Abstract

The JmjC histone lysine demethylases (KDMs) are epigenetic regulators involved in the removal of methyl groups from post-translationally modified lysyl residues within histone tails, modulating gene transcription. These enzymes require molecular oxygen for catalytic activity and, as 2-oxoglutarate (2OG)-dependent oxygenases, are related to the cellular oxygen sensing HIF hydroxylases PHD2 and FIH. Recent studies have indicated that the activity of some KDMs, including the pseudogene-encoded KDM4E, may be sensitive to changing oxygen concentrations. Here, we report detailed analysis of the effect of oxygen availability on the activity of the KDM4 subfamily member KDM4A, importantly demonstrating a high level of O2 sensitivity both with isolated protein and in cells. Kinetic analysis of the recombinant enzyme revealed a high KMapp(O2) of 173 ± 23 μM, indicating that the activity of the enzyme is able to respond sensitively to a reduction in oxygen concentration. Furthermore, immunofluorescence experiments in U2OS cells conditionally overexpressing KDM4A showed that the cellular activity of KDM4A against its primary substrate, H3K9me3, displayed a graded response to depleting oxygen concentrations in line with the data obtained using isolated protein. These results suggest that KDM4A possesses the potential to act as an oxygen sensor in the context of chromatin modifications, with possible implications for epigenetic regulation in hypoxic disease states. Importantly, this correlation between the oxygen sensitivity of the catalytic activity of KDM4A in biochemical and cellular assays demonstrates the utility of biochemical studies in understanding the factors contributing to the diverse biological functions and varied activity of the 2OG oxygenases.
Introduction
The organization of genetic material into chromatin enables not only the compaction of DNA in the nucleus but also the dynamic regulation of gene transcription. Changes to chromatin structure affect the accessibility of the DNA to the transcriptional machinery and are mediated by hundreds of chromatin-interacting proteins in response to various stimuli. One such family of chromatin-interacting proteins are the Jumonji-C-domain-containing histone lysine demethylases (JmjC-KDMs), which catalyze the removal of methyl groups from Nε-methylated lysyl residues on histone tails, one of numerous known post-translational modifications.1 Lysyl residues at positions 4, 9, 27, 36, and 79 of histone 3 (H3) may be mono-, di- or tri-Nε-methylated, with methylation at residues H3K4 and H3K36 generally associated with active genes and methylation at residues H3K9 and H3K27 with transcriptional repression. The KDM4 subfamily of enzymes catalyze demethylation of tri- and dimethylated H3K9 (H3K9me2/3), H3K36me2/3 marks as well as mono- and dimethylated histone 1.4 lysine 26 (H1.4K26me1/2).2,3
The JmjC-KDMs are members of the Fe2+ and 2-oxoglutarate (2OG)-dependent oxygenase superfamily, which are characterized by a conserved double-stranded β helix (DSBH) fold and share many mechanistic features.4,5 There are over 60 known human 2OG oxygenases, all of which use molecular oxygen as a co-substrate in their catalysis of a range of reactions in varied biological contexts. Some of the more well characterized members of this superfamily include the prolyl hydroxylase domain enzymes 1–3 (PHD1–3) and factor-inhibiting HIF (FIH), collectively termed the HIF hydroxylases. These enzymes are responsible for the regulation of hypoxia inducible factor (HIF), a transcription factor which is considered the master regulator of the cellular hypoxic response.6 In normoxia, the PHD enzymes catalyze hydroxylation of HIF at two prolyl residues in its oxygen-dependent degradation domain, targeting HIF for proteasomal degradation, while FIH catalyzes hydroxylation of an asparaginyl residue in the C-terminal activation domain of HIF, ablating its interaction with transcriptional coactivators.7 The HIF hydroxylases act as oxygen sensors such that in moderate hypoxia, their activity is inhibited, allowing HIF to activate hundreds of target genes8 such as those encoding for erythropoietin (EPO), glycolytic enzymes, and vascular endothelial growth factor (VEGF), enabling the cell to adapt to conditions of low oxygen.7,9,10
There is substantial evidence supporting the proposal that the 2OG oxygenases act via a consensus mechanism involving 2-electron oxidation of 2OG to form an Fe4+-oxo intermediate, followed by oxidation of the primary substrate (Figure S1).11 The 2OG oxygenases have a catalytic requirement for molecular oxygen, which has prompted kinetic analyses of this co-substrate for some members of the superfamily. The collagen prolyl hydroxylase CP4H and the plant phytanoyl-CoA hydroxylase mPAHX have relatively low KMapp(O2) values of 40 and 93 μM, respectively,12,13 suggesting a low degree of sensitivity to oxygen availability in vitro. In contrast, PHD2 activity is highly sensitive to O2 availability, with reported KMapp(O2), values ranging between 230 and 1746 μM12−15 and a slow reaction with oxygen in pre-steady state kinetic experiments.15 These features, along with the rapid degradation of HIF, are proposed to underlie its importance as a cellular oxygen sensor. FIH has a KMapp (O2) of 110 μM,16 implying a reduced dependence on oxygen availability and thus a potentially less significant role in oxygen sensing. Cellular studies have also demonstrated that PHD2-catalyzed HIF prolyl hydroxylation is reduced to a greater degree upon reduction of oxygen availability than HIF asparaginyl hydroxylation catalyzed by FIH.17 Taken together, these results indicate that, although all 2OG oxygenases use oxygen as a co-substrate, the sensitivity of their activity with respect to oxygen concentration varies widely across the superfamily. This raises the question as to whether other members of the 2OG oxygenase superfamily may also have an “oxygen-sensing” capacity or whether the HIF hydroxylases, and particularly PHD2, are uniquely sensitive to changes in oxygen availability.
JmjC-KDM levels and activity have been reported to be influenced by hypoxia, as has histone methylation status. Our recent biochemical analysis of KDM4E revealed that its activity displays a graded response to oxygen over a physiologically relevant range of oxygen concentrations (0–21%).18 Additionally, a range of KMapp(O2) values for members of the KDM4 subfamily has been reported.19 Some JmjC-KDMs are known to be HIF target genes and upregulated in hypoxia in various cell lines (KDM3A, KDM4B, KDM4C, KDM5B, KDM5C, and KDM6B),20−26 while under hypoxia, KDM3A and KDM4C have been found to interact or cooperate with HIF at gene promoters and enhance the induction of HIF target genes in HCT116 and HeLa cell lines, respectively.27,28 Ectopically expressed KDM4B demonstrated a loss of cellular demethylase activity in HeLa cells incubated in hypoxia (0.2% O2), whereas minimal effects on the activity of KDM3A were observed, suggesting differential oxygen sensitivity within the HIF-regulated JmjC-KDMs.21 Furthermore, hypoxia has been found to induce changes to global (and, in some cases, gene-specific) histone lysine methylation status in numerous cell lines, with consequent changes to gene expression profiles.23,29−32 While the impact of hypoxia on chromatin structure is not well-understood, hypoxia in combination with nutrient depletion was shown to result in chromatin compaction,33 and the potential role of chromatin as an oxygen sensor has been postulated.34
These studies on the oxygen-dependence of JmjC-KDMs and global changes in histone lysine methylation status in hypoxic cells and tissues suggest that the activity of the JmjC-KDMs may be differentially modulated by oxygen availability. This led us to investigate the effect of a range of oxygen concentrations on the activity of KDM4A to assess whether this enzyme has the capacity to function as an oxygen sensor, both kinetically and in a cellular context. KDM4A was chosen for investigation as it has been implicated in numerous hypoxic disease states, including breast, prostate, and lung cancers,35 and cardiac hypertrophy,36 and is a close homologue of KDM4E. Here, we report that the activity of both recombinant KDM4A and KDM4A overexpressed in cells displays sensitivity over physiologically relevant oxygen concentrations. This reveals the potential for the JmjC-KDMs to play a key role in fine-tuning transcriptional regulation in an oxygen-dependent manner.
