Abstract
Background
Signal peptide peptidases play an important role in the removal of remnant signal peptides in the cell membrane, a critical step for extracellular protein production. Although these proteins are likely a central component for extracellular protein production, there has been a lack of research on whether protein secretion could be enhanced via overexpression of signal peptide peptidases.
Results
In this study, both nattokinase and α-amylase were employed as prototypical secreted target proteins to evaluate the function of putative signal peptide peptidases (SppA and TepA) in Bacillus licheniformis. We observed dramatic decreases in the concentrations of both target proteins (45 and 49%, respectively) in a sppA deficient strain, while the extracellular protein yields of nattokinase and α-amylase were increased by 30 and 67% respectively in a strain overexpressing SppA. In addition, biomass, specific enzyme activities and the relative gene transcriptional levels were also enhanced due to the overexpression of sppA, while altering the expression levels of tepA had no effect on the concentrations of the secreted target proteins.
Conclusions
Our results confirm that SppA, but not TepA, plays an important functional role for protein secretion in B. licheniformis. Our results indicate that the sppA overexpression strain, B. licheniformis BL10GS, could be used as a promising host strain for the industrial production of heterologous secreted proteins.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-017-0688-7) contains supplementary material, which is available to authorized users.
Keywords: Bacillus licheniformis, Signal peptide peptidase, Protein secretion, sppA, tepA
Background
Heterologous expression is an effective strategy to improve the production of secreted proteins. Many industrial enzymes (proteases, α-amylase, etc.) have been produced by heterologous expression in bacteria and fungi [e.g. Escherichia coli, Bacillus species, and Saccharomyces] [1–3]. Compared to other expression systems, Bacillus species are regarded as promising host strains with numerous advantages including: non-toxicity, convenience for gene modification and high yields of target proteins [4–6]. In particular, Bacillus licheniformis has been shown to be an effective host strain for protein production in previous studies [7, 8].
The genetic engineering of host strain to improve production is regarded as an efficient tactic due to its universality and efficiency [9, 10]. A number of examples of this effectiveness are available. For example, B. licheniformis BL10 was constructed as a highly efficient host strain for protein production by knocking out ten genes coding for extracellular proteases (Mpr, Vpr, AprX, Epr, Bpr, WprA, AprE, BprA), flagellin and amylase in B. licheniformis WX-02. Using this engineered B. licheniformis BL10, a previous study has demonstrated that nattokinase activity could be increased by 39% [7]. Also, the translocation elements (SecA, SecDF etc.), post-translocation chaperone PrsA and signal peptidase I have been engineered for production of recombinant protective antigen, α-amylase, subtilisin and Dsrs [9, 11–14].
In general, secreted proteins are extracellularly transported through three steps: protein synthesis, translocation, and release. Signal peptides play an important role in protein secretion, these peptide sequences are cleaved from the mature protein by signal peptidase I during the late stage of the secretion process. The cleaved remnant signal peptides are left in the cell membrane and could affect protein transportation [15–17]. Signal peptide peptidase (SPP) is a membrane-bound enzyme that uses a serine/lysine catalytic dyad mechanism to cleave the remnant signal peptides in the cellular membrane and aids in protein secretion [18, 19]. TepA and SppA have been previously identified as putative signal peptide peptidases in B. subtilis, and the concentrations of secreted target proteins decreased dramatically for a sppA-null strain, which suggested that SppA was involved in protein secretion in B. subtilis [18]. Despite the central role for SppA in protein secretion, prior to our study, there have been no studies focused on the improvement of protein secretion via overexpression of SPP.
In this study, both nattokinase and α-amylase were selected as prototype target proteins for secretion to evaluate the function of SppA and TepA in B. licheniformis BL10, derived from the native strain B. licheniformis WX-02 [7]. Our results identified and confirmed that SppA is the main SPP for protein secretion, and the concentrations of total extracellular proteins and target proteins increased significantly for a strain overexpressing sppA. As a result of these studies, the host strain B. licheniformis BL10GS was successfully constructed as a highly efficient platform for protein secretion.
