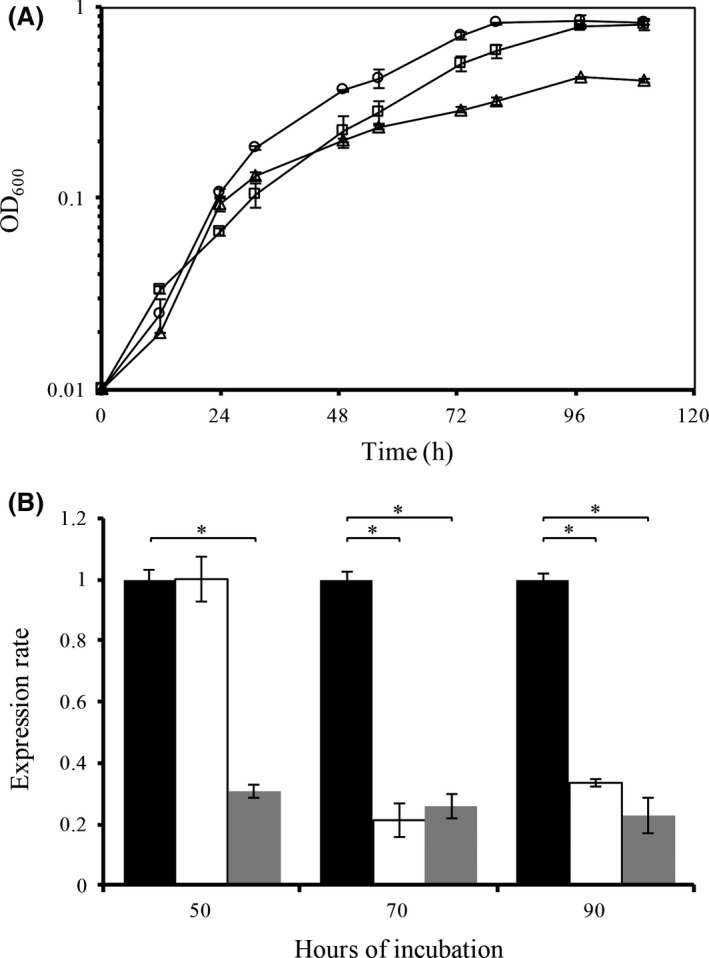
Effect of temperature on mRNA levels of LotP.
A. L. crescens growth curve: 25°C (circles, ○), 30°C (squares, □) and 32°C (triangles, ▵). Plotted values represent the mean ± SD of three biological replicates.
B. The mRNA transcripts of B488_12950 (encoding a LotP homologue) were quantified by qRT‐PCR. The expression rate was normalized using the expression of the L. crescens 16S rRNA as internal standard using the primers depicted in Table 4. Samples were obtained at 50, 70 and 90 h of incubation. The temperature of incubation for each case was 25°C (black bars), 30°C (white bars) and 32°C (grey bars). Expression rate values represent the mean normalized fold change ± SD as compared with the control, performed in quadruplicates. Statistical significance was determined using a Student's t‐test (*P < 0.05). All the assays were conducted in a rotary shaker at 200 rpm.
LotP is highly induced by conditions with high osmolarity in L. crescens
In search of potential regulatory sequences controlling LotP expression, a direct analysis of the DNA region immediately upstream of the lotP gene was performed using the binding motif determined by Pagliai et al. (2014). A putative binding motif for LdtR, a recently characterized transcription factor, was found 117 base pairs upstream of the LotP translational start codon (Fig. 8A). LdtR is a MarR‐family transcriptional regulator governing the expression of several L. asiaticus genes. The LdtR regulon modulates the expression of a subset of proteins facilitating osmotic tolerance in L. asiaticus. The predicted DNA binding region was amplified, labelled with biotin and used in electrophoretic mobility shift assay (EMSA) as the LdtR target. The binding analysis showed that LdtR recognizes and binds the DNA sequence predicted via in silico analysis (Fig 8B). These results confirm that LotP is a member of the LdtR regulon.
Figure 8.
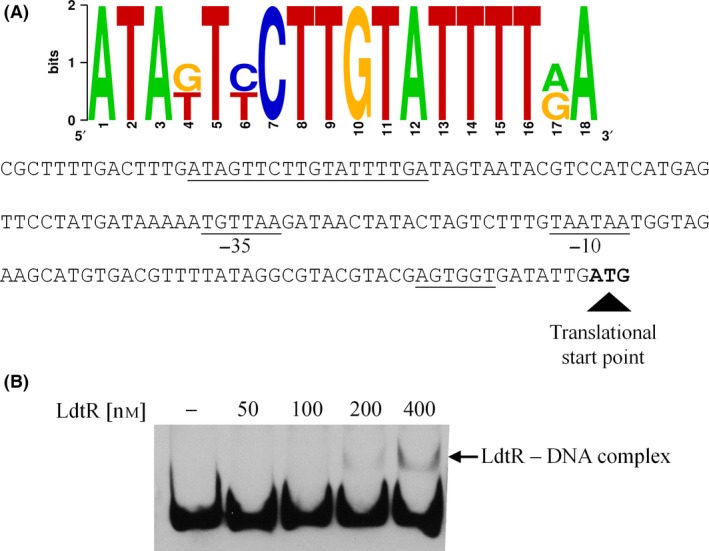
LdtR binds in the LotP promoter A. The LdtR binding sequence was estimated by RegPredict software using a known position weight matrix based on data previously published (Pagliai et al., 2014).