Results and Discussion
Dependence of KDM4A Activity on Oxygen Availability in Vitro
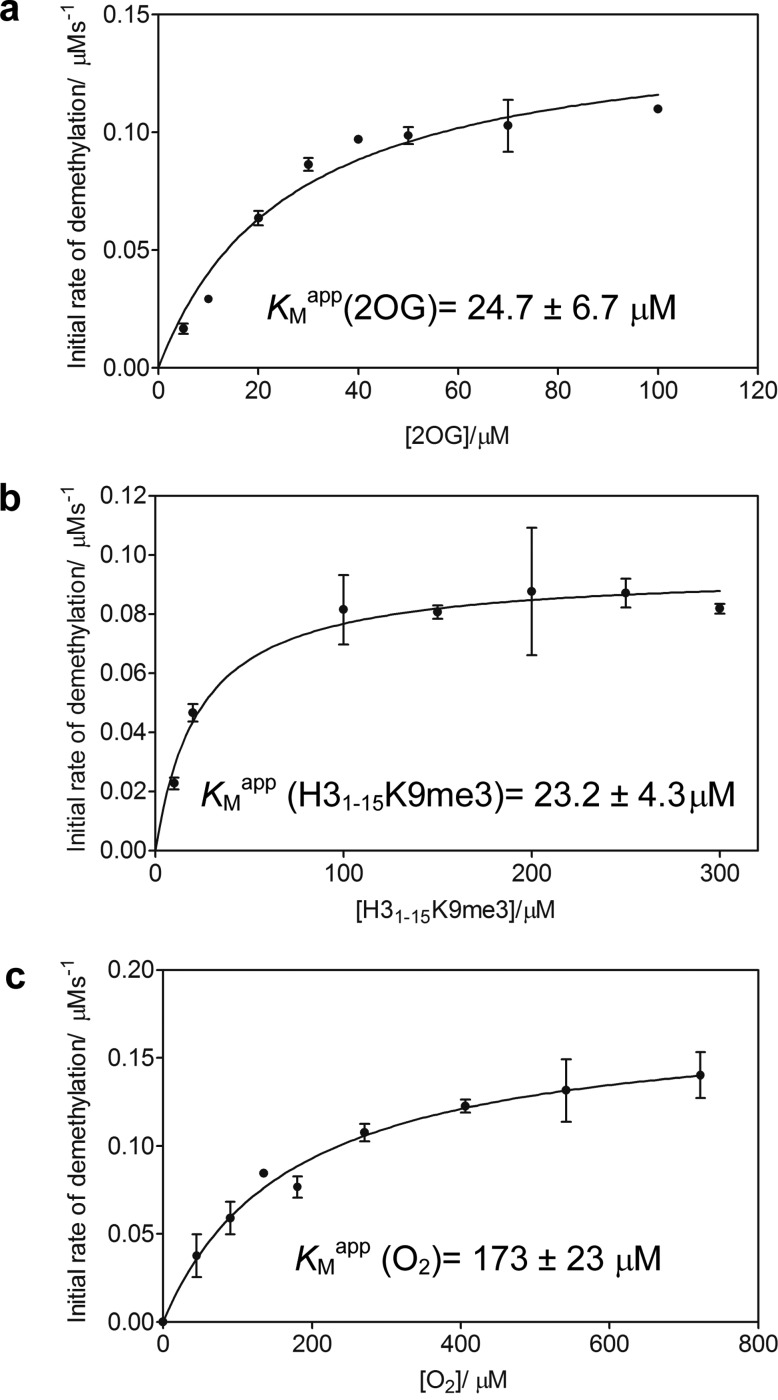
We initially assessed the sensitivity of isolated KDM4A demethylase activity at different oxygen concentrations by evaluating the KMapp(O2) of recombinant KDM4A1–359, which comprises the Jumonji-N and Jumonji-C domains required for catalytic activity.37 As a prelude, steady-state kinetic analysis of KDM4A1–359 with respect to 2OG and a 15mer histone peptide substrate H31–15K9me338 was first conducted using a matrix assisted laser desorption / ionization time-of-flight mass spectrometry (MALDI-TOF-MS) assay. KMapp values of 24.7 ± 6.7 and 23.2 ± 4.3 μM for 2OG and H31–15K9me3 peptide substrate, respectively, were determined (Figure 1a, b) and found to be consistent with reported KMapp values obtained using other assay methods.38,39 We were then able to determine excess peptide and 2OG cosubstrate concentrations (200 and 100 μM, respectively) for use in the oxygen dependence assay to ensure saturating conditions of substrate and cosubstrate.
Figure 1.
Kinetic analysis of recombinant KDM4A1–359 using a MALDI-TOF-MS-based assay. KMapp values for (a) 2OG and (b) H31–15K9me3 for KDM4A1–359 were initially determined. (c) Steady-state determination of KDM4A1–359 kinetics with respect to oxygen revealed a KMapp(O2) of 173 ± 23 μM. An approximately 5–10 fold excess of KMapp 2OG and H31-15K9me3 peptide was used to ensure saturating conditions of all (co)substrates at 37 °C. GraphPad Prism was used to fit data to a Michaelis-Menten model and evaluate kinetic parameters. A total of three biological repeats using different preparations of KDM4A1–359 with n = 3 technical repeats were performed; error bars denote standard deviation (sd).
We then investigated the oxygen dependence of the reaction of KDM4A1–359 with H31–15K9me3 peptide using a method described previously.15 Briefly, a mass flow controller (MFC, Brooks Instruments) was used to regulate the proportion of O2 and N2 used to purge sealed glass vials in which the reaction was performed. Analysis of the initial rate of reaction at oxygen concentrations between 0 and 80% (0–722 μM Figure S2) yielded a KMapp(O2) of 173 ± 23 μM for KDM4A1–359 (Figure 1c). This value lies between reported KMapp(O2) values of the HIF hydroxylases FIH and PHD2, as determined using the same assay (110 and >450 μM, respectively)15,16 and is significantly higher than reported values for other 2OG oxygenases.12,13,32 This suggests that KDM4A activity may be responsive to changes in a physiologically relevant range of oxygen concentrations, thereby possessing the biochemical potential to act as an oxygen sensor.
Loss of H3K9me3 in Cells Overexpressing KDM4A is Dependent on Oxygen Availability
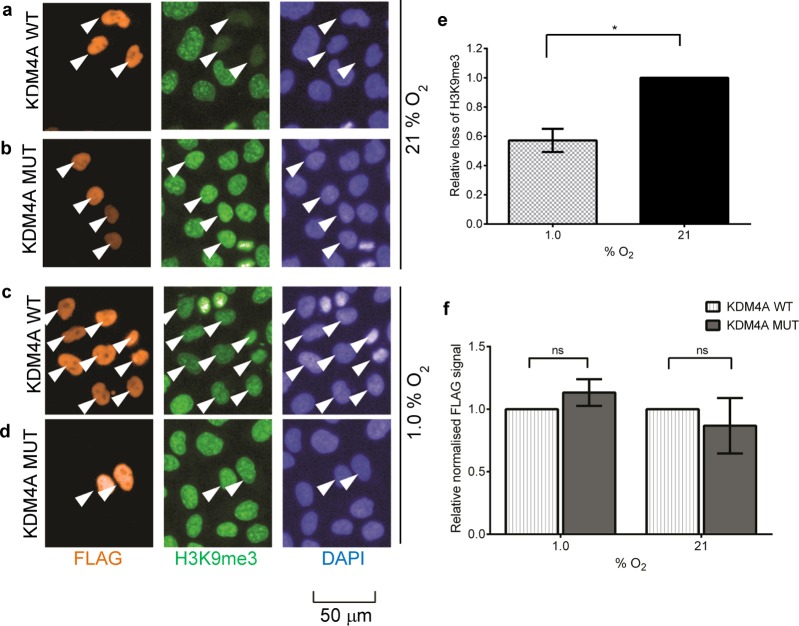
Given that the differences in the in vitro oxygen sensitivity of the HIF hydroxylases15,16 appeared to correlate with their O2 sensitivities in cells,17 we next investigated the effect of hypoxia on the cellular activity of KDM4A to establish whether a similar relationship between the in vitro and cellular oxygen dependence of the enzyme was apparent. Furthermore, some previous biochemical studies have reported KM(O2) values that vary with peptide length;13 hence, it was important to assess the oxygen dependence of the enzyme against its endogenous substrate. HeLa cells were transiently transfected with full-length wild-type (WT) N-terminally FLAG-tagged KDM4A or the inactive variant KDM4A H188A2 (MUT) and incubated in either 1 or 21% O2 for 24 h. Cells were stained for immunofluorescence analysis of FLAG and H3K9me3 levels, with 4′,6-diamidino-2-phenylindole (DAPI) staining verifying that ectopically expressed KDM4A was located in the nucleus (Figure 2a–d). Nuclear FLAG fluorescence signal was used to select a population of highly transfected cells whose H3K9me3 levels were then quantified (Figure 2e). Similar expression of both WT and MUT enzymes was observed (Figure 2f). As expected, MUT-transfected cells displayed no changes in H3K9me3 levels compared to untransfected cells in either normoxia or hypoxia (Figure 2b,d). To quantify loss of H3K9me3 in WT-transfected cells, average H3K9me3 fluorescence signal (RFU) was normalized against that in MUT-transfected cells incubated at the same oxygen concentration (Figure S4). In normoxia, the cells overexpressing WT KDM4A showed a pronounced decrease of 55 ± 11% in H3K9me3 staining relative to H3K9me3 fluorescence in MUT-transfected cells, indicating demethylase activity (Figure 2a, b). In hypoxia, KDM4A WT-transfected cells retained significantly higher levels of H3K9me3 compared with normoxic cells (Figure 2a, c) with a percentage loss of 33 ± 11% relative to the MUT control. This was attributed to an approximately 40% reduction in KDM4A activity at 1% oxygen relative to that at 21% oxygen (P ≤ 0.05, Figure 2e), suggesting that cellular KDM4A catalytic activity is sensitive to reduction in oxygen availability.
Figure 2.
Hypoxia-dependent alteration of H3K9me3 levels in HeLa cells overexpressing FLAG-KDM4A. (a–d) Selected images from immunofluorescence analysis of the effect of hypoxia on the cellular activity of KDM4A. HeLa cells were transiently transfected with FLAG-tagged full length KDM4A WT (a,c) or catalytically inactive KDM4A MUT (b,d) and then incubated for 24 h at 21% (a,b) or 1% O2 (c,d). Images were processed and analyzed at the same time using the same fluorescence intensity signal window. (e) Calculation of relative loss of H3K9me3 in normoxia (21% O2) and hypoxia (1.0% O2) reveals an approximately 40% reduction in hypoxia. Error bars denote sd; N = 3; a paired t test was performed using GraphPad Prism; P ≤ 0.05. For the calculation method, see Figure S4. (f) Expression of transiently expressed FLAG-tagged KDM4A WT and KDM4A MUT is comparable in normoxia and hypoxia. Average FLAG fluorescence signal in KDM4A MUT-overexpressing cells was normalized against that in KDM4A WT-overexpressing cells in the cell populations selected for analysis (n > 50 cells). No significant differences in FLAG signal were apparent. Error bars denote sd; N = 3; two-way ANOVA was performed using GraphPad Prism; P ≤ 0.05.
Cellular KDM4A Activity Demonstrates a Graded Response to Hypoxia
We next assessed the cellular KDM4A demethylase activity across a range of physiologically relevant oxygen concentrations to ascertain its sensitivity to cellular oxygen availability. To facilitate this and to test whether KDM4A activity can also be modulated by hypoxia in a different cell line, a U2OS cell line stably expressing doxycycline-inducible FLAG-tagged WT KDM4A (full length) (herein termed U2OS F-KDM4A cells) was used (Figure S3).