Methods
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study were listed in Table 1. B. licheniformis BL10 was employed as the original strain for gene modification, and E. coli DH5α was served as the host strain for plasmid construction. The expression vector pHY300PLK was used for constructing protein expression vectors, and T2(2)-Ori was applied for constructing the gene knockout vectors and integrating expression vectors in this study.
Table 1.
The strains and plasmids used in this study
| Strains | Relevant properties | Source of reference |
|---|---|---|
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169 (f 80 lacZΔM15) hsd R17 recA1 gyrA96 thi1 relA1 | [7] |
| Bacillus licheniformis | ||
| WX-02 | Polyglutamate productive strain (CCTCC M208065) | CCTCC |
| BL10 | WX-02(Δmpr, Δvpr, ΔaprX, Δepr, Δbpr, ΔwprA, ΔaprE, ΔbprA, Δhag, ΔamyL) | [7] |
| BL10/pHY-amyL | BL10 harboring pHY-amyL | This study |
| BL10/pP43SacCNK | BL10 harboring pP43SacCNK(CCTCC M2014253) | CCTCC |
| BL10T | BL10(ΔtepA) | This study |
| BL10S | BL10(ΔsppA) | This study |
| BL10T/pHY-amyL | BL10T harboring pHY-amyL | This study |
| BL10S/pHY-amyL | BL10S harboring pHY-amyL | This study |
| BL10T/pP43SacCNK | BL10T harboring pP43SacCNK | This study |
| BL10S/pP43SacCNK | BL10S harboring pP43SacCNK | This study |
| BL10GT | Overexpression of tepA in BL10 | This study |
| BL10GS | Overexpression of sppA in BL10 | This study |
| BL10GT/pHY-amyL | BL10GT harboring pHY-amyL | This study |
| BL10GS/pHY-amyL | BL10GS harboring pHY-amyL | This study |
| BL10GT/pP43SacCNK | BL10GT pP43SacCNK | This study |
| BL10GS/pP43SacCNK | BL10GS pP43SacCNK | This study |
| Plasmids | This study | |
| T2(2)-ori | Bacillus knockout vector; Kanr | [7] |
| T2-sppA | T2(ori)-sppA(A + B); to knock out sppA | This study |
| T2-tepA | T2(ori)-tepA(A + B); to knock out tepA | This study |
| T2-GsppA | T2(ori)-sppA(A + B+sppA); to over-express sppA | This study |
| T2-GtepA | T2(ori)-tepA(A + B+tepA); to over-express tepA | This study |
| pHY300PLK | E. coli and B. s shuttle vector; Ampr, Tetr | [7] |
| pP43SacCNK | PHY300PLK + Promotor-P43 (B. subtilis 168) + signal peptide of SacC (B. subtilis 168) + aprN(B. subtilis MBS 04-6) + Terminator of amyL (B. licheniformis WX-02) | [7] |
| pHY-amyL | PHY300PLK + Promotor-P43 (B. subtilis 168) + amyL (containing its own signal peptide and terminator, B. licheniformis WX-02) | This study |
Media and culture conditions
The LB medium was served as the basic medium for bacterial growth, and the corresponding titer of antibiotic (50 μg/mL ampicillin, 25 μg/mL tetracycline or 20 μg/mL kanamycin) was added into the medium if necessary. The ME medium for cell growth contains 20 g/L glucose, 10 g/L sodium citrate, 7 g/L NH4Cl, 0.5 g/L K2HPO4·3H2O, 0.5 g/L MgSO4·7H2O, 0.04 g/L FeCl3·6H2O, 0.104 g/L MnSO4·H2O, 0.15 g/L CaCl2·2H2O. The inoculum (3%) was added into 30 mL of ME media in a 250 mL flask, and was incubated at 180 rpm and 37 °C for 24 h. All the fermentation experiments were repeated at least three times.