B. EMSAs were conducted with increasing concentrations of LdtR as shown in the top of each figure. The arrow denotes the shift obtained using 200 and 400 nanomolar of purified protein.
The apparent affinity obtained was directly comparable to the affinity originally described for this LdtR promoter. This result suggests that LotP expression could be induced in response to high concentrations of osmotically active solutes. To verify the biological significance suggested by this in vitro assay, LotP expression was quantified in samples obtained from L. crescens exposed to high concentrations of NaCl and sucrose. In this strain, the LotP homologue as well as its regulatory region is highly conserved. L. crescens can tolerate up to 150 mM NaCl and 200 mM of sucrose in the culture media; therefore, the concentrations used to increase the osmolarity did not significantly affect the growth of L. crescens as previously described (Pagliai et al., 2014). The cells were grown in BM7 culture media amended with NaCl (50 and 100 mM) or sucrose (50 and 100 mM). The results obtained indicate that the rate of expression of LotP increased up to 90 ± 10 times in the presence of 100 mM sucrose and 30 ± 3 times with the same concentration of NaCl (Fig. 9). These results suggest that LotP is a member of the LdtR regulon and could play an important role in tolerance to high osmotic pressure, an environmental condition encountered in the citrus phloem sap.
Figure 9.
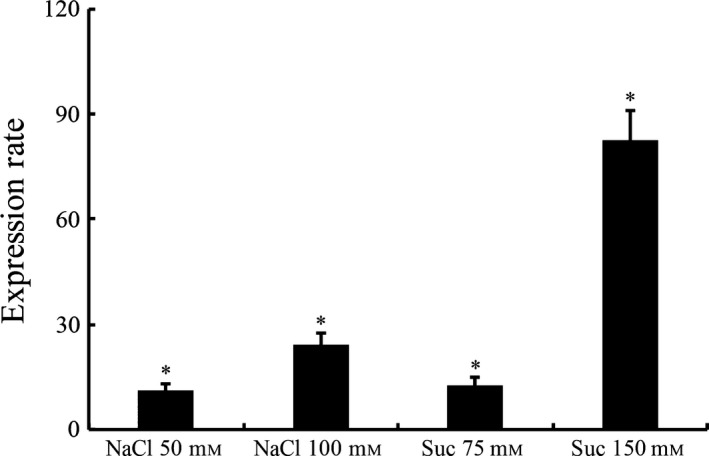
LotP is induced under high osmotic conditions. The expression level of LdtR was assessed in L. crescens growing on BM7 culture media amended with NaCl and sucrose at the indicated concentrations (x‐axis). The assays were performed in a rotary shaker at 200 rpm and 25°C. mRNA was extracted as described and the expression rate measured by qRT‐PCR. The 16S rRNA expression was used as internal standard. Expression rate values represent the mean normalized fold change ± SD as compared with the control, performed in quadruplicates. Statistical significance was determined using a Student's t‐test (*P < 0.05).
Discussion
The analysis of LotP primary structure, linear sequence, indicates that it is highly conserved and well represented in bacterial species frequently associated with plants. Structural analysis of the protein indicates that it displayed remarkable structural homology with the amino‐terminal region of the LON protease (240 amino acids long) (Duman and Löwe, 2010; Li et al., 2010). Based on these structural similarities, LotP was systemically annotated as an ATP‐dependent LON protease in all sequenced genome databases; however, the LON protease is an ATP‐dependent aminopeptidase composed by six units of 90 kDa each (760–800 amino acids) (Lin et al., 2016).
These contrast with the characteristics and native molecular mass (44 kDa) determined for LotP in solution. Additionally, LotP is missing two critical LON modules: the ATP‐binding module with a characteristic Walker motif and the proteolytic module located in the carboxy‐terminal region. The phylogenetic relationships suggest that LotP evolution belongs to a different lineage of proteins unrelated to the LON protease. Altogether, these features suggest that LotP belongs to a different family of proteins with a protein fold similar to the amino‐terminal module of the LON protease.
The structural similarities described are likely the consequence of convergent evolution. The function of the LON protease's N‐terminal module is the identification of proteins (substrates) to be hydrolysed by the proteolytic domain. Based on the similarity between the folds of the N‐terminal module of LON protease and LotP, we expected that LotP might have a similar function. In absence of direct biological data to guide our research, the immunoprecipitation assay to identify LotP protein partners was the best starting strategy. The recovery of a 60 kDa protein in the immunoprecipitation assay, later identified as GroEL (34 peptides identified by GC/MS), suggests that LotP could have a central role in the L. asiaticus–Citrus sinensis pathosystem. GroEL belongs to the chaperonin protein family, CPN60, that is an evolutionarily conserved family associated with the maintenance and refolding of intracellular proteins (Mayhew et al., 1996; Guisbert et al., 2008; Hayer‐Hartl et al., 2016). GroEL is also indispensable for several nodulating species of Rhizobiaceae which encode up to seven copies per genome (Fischer et al., 1993; Lund, 2009). In other cases, it behaves as a potent MAMP (microbial‐associated molecular pattern), triggering plant resistance mechanisms (Chaudhary et al., 2014). It has been directly associated with the modulation of several microbial–host interactions and directly connected to a baffling variety of biological roles. One of the most intriguing examples involves Buchnera aphidicola, a gram‐negative bacterium of the gammaproteobacteria group. This bacteria is an aphid endosymbiont that depends upon the overexpression of GroEL for long‐term maintenance of its relationship with the insect host (Wilcox et al., 2003). However, when B. aphidicola purified GroEL is injected into vegetable tissues, it triggers the expression of the defensive arsenal of the plant. A complete overview of the biological significance of GroEL has been compiled in two excellent reviews (Henderson et al., 2013; Kupper et al., 2014). Understanding the fundamental role of GroEL in this system could shed light on the biological meaning of LotP. In L. asiaticus, the transcription of GroEL slightly increases when the bacteria is transferred to the plant (Yan et al., 2013). To minimize the plant response to L. asiaticus, GroEL should be silenced or ‘protected’ to evade the plant defence system. We hypothesize that LotP's biological role could be to minimize the plant's immune response, protecting L. asiaticus GroEL. In this way, the plant's resistance will be hampered, permitting infection by L. asiaticus. As of yet, this question remains to be properly answered.
Recombinant strains of E. coli expressing L. asiaticus LotP are more sensitive to high temperatures and the bacterial growth is directly affected. In the bacterial strains where the main target of LotP is lower (strain JW4103), LotP interacts with a subset of stress‐related proteins and impedes cellular growth at 42°C. The results observed in L. crescens correlate with those obtained in E. coli. The transcriptional repression of LotP in L. crescens incubated at elevated temperatures clearly indicates that LotP is not enhancing the protective role of GroEL when cells are exposed to high temperatures. Although L. crescens encodes a functional rpoH gene, the transcriptional rate of GroEL does not change or shows a negligible decrease when the strain is cultured at 32°C. This suggests, as was reported in many primary symbionts, that the main function of these proteins in L. asiaticus is not synergistic chaperone/co‐chaperone activity (Stoll et al., 2009). Indeed, L. asiaticus is especially sensitive to elevated temperatures and controlled heat treatment of citrus trees is the only method used to transiently decrease the bacterial titre in inflected plants (Hoffman et al., 2013). Collectively, these data suggest an overall post‐translational regulatory role for LotP through protein–protein interactions. In this context, the ATPase activity described herein became critical to provide the energy necessary to release the interacting proteins according to the needs of the cell (Fenton et al., 1994). Alternatively, LotP may help the bacteria to cope with the stress produced by heat. LotP transcription is repressed when the bacterium grows at high temperatures, which will result in lower LotP synthesis releasing active GroEL when necessary. This possibility is in agreement with the evidence herein displayed. However, this is difficult to evaluate due to the narrow range of growth temperature for L. crescens and the impossibility to culture L. asiaticus in a laboratory setting.
The gene encoding LotP is one of the few genes in L. asiaticus that is highly transcribed in mRNA samples obtained from infected citrus. The transcriptional rate increased nine times when compared with samples collected from infected psyllids (Yan et al., 2013). The in silico identification of highly conserved LdtR binding sequences along with the associated in vitro DNA binding assays suggests a direct regulation of LotP by this recently characterized transcription factor (Pagliai et al., 2014, 2015). The exceptionally high expression (up to 90 times) obtained when L. crescens is cultured under hyperosmotic conditions confirmed that LotP belongs to the LdtR regulon. It may, directly or indirectly, help the cells to tolerate the high osmotic pressure of the phloem sap. The inability to culture L. asiaticus hampers the possibility of directly addressing this question by using mutant strains. The biological role of GroEL on high saline and high osmotic conditions varies depending on the bacterial species analysed (Susin et al., 2006; Gunasekera et al., 2008; Rao et al., 2013). Overall, the role of GroEL cannot be directly linked to protein refolding as it can with cells growing at high temperature.
We hypothesize that the biological significance of LotP is directly related to the moonlighting activity of GroEL. The GroEL protein is extremely important in almost all insect‐vectored pathosystems studied and L. asiaticus should not be an exception. Several questions need to be properly addressed before completely understanding the true biological value of this protein–protein interaction. However, the results described herein represent one step towards improved comprehension of the complex molecular events driving the relationship between L. asiaticus and the host.
Experimental procedures
In silico analysis
Homologues of CLIBASIA_03135 were obtained using two iterations of a PSI‐BLAST search against a non‐redundant sequence database at the NCBI (www.blast.ncbi.nlm.nih.gov/). Redundant sequences were eliminated and alignments were made using the ClustalW (Thompson et al., 1994). The evolutionary history of LotP was inferred by using the maximum‐likelihood method based on the JTT matrix‐based model. The phylogenetic tree with the highest log likelihood (−15397.4801) was constructed. The initial data for the heuristic search was obtained automatically by applying neighbor‐joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model. The topology with superior log‐likelihood value was selected. The branch lengths were measured based on the number of substitutions per site. The analysis involved 96 amino acid sequences. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016). Further, the phylogenetic tree was constructed using the online tool Itol: Interactive Tree of Life (Letunic and Bork, 2016). The main LotP protein scaffold was modelled in silico using the automated mode of the SWISS‐MODEL and PHYRE2 server (Arnold et al., 2006; Kelley and Sternberg, 2009). The crystal structure of C3M65A from the N‐terminal domain of Bacillus subtilis Lon Protease was retrieved as the best template.
Strains and growth conditions
Bacterial strains and plasmids are listed in Table 2. E. coli strains were grown in either LB medium or M9 minimal medium with 0.2% glycerol as the carbon source. S. meliloti cells were grown at 30°C in Luria–Bertani (LB). When required, the media were supplemented with ampicillin (100 μg ml−1) or kanamycin (20 μg ml−1) for E. coli.
Liberibacter crescens BT‐1 was cultured at 25°C with agitation (200 rpm) in modified BM7 media, pH 6.9 (Leonard et al., 2012). This culture medium was composed of 1% brain heart infusion (Difco Laboratories, Detroit, MI), 15% fetal bovine serum (Sigma‐Aldrich, St. Louis, MO), 30% TMN‐FH insect medium (Sigma), α‐ketoglutaric acid (2 mg ml−1), ACES (10 mg ml−1) and potassium hydroxide (3.75 mg ml−1).
DNA manipulations and gene cloning
Standard methods were used for chromosomal DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, ligation and transformation (Sambrook et al., 1989). Plasmids were isolated using QIAprep® Spin Miniprep Kit (Qiagen, Valencia, CA, USA), and PCR products were purified using QIAquick® purification kits (Qiagen). The primers utilized are described in Table 4. Liberibacter asiaticus DNA was isolated from leaf tissues of infected plants using the Isolate II Plant DNA kit (Bioline, Taunton, Massachusetts, USA).
Table 4.
Oligonucleotides used in this study
| Primers | Oligonucleotide sequence (5′→3′) |
|---|---|
| CLIB_03135_Ext_Fw | CAAGGTTGCGTCGTATCTTGA |
| CLIB_03135_Ext_Rv | CCCTGATCATCTACCTGACGC |
| CLIB_03135_Fw | CAAGCTTCGTCATCATTGCAACCTATTTTCACAAT |
| CLIB_03135_Rv | TTGTATTTCCAGGGC ATGAAAATTGGTAATACAATATACA |
| CLIB_03135_NdeI_Fw | GGATCCATATGATGAAAATTGGTAATACAATATACAAA |
| CLIB_03135_NotI_Rv | TATATGCGGCCGCTTGCAACCTATTTTCACAATGA |
| CLIB_03135_KpnI_Fw | CGATGTGGTACCTATGAAAATTGGTAATACAATA |
| CLIB_03135_SalI_Rv | TTCTACGTCGACTATTGCAACCTATTTTC |
| CLIB_03720_Ext_Fw | GGTGACATTGTCCTTTTTGG |
| CLIB_03720_Ext_Rv | AGCTTCCTTCTCCCATAAGC |
| CLIB_03720_NdeI_Fw | GGAATTCCATATGGGTGTTAATACGCTTGC |
| CLIB_03720_NotI_Rv | TATATGCGGCCGCCATCATCATATCCATACCAC |
| 16SLcr_Rv | AAGGTTGAGCCTTGGGATTT |
| 16SLcr_Fw | GTTCGGAATAACTGGGCGTA |
| CLIB_03135_EMSA_Fw1 | GCAAGGTTGCGTCGTATCTT |
| CLIB_03135_EMSA_Rv_Bioa | CGGTAAGTCTTCACGATTTTTG |
a. Biotin labelled.
Protein purification
CLIBASIA_03135 was cloned into the p15TV‐L vector and transformed into E. coli DH5α. The correct cloning was confirmed by sequencing. The recombinant plasmids selected were used to transform E. coli BL21‐Star (DE3) cells (Stratagene). The cells were grown in LB broth at 37°C. His6x‐tagged protein was induced with 0.5 mM IPTG when OD600 = 0.5. After addition of IPTG, cells were incubated at 17°C overnight with shaking. The cells were harvested and the pellet was suspended in binding buffer (500 mM NaCl, 5% glycerol, 50 mM Tris pH 8.0, 0.25 mM TCEP and 5 mM imidazole) and stored at −80°C. The cells were disrupted using a French Press, and the recombinant protein was purified as described in Pagliai et al. (2014). The purified proteins were dialysed against 50 mM Tris pH 8.0, 150 mM NaCl, 10 mM MgCl2, 5% glycerol, 0.25 mM TCEP and stored in aliquots at −80°C until further use. Protein concentration was estimated using the Bio‐Rad protein assay kit (Bio‐Rad). LdtR was purified as previously described (Pagliai et al., 2014).
Size‐exclusion chromatography
A solution containing 36–40 μM of recombinant LotP was prepared using 50 mM Tris pH 8.0, 150 mM NaCl, 10 mM MgCl2 buffer. Aliquots of this preparation were injected onto a prepacked Superose 12 10/300 chromatography column (GE) equilibrated with the same buffer. The gel filtration assay was carried out at 4°C using a flow rate of 0.3 ml min−1. The eluted proteins were monitored continuously at 280 nm using a UV‐M II monitor (GE Healthcare, United Kingdom). A mixture of protein molecular weight standards, containing thyroglobulin (670 kDa), γ‐globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa) and vitamin B12 (1.36 kDa), was used in similar conditions as native molecular mass markers. The molecular weight of eluted proteins was determined using a calibration curve based on the retention time of each marker used.
Immunoprecipitation
Protein–protein interaction assays were carried out using L. crescens or E. coli cell‐free extracts as indicated. His6X antibody (Ab) was added to previously separated Protein G Dynabeads (Life Technologies (Acquired by Thermo Fisher), Waltham, Massachusetts, USA). After the Ab binds to the Dynabeads, the pearls with the Ab attached were collected using a magnet. The Ab was cross‐linked to the Dynabeads to avoid co‐elution of the antibody, and the magnetic beads were re‐purified. The beads were re‐suspended and incubated in a rotary shaker for 15 min in the presence of the recombinant protein (His‐LotP) and the bacterial lysate to favour the antigen (Ag)–antibody reaction. The final protein ratio used was 0.5 mg ml−1 His6X‐LotP and 750 μl of cell lysate with 1.5 mg ml−1 of total protein concentration. The mixture was washed in a buffer three times, separating the complex with the magnets between washes. The supernatant was removed and the Dynabead–Ab–Ag complex was transferred to a clean tube. The proteins were eluted from the magnetic beads and aliquots analysed in SDS‐PAGE. The protein bands were visualized with CBBR‐250, excised from the acrylamide gel and used in mass spectroscopy analysis.
Mass spectroscopy
All MS/MS samples were analysed using Mascot (version 2.4.1; Matrix Science, London, UK). Mascot was set up to search the NCBInr_20130403 database (selected for Bacteria, unknown version, 14961948 entries) assuming the digestion enzyme trypsin. Mascot search was set with a fragment ion mass tolerance of 0.50 Da and a parent ion tolerance of 10.0 PPM. Scaffold (version Scaffold_4.4.8; Proteome Software Inc., Portland, OR, USA) was used to validate MS/MS‐based peptide and protein identifications. The identification of the proteins was accepted if at least two peptides match a score of 80.0% by the Peptide Prophet algorithm with Scaffold delta‐mass correction.
Enzymatic activities
Phosphate released after enzymatic ATP hydrolysis was quantified using the malachite green assay in order to assess ATPase activity (Crowe et al., 2010). Protease activity was assessed by using azocasein as the substrate in the presence and absence of ATP and using proteinase K as a positive control (Secades and Guijarro, 1999).
Growth curves
Growth kinetics using E. coli strains were carried out in M9 media with 0.2% glycerol as the sole carbon source. Each strain harbouring a plasmid was cultured in the presence of the corresponding antibiotic. Individual flasks were inoculated with aliquots obtained from an overnight culture with the necessary amount of cells to reach an initial OD600 = 0.05. The inducer (10 mM l‐arabinose) was added to the culture at the beginning of the assay. The flasks were incubated at 200 rpm at the indicated temperatures (37 or 42°C). In all cases, the growth curves were performed in triplicate.
Two‐hybrid system
The protein–protein interaction between LotP and GroEL was assessed by cloning the genes encoding the interacting proteins in a two‐hybrid system (Vallet‐Gely et al., 2005). The assay was carried out using E. coli as previously described (Wrench et al., 2013). Briefly, CLIBASIA_03135 and CLIBASIA_03720 genes from L. asiaticus were PCR‐amplified and fused to the ω subunit of the RNAP by cloning the fragment into the NdeI and NotI sites of the pBRGP‐ω plasmid. The CLIBASIA_03135 and CLIBASIA_03720 genes were also fused to the zinc finger DNA‐binding protein of the murine Zif268 protein cloned in the same restriction sites of the plasmid pACTR‐AP‐Zif. The recombinant clones were selected by transformation in E. coli JM109, confirmed by sequencing and transformed into the reporter strain KDZif1ΔZ by standard methods (Sambrook et al., 1989). The recombinant E. coli cells were grown at different temperatures in MOPS media amended with 2 g l−1 glycerol as the main carbon source. The expression of the chimerical proteins in the reporter strain was induced by the addition of 20 μM IPTG at the beginning of the assay. Cell samples obtained at different culture times were permeabilized with 0.15% SDS and 1.5% chloroform in Z‐buffer (Miller, 1972). β‐Galactosidase activity was assayed using chlorophenol red‐β‐D galactopyranoside (CRPG). The substrate hydrolysis was continuously monitored, reading the absorbance at 570 nm for 10 min in a Synergy HT 96‐well plate reader (BioTek, Winooski, VT, USA). The β‐galactosidase activity, expressed as arbitrary units (AU), was calculated using the slope of the first‐order rate kinetic (V0) normalized to the cell density of each sample. The assays were performed in triplicate.
Electrophoretic Mobility Shift Assays (EMSAs)
DNA gel shift assays were performed according to the protocol described in Pagliai et al. (2014). A fragment of the CLIBASIA_03135 promoter was generated by PCR using biotin‐labelled primers (Table 4) and then purified using QIAquick spin columns (Qiagen). Briefly, the reaction for EMSA contained 1 ng of 5′‐labelled DNA probe, 50 mM Tris–HCl pH 7.2, 150 mM KCl, 10 mM MgCl2, 0.01% Triton X‐100, 12.5 ng μl−1 of an equimolar mix of Poly(dI‐dC) and Poly(dA‐dT) non‐specific DNA competitor and purified LdtR protein (0 – 500 nM). The whole reaction was incubated at 37°C for 20 min and then separated on 6% acrylamide–bisacrylamide non‐denaturing gels using 0.5× Tris–borate–EDTA buffer, pH 8.3 (TBE). Samples were subjected to electrophoresis for 2 h at 100 V using ice‐cold 0.5× TBE buffer, and then, the DNA was transferred from the polyacrylamide gel to a Hybond‐N+ membrane (GE Healthcare) by blotting at 250 mA for 45 min using a semidry transfer unit, followed by a UV cross‐link of the DNA to the membrane. The biotin‐labelled DNA was visualized using the Phototope‐Star detection kit (New England Biolabs, Ispwich, Massachusetts, USA).
qRT‐PCR studies
Liberibacter crescens cells were grown at 200 rpm in BM7 media at 25, 30 and 32°C. The cells were harvested by centrifugation at 4°C at different points of the growth curve. Total RNA was isolated using the RiboPure‐Bacteria kit (Ambion, (Owned by Thermo Fisher), Waltham, Massachusetts, USA). The cDNAs were synthesized using the iScript cDNA Synthesis kit (Bio‐Rad, Hercules, CA, USA) using random hexamers, in accordance with the manufacturer's protocol. Quantitative real‐time PCR was performed in a Bio‐Rad iCycler IQ apparatus, using the iQ SYBR Green SuperMix (Bio‐Rad). The sequences of primers for B488_12950 and 16S RNA, used to normalize the expression values, are depicted in Table 4.
Statistical analysis
Data analysis was performed using the statistical analysis software program R. A two‐tailed Student's t‐test was used to determine the statistical significance of the qRT‐PCR data. The fold changes in expression were normalized to the 16S rRNA control. The statistical significance of β‐galactosidase activities was determined with an analysis of variance (ANOVA) and Tukey's HSD post hoc test. ANOVA was completed to analyse the overall variation in mean activity. Tukey's HSD test was applied in order to specifically determine which strains exhibited the observed variation in mean activity. A P‐value < 0.05 was considered statistically significant for all analyses and α = 0.05 was implemented for Tukey's HSD testing. All assays were completed in a minimum of triplicate.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author contributions
FL, JFC and CFG designed, performed and analysed the experiments. KAP, FAP and CLG provided assistance and contributed in the preparation of the manuscript. JFC, FL and CFG wrote the paper. CFG and GLL conceived and coordinated the study. All authors reviewed the results and approved the final version of the manuscript.
Supporting information
Fig. S1. Immunoprecipitation assays.
Fig. S2. Immunoprecipitation assays.
Fig. S3. Determination of LotP native molecular weight.
Acknowledgements
We would like to acknowledge Cecilia Silva‐Sanchez for the proteomic analysis and Laurence Prunetti for helping us with the gel filtration techniques. Publication of this article was funded in part by the University of Florida Open Access Publishing Fund.
Microbial Biotechnology (2017) 10(3), 642–656
Funding information
This material is based upon work that is supported by National Institute of Food and Agriculture, U.S. Department of Agriculture http://nifa.usda.gov/ (Award Number 2015‐70016‐23029 to GL and CFG). The content is solely the responsibility of the authors and does not necessarily represent the official views of the granting agencies.
References
- Ammar, E.D. , Shatters, R.G. , and Hall, D.G. (2011) Localization of Candidatus Liberibacter asiaticus, associated with citrus huanglongbing disease, in its psyllid vector using fluorescence in situ hybridization. J Phytopathol 159: 726–734. [Google Scholar]
- Arnold, K. , Bordoli, L. , Kopp, J. , and Schwede, T. (2006) The SWISS‐MODEL workspace: a web‐based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- Bezawork‐Geleta, A. , Brodie, E.J. , Dougan, D.A. , and Truscott, K.N. (2015) LON is the master protease that protects against protein aggregation in human mitochondria through direct degradation of misfolded proteins. Sci Rep 5: 17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchart, F. , Delangle, A. , Lemoine, J. , Bohin, J.P. , and Lacroix, J.M. (2007) Proteomic analysis of a non‐virulent mutant of the phytopathogenic bacterium Erwinia chrysanthemi deficient in osmoregulated periplasmic glucans: change in protein expression is not restricted to the envelope, but affects general metabolism. Microbiology 153: 760–767. [DOI] [PubMed] [Google Scholar]
- Bové, J.M. (2006) Huanglongbing: a destructive, newly‐emerging, century‐old disease of citrus. J Plant Pathol 88: 7–37. [Google Scholar]
- Chaudhary, R. , Atamian, H.S. , Shen, Z. , Briggs, S.P. , and Kaloshian, I. (2014) GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc Natl Acad Sci USA 111: 8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chir, J.L. , Liao, J.H. , Lin, Y.C. , and Wu, S.H. (2009) The N‐terminal sequence after residue 247 plays an important role in structure and function of Lon protease from Brevibacillus thermoruber WR‐249. Biochem Biophys Res Commun 382: 762–765. [DOI] [PubMed] [Google Scholar]
- Cho, Y. , Zhang, X. , Pobre, K.F.R. , Liu, Y. , Powers, D.L. , Kelly, J.W. , et al (2015) Individual and collective contributions of chaperoning and degradation to protein homeostasis in E. coli . Cell Rep 11: 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe, E. , Valladares, R. , Vu, C. , Kuznetsova, E. , and Gonzalez, C.F. (2010) Determination of Francisella tularensis AcpB acid phosphatase substrate preferences. J Mol Microbiol Biotechnol 19: 198–203. [DOI] [PubMed] [Google Scholar]
- Ding, F. , Duan, Y. , Paul, C. , Brlansky, R.H. , and Hartung, J.S. (2015) Localization and distribution of “Candidatus Liberibacter asiaticus” in citrus and periwinkle by direct tissue blot immuno assay with an Anti‐OmpA polyclonal antibody. PLoS ONE 10: e0123939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman, R.E. , and Löwe, J. (2010) Crystal structures of Bacillus subtilis Lon protease. J Mol Biol 401: 653–670. [DOI] [PubMed] [Google Scholar]
- Fagen, J.R. , Leonard, M.T. , Coyle, J.F. , McCullough, C.M. , Davis‐Richardson, A.G. , Davis, M.J. , and Triplett, E.W. (2014) Liberibacter crescens gen. nov., sp. nov., the first cultured member of the genus Liberibacter. Int J Syst Evol Microbiol 64: 2461–2466. [DOI] [PubMed] [Google Scholar]
- Fenton, W.A. , Kashi, Y. , Furtak, K. , and Horwich, A.L. (1994) Residues in chaperonin GroEL required for polypeptide binding and release. Nature 371: 614–619. [DOI] [PubMed] [Google Scholar]
- Fischer, H.M. , Babst, M. , Kaspar, T. , Acuña, G. , Arigoni, F. , and Hennecke, H. (1993) One member of a gro‐ESL‐like chaperonin multigene family in Bradyrhizobium japonicum is co‐regulated with symbiotic nitrogen fixation genes. EMBO J 12: 2901–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert, F. , Finan, T.M. , Long, S.R. , Puhler, A. , Abola, P. , Ampe, F. , et al (2001) The composite genome of the legume symbiont Sinorhizobium meliloti . Science 293: 668–672. [DOI] [PubMed] [Google Scholar]
- Georgopoulos, C. (2006) Toothpicks, serendipity and the emergence of the Escherichia coli DnaK (Hsp70) and GroEL (Hsp60) chaperone machines. Genetics 174: 1699–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisbert, E. , Yura, T. , Rhodius, V.A. , and Gross, C.A. (2008) Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev 72: 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera, T.S. , Csonka, L.N. , and Paliy, O. (2008) Genome‐wide transcriptional responses of Escherichia coli K‐12 to continuous osmotic and heat stresses. J Bacteriol 190: 3712–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman, L.M. , Belin, D. , Carson, M.J. , and Beckwith, J. (1995) Tight regulation, modulation, and high‐level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer‐Hartl, M. , Bracher, A. , and Hartl, F.U. (2016) The GroEL‐GroES chaperonin machine: a nano‐cage for protein folding. Trends Biochem Sci 41: 62–76. [DOI] [PubMed] [Google Scholar]
- Henderson, B. , Fares, M.A. , and Lund, P.A. (2013) Chaperonin 60: a paradoxical, evolutionarily conserved protein family with multiple moonlighting functions. Biol Rev Camb Philos Soc 88: 955–987. [DOI] [PubMed] [Google Scholar]
- Hoffman, M.T. , Doud, M.S. , Williams, L. , Zhang, M.‐Q. , Ding, F. , Stover, E. , et al (2013) Heat treatment eliminates “Candidatus Liberibacter asiaticus” from infected citrus trees under controlled conditions. Phytopathology 103: 15–22. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A. , and Sternberg, M.J.E. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4: 363–371. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , and Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupper, M. , Gupta, S.K. , Feldhaar, H. , and Gross, R. (2014) Versatile roles of the chaperonin GroEL in microorganism‐insect interactions. FEMS Microbiol Lett 353: 1–10. [DOI] [PubMed] [Google Scholar]
- Lai, K.‐K. , Davis‐Richardson, A.G. , Dias, R. , and Triplett, E.W. (2016) Identification of the genes required for the culture of Liberibacter crescens, the closest cultured relative of the liberibacter plant pathogens. Front Microbiol 7: 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard, M.T. , Fagen, J.R. , Davis‐Richardson, A.G. , Davis, M.J. , and Triplett, E.W. (2012) Complete genome sequence of Liberibacter crescens BT‐1. Stand Genomic Sci 7: 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I. , and Bork, P. (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Gustchina, A. , Rasulova, F.S. , Melnikov, E.E. , Maurizi, M.R. , Rotanova, T.V. , et al (2010) Structure of the N‐terminal fragment of Escherichia coli Lon protease. Acta Crystallogr D Biol Crystallogr 66: 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.‐C. , Su, S.‐C. , Su, M.‐Y. , Liang, P.‐H. , Feng, C.‐C. , Wu, S.‐H. , and Chang, C.‐I. (2016) Structural insights into the allosteric operation of the lon AAA+ protease. Structure 24: 667–675. [DOI] [PubMed] [Google Scholar]
- Louzada, E.S. , Vazquez, O.E. , Braswell, W.E. , Yanev, G. , Devanaboina, M. and Kunta, M. (2016) Distribution of “Candidatus Liberibacter Asiaticus” above and below ground in texas citrus. Phytopathology 106: 702–709. [DOI] [PubMed] [Google Scholar]
- Lund, P.A. (2009) Multiple chaperonins in bacteria – Why so many?. FEMS Microbiol Rev 33: 785–800. [DOI] [PubMed] [Google Scholar]
- Mayhew, M. , da Silva, A.C. , Martin, J. , Erdjument‐Bromage, H. , Tempst, P. , and Hartl, F.U. (1996) Protein folding in the central cavity of the GroEL‐GroES chaperonin complex. Nature 379: 420–426. [DOI] [PubMed] [Google Scholar]
- Miller, J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harb, NY: Cold Spring Harb. Lab. Press. [Google Scholar]
- Okinaka, Y. , Yang, C.H. , Perna, N.T. , and Keen, N.T. (2002) Microarray profiling of Erwinia chrysanthemi 3937 genes that are regulated during plant infection. Mol Plant Microbe Interact 15: 619–629. [DOI] [PubMed] [Google Scholar]
- Oshima, K. , Ishii, Y. , Kakizawa, S. , Sugawara, K. , Neriya, Y. , Himeno, M. , et al (2011) Dramatic transcriptional changes in an intracellular parasite enable host switching between plant and insect. PLoS ONE 6: e23242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliai, F.A. , Gardner, C.L. , Pande, S.G. and Lorca, G.L. (2010) LVIS553 transcriptional regulator specifically recognizes novobiocin as an effector molecule. J Biol Chem 285, 16921–16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliai, F.A. , Gardner, C.L. , Bojilova, L. , Sarnegrim, A. , Tamayo, C. , Potts, A.H. , et al (2014) The transcriptional activator LdtR from “Candidatus Liberibacter asiaticus” mediates osmotic stress tolerance. PLoS Pathog 10: e1004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliai, F.A. , Gonzalez, C.F. and Lorca, G.L. (2015) Identification of a ligand binding pocket in LdtR from Liberibacter asiaticus . Front Microbiol 6: 1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, M. , Kuznetsova, E. , Sanders, S.A. , Gonzalez, C.F. , Brown, G. , Edwards, A.M. , et al (2008) High throughput screening of purified proteins for enzymatic activity. Methods Mol Biol 426, 331–341. [DOI] [PubMed] [Google Scholar]
- Rao, N. , Shashidhar, R. , and Bandekar, J.R. (2013) Comparative analysis of induction of osmotic‐stress‐dependent genes in Vibrio vulnificus exposed to hyper‐and hypo‐osmotic stress. Can J Microbiol 59: 333–338. [DOI] [PubMed] [Google Scholar]
- Roudiak, S.G. , and Shrader, T.E. (1998) Functional role of the N‐terminal region of the Lon protease from Mycobacterium smegmatis . Biochemistry 37: 11255–11263. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. , and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual. (No. Ed. 2). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Secades, P. and Guijarro, J.A. (1999) Purification and characterization of an extracellular protease from the fish pathogen Yersinia ruckeri and effect of culture condition on production. Appl Environ Microbiol 65, 3969–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt, E. , Frederiksen, W. , Garrity, G.M. , Grimont, P.A.D. , Kämpfer, P. , Maiden, M.C.J. , et al (2002) Report of the ad hoc committee for the re‐evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52: 1043–1047. [DOI] [PubMed] [Google Scholar]
- Stoll, S. , Feldhaar, H. , and Gross, R. (2009) Promoter characterization in the AT‐rich genome of the obligate endosymbiont “Candidatus Blochmannia floridanus”. J Bacteriol 191: 3747–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin, M.F. , Baldini, R.L. , Gueiros‐Filho, F. , and Gomes, S.L. (2006) GroES/GroEL and DnaK/DnaJ have distinct roles in stress responses and during cell cycle progression in Caulobacter crescentus . J Bacteriol 188: 8044–8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D. , Higgins, D.G. , and Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet‐Gely, I. , Donovan, K.E. , Fang, R. , Joung, J.K. and Dove, S.L. (2005) Repression of phase‐variable cup gene expression by H‐NS‐like proteins in Pseudomonas aeruginosa . Proc Natl Acad Sci USA 102: 11082–11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. , and Trivedi, P. (2013) Citrus huanglongbing: a newly relevant disease presents unprecedented challenges. Phytopathology 103: 652–665. [DOI] [PubMed] [Google Scholar]
- Wilcox, J.L. , Dunbar, H.E. , Wolfinger, R.D. , and Moran, N.A. (2003) Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol Microbiol 48: 1491–1500. [DOI] [PubMed] [Google Scholar]
- Wohlever, M.L. , Baker, T.A. , and Sauer, R.T. (2014) Roles of the N domain of the AAA+ Lon protease in substrate recognition, allosteric regulation and chaperone activity. Mol Microbiol 91: 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrench, A.P. , Gardner, C.L. , Siegel, S.D. , Pagliai, F.A. , Malekiha, M. , Gonzalez, C.F. , and Lorca, G.L. (2013) MglA/SspA complex interactions are modulated by inorganic polyphosphate. PLoS ONE 8: e76428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Q. , Sreedharan, A. , Wei, S. , Wang, J. , Pelz‐stelinski, K. , Folimonova, S. , and Wang, N. (2013) Global gene expression changes in Candidatus Liberibacter asiaticus during the transmission in distinct hosts between plant and insect. Mol Plant Pathol 14: 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Immunoprecipitation assays.
Fig. S2. Immunoprecipitation assays.
Fig. S3. Determination of LotP native molecular weight.