Doxycycline hyclate was added to U2OS F-KDM4A cells, which were subsequently incubated at 0.1, 1, 5 and 21% O2, for 24 h. Cells were then fixed and stained for FLAG and H3K9me3 (Figure 3a–d). At each oxygen concentration, global H3K9me3 levels were normalized to those in uninduced U2OS F-KDM4A cells incubated under the same conditions to account for endogenous responses of the mark to hypoxia. Loss of H3K9me3 fluorescence in induced cells was then used to calculate percentage demethylation of this mark by the overexpressed KDM4A and correlated with KDM4A activity (Figure 3e; for calculations, see Figure S4). The percentage loss of H3K9me3 in KDM4A induced U2OS cells at 1% O2 was relatively consistent with loss of H3K9me3 found in HeLa cells transiently transfected with KDM4A (50 ± 5% compared with ∼40%, respectively), indicating a similar effect of hypoxia on KDM4A activity in different cell lines. Overall, the data revealed a relative decrease in the demethylation of H3K9me3 consistent with a graded loss of KDM4A activity with decreasing oxygen concentration. As expected, HIF-1α stabilization was also observed for cells incubated at reduced oxygen concentrations (0.1–5.0% O2) by Western blot analysis (Figure 3f(i)).
Figure 3.
Cellular demethylase activity of KDM4A demonstrates a graded response to oxygen concentration in U2OS cells. (a–d). Representative images from immunofluorescence analysis of the oxygen sensitivity of cellular KDM4A. Doxycycline (Dox)-inducible U2OS stable cells were induced to overexpress FLAG-KDM4A WT and incubated at (a) 0.1%, (b) 1.0%, (c) 5.0%, or (d) 21% O2 for 24 h. H3K9me3 signal in induced cells (indicated by arrows) was normalized against signal in uninduced cells and correlated to enzyme activity (Figure S4). (e) Cellular KDM4A H3K9me3 demethylase activity in O2 concentrations from 0.1 to 5.0% relative to 21%. Quantification is based on three biological repeats; error bars denote sd. One-way ANOVA with Dunnett’s multiple comparison test against the result for 21% O2 was performed in GraphPad Prism. N > 50 cells were used for each experiment; cells incubated in each oxygen concentration were processed, imaged, and analyzed simultaneously. (f)(i)Western blot analysis of cell lysates from induced and uninduced U2OS F-KDM4A and parental U2OS cells (P) reveals stabilization of HIF-1α in cells incubated at 0.1–5.0% O2 compared to cells incubated at 21% O2. β-actin was used as a loading control. (ii) Multiplexed fluorescent Western blot (red, anti-KDM4A; green, anti- FLAG and anti-actin) and (g) quantitative graph showing levels of FLAG-KDM4A protein in U2OS F-KDM4A cells treated ± dox and incubated at 0.1–21% O2 for 24 h. The membrane was stained simultaneously for KDM4A, FLAG, and β-actin to enable direct quantification and comparison. P denotes parental U2OS cells, which were used to control for dox-dependent effects. All signals were normalized to the β-actin loading control. Levels of overexpressed FLAG-KDM4A were quantified relative to those in normoxic cells treated with dox. Quantitation is based on three biological repeats; error bars denote sd and two-way ANOVA with Dunnett’s multiple comparison test was performed using GraphPad Prism. (h) Hypoxia does not affect expression levels of KDM4A mRNA. Relative mRNA levels of KDM4A U2OS F-KDM4A cells ± dox incubated for 24 h in 0.1% −21% oxygen, and U2OS parental cells (P) incubated at 1.0 and 21% O2, were analyzed by RT-qPCR. A two-way ANOVA with Dunnett’s multiple comparison was performed using GraphPad Prism; error bars denote sd; N = 2; control= −dox; and 21% O2.
Interestingly, we also observed an increase in KDM4A at the protein level with decreasing oxygen concentration in U2OS F-KDM4A cells (Figure 3f(ii),g) independent of changes at the transcript level (Figure 3h), suggesting that our analysis from immunofluorescence experiments may be underestimating the effect of hypoxia on loss of KDM4A activity because of increased FLAG-KDM4A expression. This increase in KDM4A protein is consistent with a recent study by Black et al, who observed a reduction of KDM4A activity against H3K9me3 in RPE cells exposed to 1% O2 for 24 h.40 However, although chemical inhibition of JmjC-KDM activity abrogated KDM4A-mediated transient site-specific copy gain in hypoxia, hypoxic loss of activity did not have the same effect,40 suggesting that the downstream physiological effects of hypoxia on KDM4A function are somewhat subtle. Previous studies have revealed that stabilization of KDM4A in hypoxia is due to reduced interaction of the enzyme with the SKP1-Cul1-F-box ubiquitin ligase responsible for regulation of KDM4A turnover by the proteasome.41 Given the reduced catalytic activity of KDM4A in hypoxia, this stabilization may be a compensatory mechanism, as has been suggested regarding the HIF-induced upregulation of KDM4 isoforms KDM4B and KDM4C.21,22 Indeed, hypoxic stabilization of KDM4A has been shown to increase its association with chromatin,41 thereby potentially ameliorating the effects of hypoxia on its catalytic activity and rationalizing the different effects observed upon chemical and hypoxic inhibition of KDM4A.40 Although the biological implications of the observed oxygen sensitivity are yet to be elucidated, our experiments in U2OS F-KDM4A cells suggest that both KDM4A activity and protein levels can be “tuned” in varying degrees of hypoxia, which may be further indication of an oxygen sensing role for KDM4A.
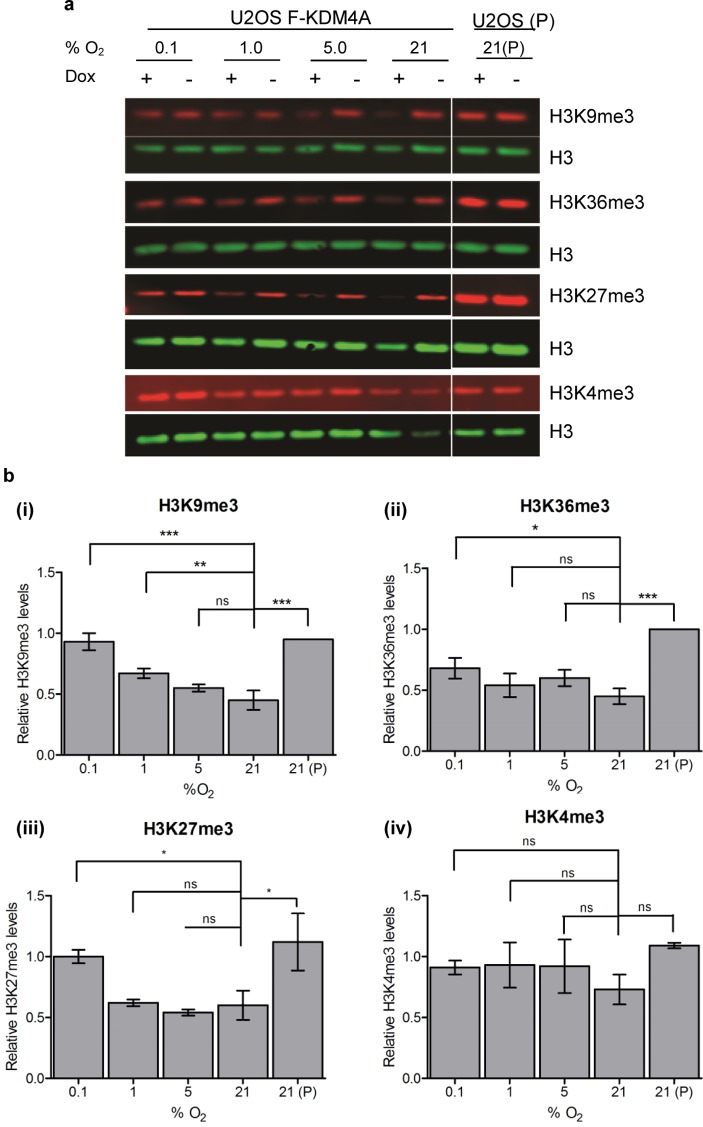
Quantitative Western blots were performed to assess the effect of hypoxia on KDM4A activity against multiple methylated lysine marks (H3K9me3, H3K36me3, H3K27me3, and H3K4me3) in total populations of both induced and uninduced U2OS F-KDM4A cells as well as a control parental U2OS cell line (U2OS (P), Figure S3). Multiplexed fluorescent analysis allowed levels of these methylation marks to be normalized against a simultaneously quantified H3 loading control for each sample (Figure 4a). Normalized values from induced samples were then compared to those in uninduced samples incubated at the same concentration of O2, and the relative loss of methylation due to overexpression of KDM4A in each condition was calculated. The Western blots confirmed that induction of KDM4A overexpression led to a significant decrease in H3K9me3 levels in normoxia (Figure 4b(i)). Moderate and graded reduction in H3K9me3 levels and, to a lesser extent, H3K36me3 levels was observed in KDM4A overexpressing cells in hypoxia (Figure 4b(ii)), displaying a similar trend to that seen in immunofluorescence experiments. Although levels of the non-KDM4A-substrate H3K4me3 mark demonstrated no significant changes on overexpression of KDM4A (Figure 4b(iv)), interestingly, analysis of the H3K27me3 mark showed loss of this mark upon overexpression of KDM4A in normoxia, with a graded response to oxygen (Figure 4b(iii)) similar to that seen for H3K9me3, although less-pronounced. H3K27me3 is not a canonical substrate for KDM4A, hence, loss of this mark cannot be directly attributed to catalytic KDM4A activity. However, KDM4A has been shown to be capable of catalyzing demethylation of a H3K27me3 peptide substrate in vitro.42 It is also possible that global demethylation of H3K9me3 and H3K36me3 affects the H3K27me3 mark through alternative pathways. There is increasing evidence to suggest that the function, selectivity, and activity of individual JmjC-KDMs is affected by the presence or absence of post-translational modifications distal to the mark upon which they are acting via disruption of necessary interactions with binding and reader domains;43,44 hence, aberrant demethylation of one lysine methylation mark may have an impact on others.
Figure 4.
Alteration of multiple H3 methylation marks by KDM4A at different oxygen concentrations. (a) Fluorescent Western blots showing levels of H3K9me3, H3K36me3, and H3K27me3 histone methylation marks in induced or uninduced U2OS F-KDM4A cells incubated at 0.1–21% O2. P denotes parental U2OS cells. The membrane was stained simultaneously for H3K9me3, H3K36me3, H3K27me3, H3K4me3, and H3, with methylation marks being normalized against the signal for H3. Representative Western blots are shown and have been cropped to show only relevant molecular weights. Western blots were imaged using Odyssey CLx fluorescent imaging system (LI-COR) and analyzed using Image Studio software (LI-COR). (b) Graphs showing quantification of (i) H3K9me3, (ii) H3K36me3, (iii) H3K27me3, and (iv) H3K4me3 levels from Western blots. Levels of trimethylated lysine marks were normalized to H3 levels and then quantitated relative to those in uninduced cells incubated at the same O2 concentration to discount any non-KDM4A mediated effect in methylation changes. Error bars denote sd and one-way ANOVA with Dunnett’s multiple comparison test was performed using GraphPad Prism; N = 3.
Aberrant histone lysine methylation and altered JmjC-KDM expression levels have been observed in hypoxic disease states such as cancer35 and cardiovascular diseases such as cardiomyopathy and cardiac hypertrophy.32,36,45 The potential involvement of the JmjC-KDMs in these hypoxic diseases highlights the importance of understanding the impact of hypoxia on their activity. Here, we demonstrate a link between the biochemical oxygen sensitivity of isolated KDM4A and the effect of cellular hypoxia on its catalytic activity, with a loss of KDM4A activity in hypoxia resulting in changes to global histone lysine methylation. A comparative study with which to assess the oxygen sensitivity of multiple demethylases using the methods described herein would further develop our understanding of the potential for differential oxygen sensitivity across the JmjC-KDMs, as has previously been observed both in cells21,46 and in vitro, particularly for the KDM4 subfamily.19 Overall, however, these results suggest that KDM4A possesses the potential to act as an oxygen sensor in the context of epigenetic regulation. While these observations are yet to be validated in an endogenous setting, importantly, they present a possible complementary mechanism to the HIF pathway for hypoxic regulation of gene expression.
Methods
Recombinant KDM4A Assays
N-terminally His6-tagged truncated KDM4A1–359 was expressed in Escherichia coli BL21(DE3) cells and purified via nickel affinity followed by size-exclusion chromatography, as reported previously.37 H31–15K9me3 peptide was synthesized using a Liberty Blue automated microwave peptide synthesizer (CEM) and purified via reverse-phase high-performance liquid chromatography (HPLC). Activity assays were performed using the following conditions: 1 μM KDM4A1–359, 200 μM H31–15K9me3, 200 μM 2OG, 100 μM l-ascorbate, and 10 μM Fe2+ in 50 mM HEPES (pH 7.5). Experiments to determine KM values of H31–15K9me3, 2OG, and O2 used these conditions but varied the concentration of the (co)substrate under investigation. For oxygen-dependence assays, the activity of KDM4A1–359 was assessed at varied oxygen concentrations. Peptide and buffer (94 μL) were equilibrated in septa-sealed glass vials for 5 min at 37 °C using a mass flow controller (Brooks Instruments) to control the ratios of O2 to N2. Sequential addition of 2OG (1 μL), l-ascorbate (1 μL), and Fe2+ (2 μL) was followed by initiation of the enzymatic reaction by addition of KDM4A1–359 (1 μL) using gastight Hamilton syringes and incubation at 37 °C for 5 min. The reaction was quenched with MeOH in a 1:1 ratio and 1 μL removed for analysis via MALDI-TOF-MS.
Stable Transfectants
The human KDM4A gene was cloned into a modified pUHD10-3 MCS vector to enable expression of an N-terminally FLAG-tagged KDM4A protein. Stable transfectants were then generated from U2OS cells bearing the reverse tetracycline responsive transactivator47 and a tetKRAB silencer construct48 by transfection with the pUHD10 FLAG vector alone or pUHD10-FLAG-KDM4A in combination with pSV2-hph (ratio 10:1) using FuGENE 6 (Roche Applied Science). A total of 24 h after transfection, cells were selected with hygromycin B (150 μgml–1). Independent clones were picked, maintained in selection media (Dulbecco’s modified Eagle’s medium supplemented with 10% Tet system-approved fetal bovine serum (FBS, Clontech), penicillin G (50 IU mL–1), streptomycin (50 μg mL–1) (Invitrogen), l-glutamine (2 mM) (Sigma), blasticidin S (5 μg mL–1) (Invitrogen), and hygromycin B (150 μg mL–1; Roche Applied Science)) and tested for doxycycline-inducible gene expression (Figure S2). Clone 1 was verified and used for experiments (Figure S3).
HeLa Cells
HeLa cells were maintained in DMEM media supplemented with 10% FBS and 5% l-glutamine and incubated at 37 °C, 5% CO2, and the required O2 concentration. Cells were transfected with pCDNA3 encoding full length, N-terminally FLAG-tagged KDM4A or KDM4A H188A variant using FuGENE HD transfection reagent (Promega) according to the manufacturer’s protocol. For immunofluorescence experiments, after 4 h, the media on the cells was changed and the cells transferred to the relevant experimental conditions.
U2OS FLAG-KDM4A Cells
U2OS FLAG-KDM4A cells were maintained in selection media throughout experiments and incubated at 37 °C, 5% CO2 and the required O2 concentration. Cells were dosed with doxycycline (1 μg mL–1) or left untreated and then transferred immediately to relevant experimental conditions.
Hypoxic Cellular Experiments
Hypoxic incubations of cells were performed in an Invivo2 400 hypoxic workstation (Ruskinn Technologies). All O2 gradient assays were performed simultaneously in hypoxic chambers set at 0.1, 1.0 and 5.0% O2, with normoxic cells maintained at 21% O2. For immunofluorescence experiments and Western blots of histone marks, cells were removed from the hypoxic stations prior to fixing or scraping, respectively. For HIF Western blots, cells were scraped in the hypoxic workstations.
Immunofluorescence
A protocol adapted from King et al, 201049 was used for immunofluorescence assays. Briefly, after 24 h of incubation at the required oxygen concentration, cells were fixed, permeabilized and stained for FLAG and H3K9me3 (see Table S1 for a list of antibodies) and DAPI (Sigma D9564) to stain DNA. Goat anti-rabbit secondary antibody conjugated to Alexa Fluor 488 (Life Technologies A11034) and goat anti-mouse secondary antibody conjugated to Alexa Fluor 594 (Life Technologies A11032) were used to fluorescently label the FLAG and H3K9me3 primary antibodies. All fixing and staining procedures were performed simultaneously for each biological experiment. Image acquisition was performed using an Operetta high content imaging system (PerkinElmer), and images were analyzed using Harmony high content imaging and analysis software (PerkinElmer). Cell nuclei were automatically identified by DAPI staining using the default parameters. Cells overexpressing high amounts of KDM4A, where FLAG-tag staining was bright, were selected for analysis of H3K9me3 levels. In HeLa cells, H3K9me3 levels were normalized against those in cells transfected with the catalytically inactive KDM4A variant, which was deemed to possess 0% activity, while in U2OS FLAG-KDM4A cells, uninduced cells were used for normalization.
Western Blots
Following hypoxic or normoxic incubation, cells were scraped on ice in SDS–urea buffer (6.7 M urea, 10 mM Tris–Cl (pH 6.8), 10% v/v glycerol, and 1% w/v SDS supplemented with 1 mM dithiothreitol and Complete Protease Inhibitor mixture (Roche Applied Science)). Lysates were then sonicated on ice for 10 s three times with a 10 s pause between cycles and snap-frozen at −80 °C. Total protein concentration was determined using a Pierce BCA assay kit (Thermo Scientific). Protein (1 μg total per well) was loaded on a NuPage 4–12% Bis–Tris gel (Life Technologies) and then transferred to a 0.45 μm nitrocellulose membrane using wet transfer at 30 V for 1 h. Membranes were blocked in SEA BLOCK blocking buffer (Thermo Scientific) overnight then probed with primary and fluorescent secondary antibodies (see Table S1) diluted in 3% SEABLOCK, PBS, and 0.1% Tween-20. Blots were imaged on an Odyssey CLx Fluorescent Imaging System (LI-COR) and analyzed using Image Studio software (LI-COR).
qPCR
RNA was isolated and purified from cells using the mirVana miRNA isolation kit (Life Technologies) according to the manufacturer’s protocol. Isolated RNA was then treated with DNase using a Turbo DNA-free kit (Invitrogen) to remove DNA before reverse transcription using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). A total of 1.5625 ng of cDNA was submitted to qPCR analysis using 300 nM primers (Sigma; see Table S2 for primer sequences) and Power SYBR Green Master Mix (Applied Biosystems) according to the manufacturer’s protocol in a CFX1000 Thermal Cycler with a CFX96 Real-Time System (Bio-Rad). Data was collected and processed using CFX Manager Software v3.1 (Bio-Rad) and analyzed using the ΔCt method.
Acknowledgments
The authors thank C. J. Schofield for valuable discussions, P. J. Ratcliffe for helpful suggestions on experimental design and Mr Ben Greenfield for establishing and calibrating the MFC assay. R.L.H., K.D., and A.K. acknowledge support from the BHF Centre of Research Excellence, Oxford (RE/08/004/23915). A.K. and E.F. are supported by Royal Society Dorothy Hodgkin Fellowships. This work was supported by the Engineering and Physical Sciences Research Council and Cancer Research UK (C8717/A18245).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.6b00958.
Figures showing a consensus mechanism, in vitro O2 calibration, validation of the U2OS F-KDM4A cell line, and calculation of KDM4A activity. Tables showing antibodies used in immunofluorescence and Western blot experiments and sequences of primers. (PDF)
Author Contributions
R.L.H. optimized and performed all in vitro and cellular experiments, analyzed the data, and wrote the manuscript. N.M. produced and validated stable U2OS cells, discussed the results, and commented on the manuscript. K.D. aided with optimization of the immunofluorescence experiments. E.F. and A.K. designed the study, discussed the results, and commented on the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705. 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Klose R. J.; Yamane K.; Bae Y.; Zhang D.; Erdjument-Bromage H.; Tempst P.; Wong J.; Zhang Y. (2006) The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442, 312–316. 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Trojer P.; Zhang J.; Yonezawa M.; Schmidt A.; Zheng H.; Jenuwein T.; Reinberg D. (2009) Dynamic Histone H1 Isotype 4 Methylation and Demethylation by Histone Lysine Methyltransferase G9a/KMT1C and the Jumonji Domain-containing JMJD2/KDM4 Proteins. J. Biol. Chem. 284, 8395–8405. 10.1074/jbc.M807818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton I. J.; McDonough M. A.; Ehrismann D.; Kershaw N. J.; Granatino N.; Schofield C. J. (2006) Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J. Inorg. Biochem. 100, 644–669. 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Hausinger R. P. (2008) Fe(II)/α-Ketoglutarate-Dependent Hydroxylases and Related Enzymes. Crit. Rev. Biochem. Mol. Biol. 39 (1), 21–68. 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- Semenza G. L. (1998) Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr. Opin. Genet. Dev. 8, 588–594. 10.1016/S0959-437X(98)80016-6. [DOI] [PubMed] [Google Scholar]
- Schofield C. J.; Ratcliffe P. J. (2004) Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 5, 343–354. 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- Myllyharju J. (2013) Prolyl 4-hydroxylases, master regulators of the hypoxia response. Acta Physiol. 208, 148–165. 10.1111/apha.12096. [DOI] [PubMed] [Google Scholar]
- Schödel J.; Mole D. R.; Ratcliffe P. J. (2013) Pan-genomic binding of hypoxia-inducible transcription factors. Biol. Chem. 394, 507–517. 10.1515/hsz-2012-0351. [DOI] [PubMed] [Google Scholar]
- Semenza G. L. (2014) Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu. Rev. Pathol.: Mech. Dis. 9, 47–71. 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- Martinez S.; Hausinger R. P. (2015) Catalytic Mechanisms of Fe(II)- and 2-Oxoglutarate-dependent Oxygenases. J. Biol. Chem. 290, 20702–20711. 10.1074/jbc.R115.648691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsilä M.; Koivunen P.; Günzler V.; Kivirikko K. I.; Myllyharju J. (2003) Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 278, 30772–30780. 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- Ehrismann D.; Flashman E.; Genn D. N.; Mathioudakis N.; Hewitson K. S.; Ratcliffe P. J.; Schofield C. J. (2007) Studies on the activity of the hypoxia-inducible-factor hydroxylases using an oxygen consumption assay. Biochem. J. 401, 227–234. 10.1042/BJ20061151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao J. H.; Kurzeja R. J. M.; Morachis J. M.; Veith H.; Lewis J.; Yu V.; Tegley C. M.; Tagari P. (2009) Kinetic characterization and identification of a novel inhibitor of hypoxia-inducible factor prolyl hydroxylase 2 using a time-resolved fluorescence resonance energy transfer-based assay technology. Anal. Biochem. 384, 213–223. 10.1016/j.ab.2008.09.052. [DOI] [PubMed] [Google Scholar]
- Tarhonskaya H.; Chowdhury R.; Leung I. K. H.; Loik N. D.; McCullagh J. S. O.; Claridge T. D. W.; Schofield C. J.; Flashman E. (2014) Investigating the contribution of the active site environment to the slow reaction of hypoxia-inducible factor prolyl hydroxylase domain 2 with oxygen. Biochem. J. 463, 363–372. 10.1042/BJ20140779. [DOI] [PubMed] [Google Scholar]
- Tarhonskaya H.; Hardy A. P.; Howe E. A.; Loik N. D.; Kramer H. B.; McCullagh J. S. O.; Schofield C. J.; Flashman E. (2015) Kinetic Investigations of the Role of Factor Inhibiting Hypoxia-inducible Factor (FIH) as an Oxygen Sensor. J. Biol. Chem. 290, 19726–19742. 10.1074/jbc.M115.653014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y.-M.; Yeoh K. K.; Lee M. K.; Eriksson T.; Kessler B. M.; Kramer H. B.; Edelmann M. J.; Willam C.; Pugh C. W.; Schofield C. J.; Ratcliffe P. J. (2011) Differential sensitivity of hypoxia inducible factor hydroxylation sites to hypoxia and hydroxylase inhibitors. J. Biol. Chem. 286, 13041–13051. 10.1074/jbc.M110.211110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Fernandez E. M.; Tarhonskaya H.; Al-Qahtani K.; Hopkinson R. J.; McCullagh J. S.; Schofield C. J.; Flashman E. (2013) Investigations on the oxygen dependence of a 2-oxoglutarate histone demethylase. Biochem. J. 449, 491–496. 10.1042/BJ20121155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella B.; Mirica L. M. (2012) Kinetic analysis of iron-dependent histone demethylases: α-ketoglutarate substrate inhibition and potential relevance to the regulation of histone demethylation in cancer cells. Biochemistry 51, 8699–8701. 10.1021/bi3012466. [DOI] [PubMed] [Google Scholar]
- Lee H.-Y.; Choi K.; Oh H.; Park Y.-K.; Park H. (2014) HIF-1-dependent induction of Jumonji domain-containing protein (JMJD) 3 under hypoxic conditions. Mol. Cells 37, 43–50. 10.14348/molcells.2014.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer S.; Kristensen M. M.; Jensen K. S.; Johansen J. V.; Staller P. (2008) The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 283, 36542–36552. 10.1074/jbc.M804578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard P. J.; Loenarz C.; Mole D. R.; McDonough M. A.; Gleadle J. M.; Schofield C. J.; Ratcliffe P. J. (2008) Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha. Biochem. J. 416, 387–394. 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- Xia X.; Lemieux M. E.; Li W.; Carroll J. S.; Brown M.; Liu X. S.; Kung A. L. (2009) Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc. Natl. Acad. Sci. U. S. A. 106, 4260–4265. 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmann S.; Bettkober M.; Zelmer A.; Seeger K.; Faigle M.; Eltzschig H. K.; Bührer C. (2008) Hypoxia upregulates the histone demethylase JMJD1A via HIF-1. Biochem. Biophys. Res. Commun. 372, 892–897. 10.1016/j.bbrc.2008.05.150. [DOI] [PubMed] [Google Scholar]
- Niu X.; Zhang T.; Liao L.; Zhou L.; Lindner D. J.; Zhou M.; Rini B.; Yan Q.; Yang H. (2012) The von Hippel-Lindau tumor suppressor protein regulates gene expression and tumor growth through histone demethylase JARID1C. Oncogene 31, 776–786. 10.1038/onc.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Ledaki I.; Turley H.; Gatter K. C.; Montero J.-C. M.; Li J.-L.; Harris A. L. (2009) Role of hypoxia-inducible factors in epigenetic regulation via histone demethylases. Ann. N. Y. Acad. Sci. 1177, 185–197. 10.1111/j.1749-6632.2009.05027.x. [DOI] [PubMed] [Google Scholar]
- Krieg A. J.; Rankin E. B.; Chan D.; Razorenova O.; Fernandez S.; Giaccia A. J. (2010) Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol. Cell. Biol. 30, 344–353. 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W.; Chang R.; Zhong J.; Pandey A.; Semenza G. L. (2012) Histone demethylase JMJD2C is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proc. Natl. Acad. Sci. U. S. A. 109, E3367–3376. 10.1073/pnas.1217394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tausendschön M.; Dehne N.; Brüne B. (2011) Hypoxia causes epigenetic gene regulation in macrophages by attenuating Jumonji histone demethylase activity. Cytokine+ 53, 256–262. 10.1016/j.cyto.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Johnson A. B.; Denko N.; Barton M. C. (2008) Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutat. Res., Fundam. Mol. Mech. Mutagen. 640, 174–179. 10.1016/j.mrfmmm.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olcina M. M.; Foskolou I. P.; Anbalagan S.; Senra J. M.; Pires I. M.; Jiang Y.; Ryan A. J.; Hammond E. M. (2013) Replication stress and chromatin context link ATM activation to a role in DNA replication. Mol. Cell 52, 758–766. 10.1016/j.molcel.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. L.; Dunne K.; Walport L. J.; Flashman E.; Kawamura A. (2015) Epigenetic regulation by histone demethylases in hypoxia. Epigenomics 7, 791–811. 10.2217/epi.15.24. [DOI] [PubMed] [Google Scholar]
- Kirmes I.; Szczurek A.; Prakash K.; Charapitsa I.; Heiser C.; Musheev M.; Schock F.; Fornalczyk K.; Ma D.; Birk U.; Cremer C.; Reid G. (2015) A transient ischemic environment induces reversible compaction of chromatin. Genome Biol. 16, 246–265. 10.1186/s13059-015-0802-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melvin A.; Rocha S. (2012) Chromatin as an oxygen sensor and active player in the hypoxia response. Cell. Signalling 24, 35–43. 10.1016/j.cellsig.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Højfeldt J. W.; Agger K.; Helin K. (2013) Histone lysine demethylases as targets for anticancer therapy. Nat. Rev. Drug Discovery 12, 917–30. 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- Zhang Q. J.; Chen H. Z.; Wang L.; Liu D. P.; Hill J. A.; Liu Z. P. (2011) The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice. J. Clin. Invest. 121, 2447–2456. 10.1172/JCI46277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. S.; Kavanagh K. L.; McDonough M. A.; Butler D.; Pilka E. S.; Lienard B. M.; Bray J. E.; Savitsky P.; Gileadi O.; von Delft F.; Rose N. R.; Offer J.; Scheinost J. C.; Borowski T.; Sundstrom M.; Schofield C. J.; Oppermann U. (2007) Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 448, 87–91. 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- Hillringhaus L.; Yue W. W.; Rose N. R.; Ng S. S.; Gileadi C.; Loenarz C.; Bello S. H.; Bray J. E.; Schofield C. J.; Oppermann U. (2011) Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J. Biol. Chem. 286, 41616–41625. 10.1074/jbc.M111.283689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R.; Yeoh K. K.; Tian Y.-M.; Hillringhaus L.; Bagg E. A.; Rose N. R.; Leung I. K. H.; Li X. S.; Woon E. C. Y.; Yang M.; McDonough M. A.; King O. N.; Clifton I. J.; Klose R. J.; Claridge T. D. W.; Ratcliffe P. J.; Schofield C. J.; Kawamura A. (2011) The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 12, 463–469. 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J. C.; Atabakhsh E.; Kim J.; Biette K. M.; Van Rechem C.; Ladd B.; Burrowes P. D.; Donado C.; Mattoo H.; Kleinstiver B. P.; Song B.; Andriani G.; Joung J. K.; Iliopoulos O.; Montagna C.; Pillai S.; Getz G.; Whetstine J. R. (2015) Hypoxia drives transient site-specific copy gain and drug-resistant gene expression. Genes Dev. 29, 1018–1031. 10.1101/gad.259796.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rechem C.; Black J. C.; Abbas T.; Allen A.; Rinehart C. A.; Yuan G.-C.; Dutta A.; Whetstine J. R. (2011) The SKP1-Cul1-F-box and leucine-rich repeat protein 4 (SCF-FbxL4) ubiquitin ligase regulates lysine demethylase 4A (KDM4A)/Jumonji domain-containing 2A (JMJD2A) protein. J. Biol. Chem. 286, 30462–30470. 10.1074/jbc.M111.273508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. T.; Walport L. J.; Hopkinson R. J.; Madden S. K.; Chowdhury R.; Schofield C. J.; Kawamura A. (2014) Studies on the catalytic domains of multiple JmjC oxygenases using peptide substrates. Epigenetics 9, 1596–1603. 10.4161/15592294.2014.983381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse B.; Helgstrand C.; Kristensen J. B. L.; Leurs U.; Cloos P. A. C.; Kristensen J. L.; Clausen R. P. (2013) Posttranslational modifications of the histone 3 tail and their impact on the activity of histone lysine demethylases in vitro. PLoS One 8, e67653. 10.1371/journal.pone.0067653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack L. R.; Yamamoto K. R.; Fujimori D. G. (2016) Opposing Chromatin Signals Direct and Regulate the Activity of Lysine Demethylase 4C (KDM4C). J. Biol. Chem. 291, 6060–6070. 10.1074/jbc.M115.696864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M.; Wagner M.; Reil J.-C.; Müller S.-A.; Tauchnitz M.; Zimmer A. M.; Lehmann L. H.; Thiel G.; Böhm M.; Backs J.; Maack C. (2013) HDAC4 controls histone methylation in response to elevated cardiac load. J. Clin. Invest. 123, 1359–1370. 10.1172/JCI61084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. Y.; Yang E. G.; Park H. (2013) Hypoxia enhances the expression of prostate-specific antigen by modifying the quantity and catalytic activity of Jumonji C domain-containing histone demethylases. Carcinogenesis 34, 2706–2715. 10.1093/carcin/bgt256. [DOI] [PubMed] [Google Scholar]
- Gossen M.; Freundlieb S.; Bender G.; Müller G.; Hillen W.; Bujard H. (1995) Transcriptional activation by tetracyclines in mammalian cells. Science 268, 1766–1769. 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Deuschle U.; Meyer W. K.; Thiesen H. J. (1995) Tetracycline-reversible silencing of eukaryotic promoters. Mol. Cell. Biol. 15, 1907–1914. 10.1128/MCB.15.4.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King O. N. F.; Li X. S.; Sakurai M.; Kawamura A.; Rose N. R.; Ng S. S.; Quinn A. M.; Rai G.; Mott B. T.; Beswick P.; Klose R. J.; Oppermann U.; Jadhav A.; Heightman T. D.; Maloney D. J.; Schofield C. J.; Simeonov A. (2010) Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors. PLoS One 5, e15535. 10.1371/journal.pone.0015535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.