The medium for α-amylase production was supplied as follow: 5 g/L corn starch, 5 g/L yeast extract, 10 g/L peptone, 12 g/L sodium citrate, 1 g/L K2HPO4·3H2O, 0.5 g/L MgSO4·7H2O, 0.15 g/L CaCl2·2H2O, pH 7.2; The medium for nattokinase production containing 20 g/L glucose, 10 g/L soy peptone, 10 g/L peptone, 10 g/L corn starch, 15 g/L yeast extract, 10 g/L sodium chloride, 3 g/L K2HPO4·3H2O, 6 g/L (NH4)2SO4, pH 7.2. The inoculum (3%) was added into 30 mL of fermentation media in a 250 mL flask, and was incubated at 180 rpm and 37 °C for 48 h. All the fermentation experiments were repeated at least three times.
Gene knockout in B. licheniformis
The genes sppA and tepA were deleted individually in B. licheniformis BL10 according to our previously reported method [20, 21], and the construction procedure of the sppA deficient strain was described briefly as following. First, the upstream (a) and downstream (b) regions of sppA were amplified respectively by the corresponding primers sppA-KF1/sppA-KR1 and sppA-KF2/sppA-KR2 listed in Additional file 1: Table S1, and fused by splicing overlap extension (SOE)-PCR using the primers sppA-KF1/sppA-KR2. The fused fragments were cloned into T2(2)-Ori at the restriction sites XbaI/SacI, diagnostic PCR and DNA sequencing confirmed that the vector, named T2-sppA, was constructed successfully (Fig. 1).
Fig. 1.
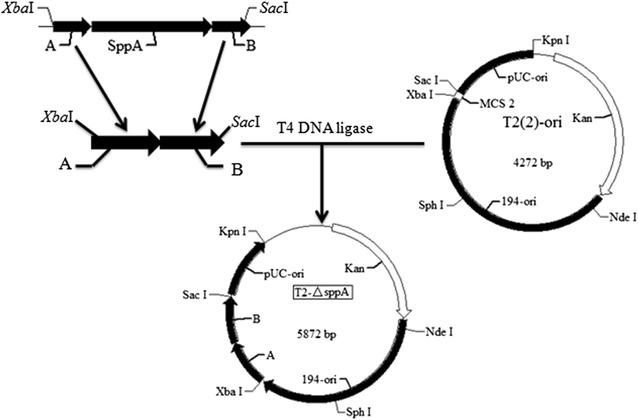
The construction procedure of gene knockout vector. A and B represented the upstream and downstream homologous arms of sppA, respectively
Then, the vector T2-sppA was electro-transferred into B. licheniformis BL10 and was verified by diagnostic PCR and plasmids extractions [22, 23]. The positive transformants were cultivated in the LB liquid medium with 20 μg/mL kanamycin at 45 °C, and subcultured for three generations, then transferred into kanamycin-free LB medium at 37 °C for another six generations. The cells were then plated on LB agar medium with or without kanamycin and incubated for 20 h, respectively. The primers sppA-KYF/sppA-KYR were used for PCR to verify the double crossover strains, and DNA sequencing confirmed that the sppA deficient strain (B. licheniformis BL10S) was constructed successfully. The tepA deficient strain (B. licheniformis BL10T) was constructed using methods similar to those employed for the construction of B. licheniformis BL10S.
Construction of the protein expression vector
The nattokinase expression vector used in this study was obtained from our previously reported research [7], and the α-amylase expression vector was constructed according to the following method. P43 promoter was from B. subtilis 168. The gene amyL (FJ556804.1) coding for α-amylase (containing its own signal peptide and terminator) was amplified from the genome DNA of B. licheniformis WX-02 with the corresponding primers (Additional file 1: Table S1), purified and fused by SOE-PCR, and inserted into the expression vector pHY300PLK at the restriction sites EcoRI and XbaI, plasmid extraction and DNA sequencing have confirmed that this new plasmid was constructed successfully, named pHY-amyL. The green fluorescent protein (GFP) expression vector (pHY-GFP) was constructed by using a similar method.
Gene integration in B. licheniformis
To develop strains to over express sppA and tepA, the individual genes sppA and tepA mediated by P43 promoter were integrated into the genome of B. licheniformis by the previous reported method [8]. The resulting strains B. licheniformis BL10GS, capable of overexpressing sppA and B. licheniformis BL10GT, capable of overexpressing tepA, were verified by PCR and used for further assays.
Enzyme activity assay
Two proteins, nattokinase and α-amylase, were served as the target proteins to evaluate the efficiency of host strains constructed in this research, and the methods for activity assay were described previously [8].
α-amylase activity assay
The mixture of 20 mL soluble starch solution (2 g/mL) and 5 mL 0.12 M phosphate buffer (pH 6) was added into the 50 mL centrifuge tube, and incubated at 60 °C for 8 min. The fermentation supernatant at a volume of 1 mL was added into the tube and incubated at 60 °C for another 10 min. Then, the 1-mL reaction solution was transferred into a new centrifuge tube containing 0.5 mL 0.1 mol/L HCl and 5 mL dilute iodine solution (266 mM KI and 0.35 mM I2), and the absorbance was detected under 660 nm wavelength. The mixture containing 0.5 mL 0.1 mol/L HCl and 5 mL dilute iodine solution was served as the control. The α-amylase activity was calculated by the standard curve made from the different concentrations of starch solution (0.00, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 0.90, 1.00 mg/mL). One Unit of α-amylase activity (U) was defined as the amount of enzyme for catalyzing 1 mg starch to release reducing sugar (glucose) per minute at 60 °C (pH 6) [24].
Nattokinase activity assay
The nattokinase activity was measured according to our previous reported method [8]: the mixture of 0.4 mL fibrinogen solution (0.72%, w/v) and 1.4 mL Tris–HCl (50 mM, pH 7.8) was incubated in a test tube at 37 °C for 10 min, followed by adding 0.1 mL thrombin solution (20 U/mL) to form the fibrin at 37 °C for 10 min. Then, the diluted sample solution (D) at the volume of 0.1 mL was added into the tube and shaken at 37 °C for 60 min at the interval of every 15 min, followed by adding 2 mL trichloroacetic acid (TCA) solution (0.2 M) to stop the reaction (AT). As for the control group, the mixture of 0.1 mL sample solution and 2 mL TCA solution (0.2 M) was used after incubating at 37 °C for 60 min (AB). Finally, all mixtures were centrifuged at 12000g for 10 min, and the absorbance of the supernatant was determined at 275 nm. One unit of nattokinase activity (1 FU) was defined as the amount of enzyme leading to the 0.01 increase for A275 in 1 min, and the formula was as follows:
Determination of the total extracellular protein and target protein concentrations
To determine the extracellular protein concentration, the volume of 100 μL fermentation broth was mixed with an equal volume of Coomassie Brilliant Blue G-250 and the absorbance was measured at 595 nm. The concentration of total extracellular protein was calculated by the standard curve made from different concentrations of bovine serum albumin (BSA) [25]. Meanwhile, the cells were washed three times with physiological saline, followed with sonication (pulse: 1 s on; 2 s off; total 4 min) to disrupt cells, and the concentration of intracellular protein was determined with the similar method.
For determining the target protein concentration, the mixture of 900 μL fermentation broth with 100 μL TCA (6.12 M) was maintained at 4 °C overnight, and centrifuged at 13,000g for 10 min. The precipitate was washed three times with ethanol and dried, and re-dissolved in a solution containing 2 M thiourea and 8 M urea. The sample was then mixed with equal volume of 2× SDS-PAGE loading buffer, and the volume of 10 μL solution was subjected to SDS-PAGE analysis. BSA was used as the standard. Protein concentration of the sample was calculated by comparing the area and pixel counts of the bands imaged from the gel with that of the standard, using a Bio-Rad GS-800 calibrated densitometer and Quantity One software [26].
Analysis of transcription level
The total RNA of the strain was extracted by the TRIzol® Reagent (Invitrogen, USA) according to our previously reported method [20], and contaminant DNA was digested by the Rnase-free DNase I enzyme (TaKaRa, Japan). The RevertAid First Strand cDNA Synthesis Kit (Thermo, USA) was applied to amplify the first stand of cDNA. The primers in the (seeing the Additional file: Table S2) were used for amplifying the corresponding genes in the 10 μL Real-Time PCR mixture (containing 5 μL SYBR® Select Master Mix, 0.5 μL primers, 1 μL cDNA, 3.5 μL DEPC water), and 16S rRNA from B. licheniformis BL10 was used as the reference gene to normalize the data. The Vii7 Real-Time PCR system (ABI, USA) was used for the Real-Time PCR reaction (95 °C for 3 min; 40 cycles of 95 °C for 30 s, 60 °C for 30 s). The transcriptional levels for genes in the recombinant strain were compared with those of the control strain after normalization to the reference gene 16S rRNA, and the experiments were performed in triplicate.
Statistical analyses
All samples were analyzed in triplicate, and the data were presented as the mean ± the standard deviation for each sample point. All data were conducted to analyze the variance at P < 0.05 and P < 0.01, and a t test was applied to compare the mean values using the software package Statistica 6.0 [27].
Results
Establishment of the signal peptide peptidase gene deficient strains
Based on the genome sequence and annotation of B. licheniformis WX-02, two genes (sppA and tepA) were predicted to function as SPPs in B. licheniformis WX-02 [28]. In order to investigate the function of these gene products, sppA and tepA were deleted in the parent strain B. licheniformis BL10, respectively. Figure 1 shows the construction procedure of the sppA knockout vector, and the bands amplified from mutants had the same length of the homologous arms without that of target genes (Additional file 1: Figure S1), confirming that sppA was deleted successfully. The new strain was named B. licheniformis BL10S. Similarly, tepA was also deleted using a similar method, and the resultant strain was named B. licheniformis BL10T.
Effects of signal peptide peptidase deficiency on the total extracellular protein secretion
The sppA and tepA deficient strains, as well as the parental strain BL10, were cultivated in ME media for 24 h, respectively, and the concentrations of total extracellular proteins were assayed to evaluate the importance of these genes on total extracellular protein secretion. As shown in Fig. 2a, the yield of total extracellular proteins decreased by 35% in the sppA deficient strain, compared to BL10. Meanwhile, there was no difference for the concentrations of total extracellular proteins produced by the tepA deficient strain and BL10, and these results were positively correlated with the SDS-PAGE results in Fig. 2b, which suggested that sppA gene product had the strongest effect on protein secretion in B. licheniformis.
Fig. 2.
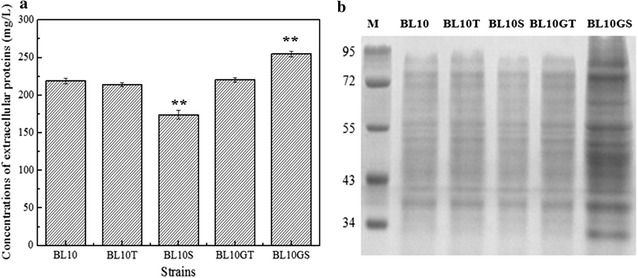
Effects of deficiency or over-expression of sppA and tepA on the extracellular secretion. a The concentrations of total extracellular proteins of different strains; b SDS-PAGE analysis of the extracellular proteins from different host strains. M protein marker (95, 72, 55, 43, 34 kDa); Lane 1 B. licheniformis BL10; