Abstract
Taiwan's National Laboratory System is one of the action packages of the Global Health Security Agenda, which was launched by the World Health Organization (WHO) to promote health security as an international priority and to encourage progress toward full implementation of the WHO International Health Regulations (IHR) 2005. The mission of each national laboratory system is to conduct real-time biosurveillance and effective laboratory-based diagnostics, as measured by a nationwide laboratory system able to reliably conduct diagnoses on specimens transported properly to designated laboratories from at least 80% of the regions in the country. In Taiwan, the national laboratory system for public health is well-established and coordinated by the Taiwan Centers for Disease Control (CDC), which is the government authority in charge of infectious disease prevention and intervention. Through the national laboratory system, Taiwan CDC effectively detects and characterizes pathogens that cause communicable diseases across the entire country, including both known and novel threats, and also conducts epidemiologic analyses of infectious diseases. In this article, we describe the national laboratory system for public health in Taiwan. We provide additional information on the national influenza laboratory surveillance network to demonstrate how our national laboratory systems work in practice, including descriptions of long-term seasonal influenza characterization and successful experiences identifying novel H7N9 and H6N1 influenza viruses.
Keywords: : Global Health Security Agenda, National laboratory system, Influenza surveillance, H7N9- and H6N1-infected human cases
The Global Health Security Agenda (GHSA) encourages collaboration on the national and international levels to promote health security. There are 11 action packages spanning a prevent-detect-response framework to facilitate regional and global collaboration toward the objectives of GHSA.1 The National Laboratory System is one of the GHSA action packages (Detect-1) whose goals are to conduct real-time biosurveillance and effective laboratory-based diagnostics, as measured by a nationwide laboratory system able to reliably conduct diagnoses on specimens transported to designated laboratories from at least 80% of the regions in the country. Laboratory testing for detection of country-specific priority diseases, the specimen referral and transport system, use of effective point-of-care and laboratory-based diagnostics, and a well-organized laboratory quality system are the 4 essential components of an effective national laboratory system.
Influenza testing is one of 6 core capabilities that are critical for effective national laboratory systems.1 Diseases caused by the highly infectious influenza virus occur annually, with seasonal variation in countries located in temperate regions. During seasonal epidemics, 5% to 15% of the worldwide population is typically infected, resulting in 3 to 5 million cases of severe illness and 250,000 to 500,000 deaths every year around the world.2,3 Influenza viruses are genetically diverse because of their high mutation rates, frequent reassortments among genomic segments, and tendency to jump between hosts.4 They undergo continuing evolution through antigenic drift and antigenic shift, the 2 major processes responsible for the generation of a newly emergent influenza virus.5,6 Therefore, not only early detection of the presence of novel influenza strains, but also continuous and comprehensive virus monitoring are critical for disease intervention and early warning of pandemics like those that occurred previously in 1918, 1957, 1968, and 2009.7
In 2016, the Taiwan Centers for Disease Control (TCDC) conducted an International Health Regulations (IHR) Joint External Evaluation in support of the goals of the GHSA and to contribute to efforts to promote global health issues. The evaluation of the country's global health security capabilities was completed with assistance from the University of Pittsburgh Medical Center (UPMC) Center for Health Security.8
In this article, we describe the national laboratory system of public health in Taiwan, including the frameworks of tiered laboratories and specimen referral systems for disease diagnoses. We also illustrate the national influenza laboratory surveillance network in Taiwan, which has been well developed to monitor the long-term circulating and evolutionary trends of influenza viruses and to detect any novel or unknown virus strains that may have the potential to affect public health. Examples of our work in long-term seasonal influenza characterization and previous success in the identification of avian-origin H7N9 and H6N1 influenza viruses are provided to illustrate how our national laboratory system works.
Taiwan's National Laboratory System
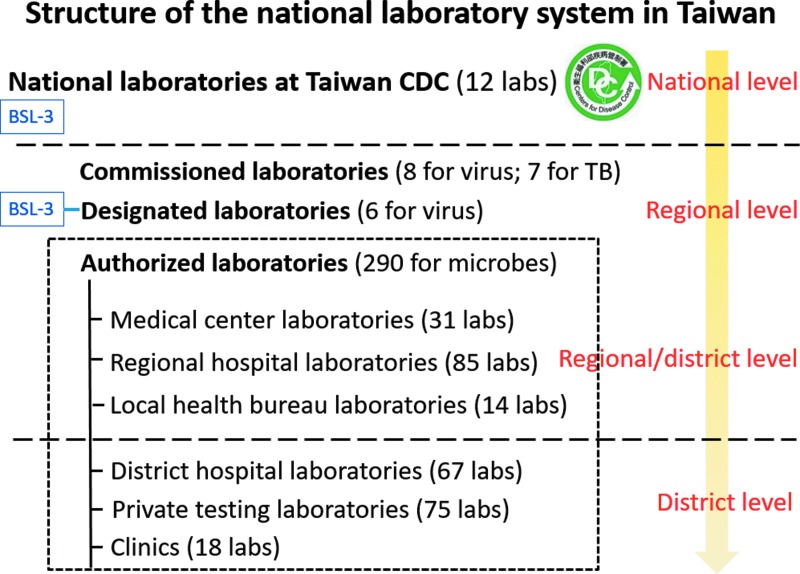
The national laboratory system for public health in Taiwan is well established and is coordinated by TCDC to detect communicable diseases, especially those that are notifiable to the government under the Communicable Disease Control Act. There are 1,251 microbiological laboratories in a tiered system that addresses medical, agricultural, food, and environmental needs. This tiered system includes 31 national labs, 23 local health bureau labs (regional level), 405 hospital labs (regional level), 530 university and research institute labs (district level), and 262 private labs (district level). Among them, numerous laboratories are involved in testing for various notifiable diseases and/or monitoring infectious pathogens; of these, 12 are national labs in TCDC, 7 are designated labs (national or regional level), 16 are commissioned labs (regional level), and 289 are authorized labs (regional and district levels) that are distributed over the country in medical centers, regional hospitals, local health bureaus, district hospitals, private testing institutes, and clinics (Figure 1). There is also a national BSL-4 laboratory in northern Taiwan affiliated with the National Defense Medical Center that serves as a designated lab. The national labs serve as reference centers for the fields of virology, bacteriology, parasitology, mycology, and vector biology (Figure 2). Designated, commissioned, and authorized labs in regional and district levels are dedicated to routine disease diagnosis in the country.
Figure 1.
Tiered structure of the national laboratory system of public health in Taiwan. Laboratories are distributed into 3 tiers (national, regional, and district levels). Commissioned laboratories help diagnose viral and bacterial (tuberculosis) diseases in the community, collect viral and bacterial isolates, and send these isolates to national laboratories at TCDC for pathogen surveillance. Designated and authorized laboratories help diagnose notifiable diseases. National laboratories at TCDC and designated laboratories have BSL-3 facilities for conducting high-risk pathogen diagnosis.
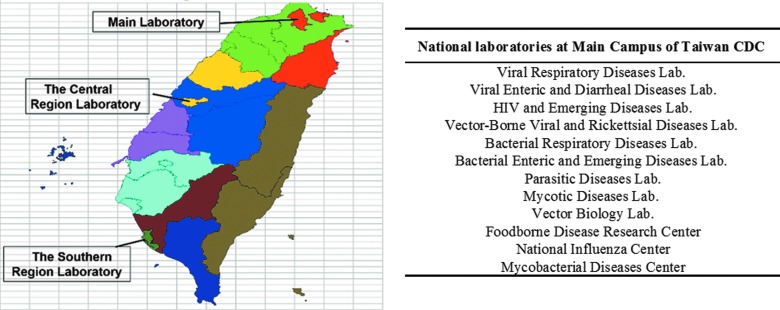
Figure 2.
National laboratories at TCDC. Within the national laboratory system of public health in Taiwan, there are 12 national laboratories at TCDC, including those for virology, bacteriology, parasitology, mycology, and vector biology, which are located at the main campus of TCDC in Taipei. There are also 2 TCDC laboratories at regional campuses in central and southern Taiwan that serve as reference laboratories for disease diagnosis and pathogen characterization.
Membership at each tiered level in the national laboratory system must be approved by the government through a quality management system based on lab diagnostic capabilities and qualities as defined in the national guidelines issued by TCDC. The national labs attached to TCDC have been accredited since 2011. For designated and contract commissioned laboratories, document examination or onsite inspection is required to ensure their diagnostic quality, while designated labs should additionally have their own BSL-3 lab or BSL-4 facilities. Authorized laboratories must provide TCDC the results of proficiency tests for specific pathogen diagnosis every 2 years; reevaluation of diagnostic capability and the quality program must be performed by each laboratory every 4 years to assure the renewal of authorization.
In addition to the tiered laboratory diagnosis infrastructure, the national laboratory system of Taiwan also includes a well-connected specimen referral network for each of the tests necessary to detect and confirm etiologies of communicable diseases in line with standardized regulations for specimen collection, packaging, and transport issued by TCDC. After a report of a suspicious case by practitioners in local clinics or hospitals, specimens are collected from the patient and are tested in the same hospital if the lab of that hospital has been authorized. Otherwise, these specimens are sent by contract couriers to the laboratories at regional or district levels of the tiered network for pathogen diagnosis. Specimens that require advanced or specialized diagnosis may be sent on to national reference laboratories by contract couriers. Courier contracts for specimen transport are co-funded annually by TCDC and local health bureaus.
Laboratory Testing of Notifiable Diseases
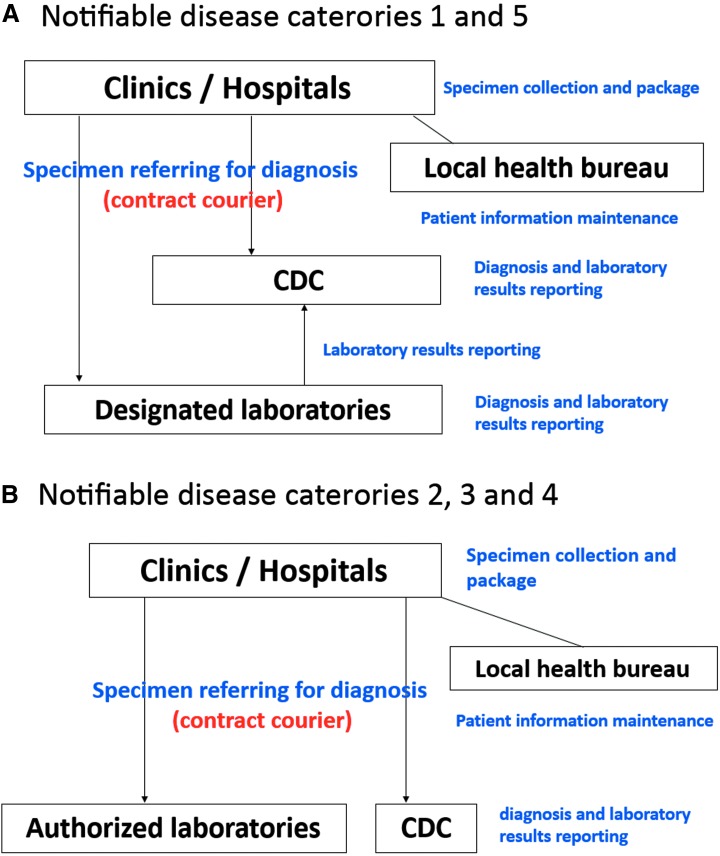
The Communicable Disease Control Act specifies a number of diseases that must be reported to the government. These notifiable diseases are divided into 5 categories based on their degree of risks and hazards, such as case fatality rate, incidence rate, and transmission speed.9 The laboratory testing required to confirm each disease is conducted through 2 specimen referral and diagnosis networks regulated by law. In terms of notifiable diseases belonging to categories 1 and 5 (12 diseases)—highly contagious, newly emerging infectious diseases that could result in a substantial impact on the health of the human population, such as avian influenza, Zika, and Middle East respiratory syndrome coronavirus (MERS)—clinical specimens from suspicious patients are collected and packaged in local clinics and hospitals and sent to designated or TCDC laboratories for diagnosis. For positive specimens tested by designated laboratories, final confirmation by TCDC is required (Figure 3A). A similar system has been established for diagnosis of the 54 diseases belonging to categories 2, 3, and 4, with a slight difference. Once the clinical specimens from clinics and hospitals are collected and packaged, they are sent to authorized or TCDC laboratories. However, each authorized laboratory has the authority to announce positive results without TCDC confirmation (Figure 3B). Personal and epidemiologic information for each suspected or laboratory-confirmed patient is maintained by the local health bureaus through a web-based system. Within a designated turnaround time based on the diagnostic methods used, local practitioners can access laboratory results online via the National Notifiable Disease Surveillance System.
Figure 3.
Specimen referral and diagnosis networks of notifiable diseases in Taiwan. The networks for notifiable diseases belonging to (A) categories 1 and 5 as well as to (B) categories 2, 3, and 4 are illustrated. (A) Clinical specimens of patients with suspected highly infectious newly emerging (categories 1 or 5) diseases are required to be collected and packaged from local clinics and hospitals and sent to designated or TCDC laboratories for diagnosis. Positive results detected by designated laboratories are confirmed by TCDC before final report announcement. (B) Regional and district authorized laboratories serve as diagnostic agencies for categories 2, 3, and 4 diseases. However, each laboratory has the authority to announce positive results without TCDC confirmation. All the personal and epidemiologic information of each suspected or laboratory-confirmed patient is maintained by the local health bureaus through a web-based system.
National Laboratory Influenza Surveillance Network
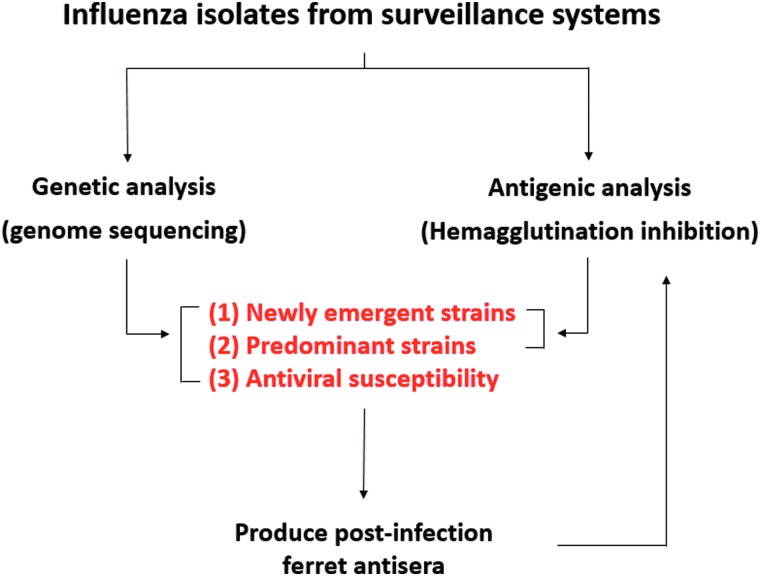
Seasonal epidemics of influenza occur globally on an annual basis. Vaccination against influenza viruses is an important disease prevention method. However, the continuously evolving nature of the virus reduces vaccine effectiveness; therefore, antigens for global seasonal influenza vaccines must be reformulated annually.10,11 To capture the dynamic pattern of the circulating influenza virus strains in a real-time manner, the national laboratory surveillance network of influenza viruses in Taiwan, which monitors the country's outpatients with influenzalike illness in the community and hospitalized patients with severe complications, was established and coordinated by TCDC. The regional commissioned laboratories are located in the northern (n = 3), central (n = 2), southern (n = 2), and eastern (n = 1) parts of Taiwan, and they continually conduct influenza testing for clinical specimens taken from ill people in the community and collect viral isolates from infected outpatients. Viruses infecting inpatients with severe complications must be detected and isolated by authorized and TCDC laboratories, as severe complicated influenza is a category 4 notifiable disease in Taiwan. Isolated influenza viruses are sent to the National Influenza Center (NIC) at TCDC for further analysis by detailed virologic assays on viral genetic and antigenic properties to monitor types and subtypes, lineage, and genetic and antigenic changes of circulating viruses, to detect antiviral resistance, and to identify novel influenza viruses (Figure 4).
Figure 4.
Virologic analyses on influenza viruses circulating in the community through the national influenza laboratory surveillance network. Influenza isolates collected from regional commissioned laboratories are sent to the national influenza center at TCDC for characterization of viral genetic and antigenic properties. Combined data from genomic sequencing and hemagglutination inhibition assays are used to identify the predominant and new variant virus strains; the antiviral susceptibility pattern of a virus is also inferred from sequences of a particular viral gene segment. Post-infection ferret sera are prepared routinely against both predominant and variant strains, which are used as reagents in antigenic analysis by hemagglutination inhibition assay.
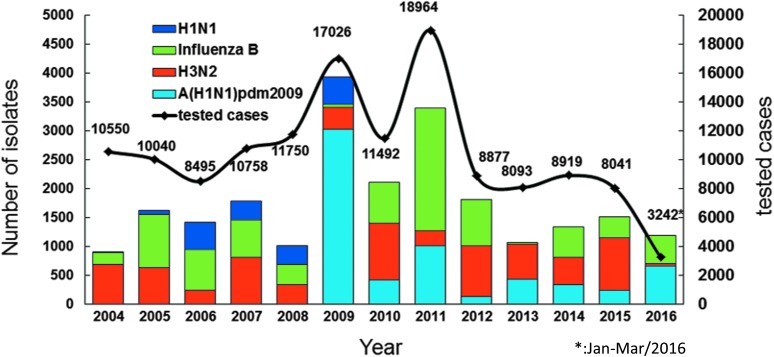
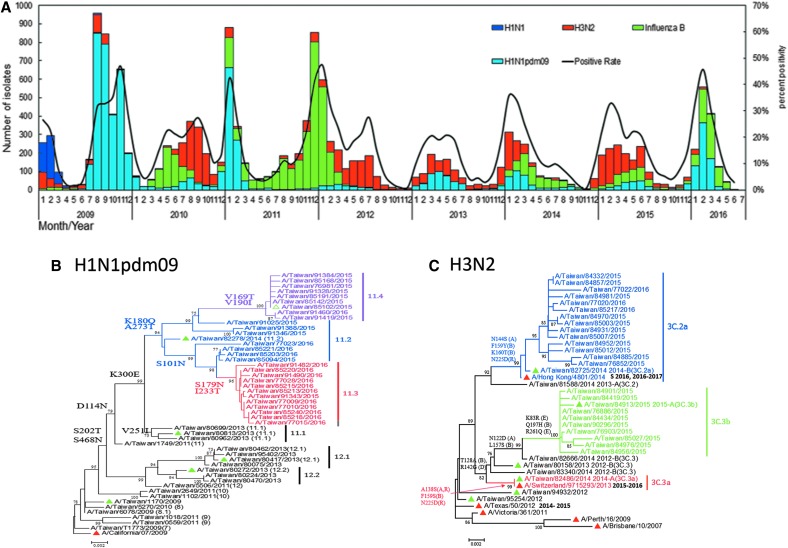
Through the national influenza laboratory surveillance network, from 2004 to 2016, more than 8,000 outpatient specimens per year could be stably collected and tested and more than 1,000 influenza viruses per year were isolated and sent to TCDC for virologic analysis (Figure 5). Based on the long-term monitoring of collected isolates, TCDC can delineate the epidemiologic pattern of circulating viruses in a real-time manner, which clearly shows annual peaks in winter dominated by various virus types or subtypes; summer influenza peaks are usually observed as well (Figure 6A). In addition, phylogenetic analysis of viral surface hemagglutinin (HA) genes is used to explore the genetic relationship between domestic circulating viruses and annual global vaccine strains (Figures 6B-E). Combined with data on viral antigenic characterization determined by a hemagglutination inhibition (HI) assay, the vaccine mismatch rate of the H3N2 subtype in Taiwan is higher than those of the H1N1pdm09 and influenza B viruses (Table 1). This supports past research showing that a higher genetic mutation rate was observed among the H3N2 viruses.12,13 Furthermore, successful examples of the laboratory surveillance system to promote health security include the timely detection of a new variant of H3N2 strain that caused a mismatch in the 2009-10 northern hemisphere winter vaccine, and also the research data reporting virologic factors that may have contributed to the previous large epidemic waves of A(H1N1)pdm09 or influenza B viruses that occurred in Taiwan.14-16
Figure 5.
National laboratory surveillance capacity on influenza viruses. The number of cases tested by regional commissioned laboratories and influenza isolates sent to TCDC are indicated on the right X and left X axes, respectively. From 2004 to 2016, more than 8,000 specimens per year from outpatients were tested and more than 1,000 influenza viruses per year were isolated and sent to TCDC for virologic analyses.
Figure 6.
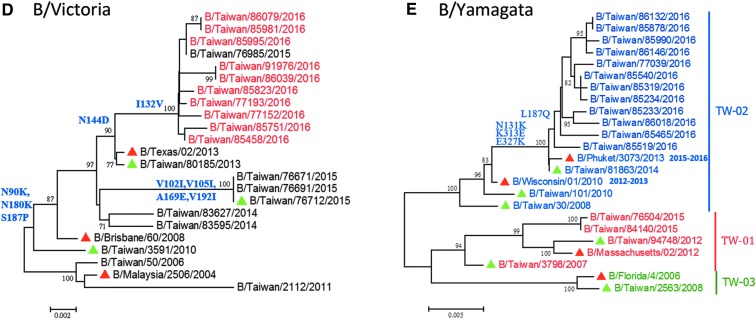
Laboratory surveillance of influenza viruses in Taiwan. (A) Circulation of influenza viruses in Taiwan from 2009 to 2016. The monthly distributions of the influenza isolates, including A(H1N1), A(H3N2), A(H1N1)pdm09, and influenza B viruses are shown as bars with different colors. Positive rates of confirmed cases are also shown in the line chart. (Data collected from January to March 2016.) (B-E) Phylogenetic relationships of HA genes of A(H1N1)pdm09 (B), A(H3N2) (C), B/Victoria lineage (D), and B/Yamagata lineage (E) viruses circulating in Taiwan. The phylogenetic trees were constructed using the neighbor-joining method with 1,000 bootstrap replications. Branch values of more than 70 and the major amino acid signatures of each genotype are indicated. WHO-recommended vaccine strains are shown as red triangles; domestic strains that are used to produce post-infection ferret antisera are shown as green triangles.
Table 1.
Comparison Between Circulating and Vaccine Strains During Influenza Seasons 2009-2016 in Taiwan
| Influenza Season | |||||||
|---|---|---|---|---|---|---|---|
| Viruses | 2009-10 | 2010-11 | 2011-12 | 2012-13 | 2013-14 | 2014-15 | 2015-16 |
| Circulating strainsa | |||||||
| A(H1N1)pdm09 | CA09a | CA09 | CA09 | CA09 | CA09 | CA09 | CA09 |
| A(H3N2) | PE09 | PE09 | VI11 | VI11 | VI11, TE12 | SW13, HK14 | SW13, HK14 |
| Influenza B | — | — | MA12(Y) | — | WI10(Y), MA12(Y), BR08(V) | MA12(Y), PH13(Y), BR08(V) | PH13(Y), BR08(V) |
| Vaccine strains | |||||||
| A(H1N1)pdm09 | BR07(H1) | CA09 | CA09 | CA09 | CA09 | CA09 | CA09 |
| A(H3N2) | BR07(H3) | PE09 | PE09 | VI11 | VI11 | TE50 | SW13 |
| Influenza B | BR08(V) | BR08(V) | BR08(V) | WI10(Y) | MA12(Y) | MA12(Y) | PH13(Y) |
The circulating strain in bold type indicates its antigenic property is significantly different from that of the contemporary global vaccine strain of the same subtype.
Monitoring of viral drug susceptibility against neuraminidase inhibitors is also conducted routinely through the surveillance system by analyzing NA gene sequences of circulating influenza strains. Our results of long-term oseltamivir-resistance surveillance in seasonal H1N1 viruses during the influenza seasons of 2005 to 2009 in Taiwan show that viruses carrying the H275Y substitution in their NA genes were sporadically detected in 2007-08 and had quickly reached 100% in 2008-09, consistent with the global situation.17 After the 2009 pandemic, oseltamivir-resistant H1N1pdm09 viruses with the NA H275Y markers were also sporadically identified from hospitalized patients.18 Detailed epidemiologic and virologic analyses revealed that all the viral resistance was obtained after Tamiflu treatment; no further viral transmission between infected patients was found. Notably, no oseltamivir-resistant H3N2 and influenza B viruses have been detected during recent years.
Success Stories and Examples
Aquatic birds are natural hosts for influenza A viruses, and they are thought to be the major source of most influenza viruses in other animal species.19 In addition to the H1N1 and H3N2 viral subtypes that have caused sustained epidemics in the human population,20 viruses originating in animal species, such as poultry and pigs, may cross species barriers and infect humans in a severe manner.7,21-24 After the large pandemic outbreaks caused by the swine-origin influenza A(H1N1)pdm09 viruses occurring globally in 2009, human infections caused by zoonotic viral subtypes have occurred. Below we describe examples of how the national influenza surveillance system has been used successfully to identify novel avian-origin influenza viruses from infected human cases, including the 4 imported H7N9-infected cases and 1 domestic H6N1-infected case identified in 2013 and 2014.25,26
Cases of H7N9 Infection
The novel avian-origin H7N9 virus was first detected in March 2013 in China, and it has subsequently caused 4 epidemic waves that resulted in 798 laboratory-confirmed human cases as of August 17, 2016.27 In Taiwan, 4 H7N9 human cases imported from China have been identified, 2 in 2013 and 2 in 2014. They were identified by the designated regional and/or TCDC laboratories. All 4 cases had severe illness with pneumonia and required admission to the intensive care unit; 1 patient died.25 Sputum and throat swab specimens were collected and diagnosed for each patient, and we determined that the viral load of the H7N9 virus was much higher in the lower respiratory tract than in the upper respiratory tract, since 2 of the 4 confirmed cases had H7N9-positive results detected only from sputum specimens.28 This trend is similar to the tissue tropism observed from H5N1 viral infections.29,30 In addition to virus detection, full-genome sequence analysis of the 4 virus strains was also conducted by TCDC; the results were released immediately to the Global Initiative on Sharing All Influenza Data (GISAID) database. These H7N9 virus strains were also shared with the WHO Collaborating Centers in the United States (US Centers for Disease Control and Prevention) and Japan (Japan National Institute of Infectious Diseases).
To evaluate possible risks of the novel H7N9 viruses to public health, viral genetic markers that may be related to human adaptation and their antigenic properties were analyzed by TCDC. All 4 H7N9 viruses are antigenically similar to the A/Anhui/1/2013 vaccine candidate strain. However, they harbor markers related to enhanced polymerase activity (the 627K or 701N in the polymerase basic protein 2) and high-affinity binding of virus to human receptors (the 226L in the hemagglutinin protein), suggesting their better replication and infection capabilities in human population.25 One virus also has markers that may confer oseltamivir resistance (the 292K in the neuraminidase protein). Furthermore, the phylogenetic relationship of viral full-genome sequences reveals that these 4 viruses were generated through various reassortment events in China, indicating their complex evolutionary history.25 These findings are consistent with the initial viral characteristics reported by other research groups at the early stage of H7N9 outbreaks in China.31
Identification of the First Global H6N1 Infection
Identification of the first global human infection with the influenza A(H6N1) virus in 2013 in Taiwan highlights the important role of the national influenza laboratory surveillance network.26 Avian influenza A(H6N1) virus is a common virus isolated from migrating birds and domestic poultry in many countries.32,33 However, human infection with this subtype had not been reported prior to the confirmation of H6N1 infection in a 20-year-old patient with influenzalike illness by TCDC in May 2013. On May 8, 2013, an influenza A virus isolated from the inpatient's throat swab specimen was identified by a regional authorized laboratory using an immunofluorescence assay. However, subsequent analyses by subtype-specific PCR assays showed negative results for human H1N1pdm09 and H3N2 viruses.
On May 20, this unsubtyped influenza virus strain was sent to TCDC. After results of H1, H3, H5, H7, and H9 molecular diagnoses were all negative, the virus was further identified as the H6N1 subtype based on sequence analysis of its hemagglutinin and neuraminidase genes. Subsequently, TCDC developed the molecular diagnostic method and conducted enhanced active surveillance. From June 10 to August 13, a total of 125 individuals with influenzalike illness or pneumonia were reported; all of their respiratory specimens tested negative for H6N1. Six symptomatic contacts were identified, and serological analyses of the paired serum specimens collected from those contacts also showed undetectable antibody titers (≦10) against the H6N1 virus, while titers of the first and second serum specimens of the index case were 40 and 80, respectively.26 Therefore, no additional H6N1-infected cases were identified.
The full-genome sequences of the H6N1 strain were analyzed by TCDC and released to the GISAID database promptly. Since then, TCDC and Taiwan Animal Health Research Institute (AHRI) have collaborated closely to conduct a comprehensive risk assessment for this virus strain. After gathering the sequences of chicken H6N1 viruses circulating in Taiwan from AHRI, results of phylogenetic analyses showed that the human H6N1 virus was highly homologous to domestic chicken H6N1 viruses, especially to the strain isolated in the same year (2013). Since all 8 viral gene segments could be traced back to several older chicken H6N1 viruses, we propose that the human H6N1 virus was generated through multiple reassortment events in chickens in Taiwan and then infected the human case. Furthermore, genetic analysis showed that the virus has the marker related to high-affinity binding of viruses to human receptors (the 228S in the hemagglutinin protein), suggesting its better adaptation to the human population.26
Conclusion and Discussion
The national public health laboratory system in Taiwan is a tiered framework that has been externally evaluated, demonstrating capabilities for testing various communicable and notifiable diseases, including the priority diseases designated by WHO. The functionality of the tiered lab system is supported by the tightly connected specimen referral network covering the entirety of Taiwan and the reliable diagnostic capabilities of a well-organized laboratory quality management system. Similar to the global influenza surveillance and response system (GISRS) operated by the WHO, the national laboratory influenza surveillance system in Taiwan continuously monitors the evolution and antiviral susceptibility of influenza viruses to evaluate possible risks caused by their dynamically changing nature and provides laboratory diagnostics to maintain the sustainable detection capacity of the lab system. It also serves as a country-wide alert system for the emergence of novel influenza viruses with pandemic potential, particularly given recent successful identification of H7N9- and H6N1-infected human cases.
The unpredictability of the circulating influenza viruses highlights the importance of intensive nationwide surveillance of this evolving pathogen. These surveillance efforts assist in early detection of such viruses and allow government authorities to engage in appropriate actions such as containment and decontamination for disease control. In addition to influenza, there are also comprehensive surveillance networks for viral, bacterial, parasitic, and fungal diseases that have been incorporated into the national laboratory system to characterize both known and novel pathogens causing epidemic disease throughout the country. On the other hand, potential threats to public health caused by zoonotic pathogens are continuously increasing. Close working relationships between public health and animal health laboratories should be encouraged to achieve the “One Health” goal of the GHSA.
Acknowledgments
The authors thank the members of the national influenza laboratory surveillance networks for their great efforts, including colleagues working at TCDC, local health bureaus, and laboratories in the tiered levels of the national laboratory system. Comments and English editing on this manuscript kindly provided by Dr. Tara Kirk Sell and Matthew Shearer at the Johns Hopkins Center for Health Security are also appreciated.
References
- 1.Global Health Security Agenda action packages. https://www.cdc.gov/globalhealth/healthprotection/ghs/pdf/ghsa-action-packages_24-september-2014.pdf Accessed February9, 2017
- 2.Stöhr K. Influenza—WHO cares. Lancet Infect Dis 2002;2(9):517. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. WHO Fact sheet N°211. Influenza. Revised March 2003. http://www.who.int/mediacentre/factsheets/2003/fs211/en/ Accessed February9, 2017
- 4.Horimoto T, Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol 2005;3(8):591-600 [DOI] [PubMed] [Google Scholar]
- 5.Nelson MI, Holmes EC. The evolution of epidemic influenza. Nat Rev Genet 2007;8(3):196-205 [DOI] [PubMed] [Google Scholar]
- 6.Nicholson KG, Wood JM, Zambon M. Influenza. Lancet 2003;362(9397):1733-1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009;459(7249):931-939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.UPMC Center for Health Security. IHR Joint External Evaluation of Taiwan. 2016. http://www.centerforhealthsecurity.org/our-work/pubs_archive/pubs-pdfs/2016/IHR_JEE_Final_Report_Taiwan_FINAL.pdf Accessed February9, 2017
- 9.Taiwan Ministry of Health and Welfare. Communicable Disease Control Act. Amended in 2015. http://law.moj.gov.tw/Eng/LawClass/LawContent.aspx?PCODE=L0050001 Accessed February9, 2017
- 10.Carrat F, Flahault A. Influenza vaccine: the challenge of antigenic drift. Vaccine 2007;25(39-40):6852-6862 [DOI] [PubMed] [Google Scholar]
- 11.Gerdil C. The annual production cycle for influenza vaccine. Vaccine 2003;21(16):1776-1779 [DOI] [PubMed] [Google Scholar]
- 12.Ghedin E, Sengamalay NA, Shumway M, et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 2005;437(7062):1162-1166 [DOI] [PubMed] [Google Scholar]
- 13.Holmes EC, Ghedin E, Miller N, et al. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol 2005;3(9):e300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang JR, Huang YP, Chang FY, et al. Phylogenetic and evolutionary history of influenza B viruses, which caused a large epidemic in 2011-2012, Taiwan. PLoS One 2012;7(10):e47179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang JR, Huang YP, Chang FY, et al. New variants and age shift to high fatality groups contribute to severe successive waves in the 2009 influenza pandemic in Taiwan. PLoS One 2011;6(11):e28288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang JR, Lin CH, Chen CJ, et al. A new antigenic variant of human influenza A (H3N2) virus isolated from airport and community surveillance in Taiwan in early 2009. Virus Res 2010;151(1):33-38 [DOI] [PubMed] [Google Scholar]
- 17.Yang JR, Lin YC, Huang YP, et al. Reassortment and mutations associated with emergence and spread of oseltamivir-resistant seasonal influenza A/H1N1 viruses in 2005-2009. PLoS One 2011;6(3):e18177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang JR, Huang YP, Lin YC, et al. Early findings of oseltamivir-resistant pandemic (H1N1) 2009 influenza A viruses in Taiwan. Antiviral Res 2010;88(3):256-262 [DOI] [PubMed] [Google Scholar]
- 19.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev 1992;56(1):152-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med 2000;51:407-421 [DOI] [PubMed] [Google Scholar]
- 21.Claas EC, Osterhaus AD, van Beek R, et al. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 1998;351(9101):472-477 [DOI] [PubMed] [Google Scholar]
- 22.Fouchier RA, Schneeberger PM, Rozendaal FW, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A 2004;101(5):1356-1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peiris M, Yuen KY, Leung CW, et al. Human infection with influenza H9N2. Lancet 1999;354(9182):916-917 [DOI] [PubMed] [Google Scholar]
- 24.Lindstrom S, Garten R, Balish A, et al. Human infections with novel reassortant influenza A(H3N2)v viruses, United States, 2011. Emerg Infect Dis 2012;18(5):834-837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang JR, Kuo CY, Huang HY, et al. Characterization of influenza A (H7N9) viruses isolated from human cases imported into Taiwan. PLoS One 2015;10(3):e0119792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei SH, Yang JR, Wu HS, et al. Human infection with avian influenza A H6N1 virus: an epidemiological analysis. Lancet Respir Med 2013;1(10):771-778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization. Human infection with avian influenza A(H7N9) virus—China. August 17, 2016. http://www.who.int/csr/don/17-august-2016-ah7n9-china/en/ Accessed February9, 2017
- 28.Chang SY, Lin PH, Tsai JC, Hung CC, Chang SC. The first case of H7N9 influenza in Taiwan. Lancet 2013;381(9878):1621. [DOI] [PubMed] [Google Scholar]
- 29.Peiris JS, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev 2007;20(2):243-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sirinonthanawech N, Uiprasertkul M, Suptawiwat O, Auewarakul P. Viral load of the highly pathogenic avian influenza H5N1 virus in infected human tissues. J Med Virol 2011;83(8):1418-1423 [DOI] [PubMed] [Google Scholar]
- 31.Cui L, Liu D, Shi W, et al. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun 2014;5:3142. [DOI] [PubMed] [Google Scholar]
- 32.Cheung CL, Vijaykrishna D, Smith GJ, et al. Establishment of influenza A virus (H6N1) in minor poultry species in southern China. J Virol 2007;81(19):10402-10412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spackman E, Stallknecht DE, Slemons RD, et al. Phylogenetic analyses of type A influenza genes in natural reservoir species in North America reveals genetic variation. Virus Res 2005;114(1-2):89-100 [DOI] [PubMed] [Google Scholar]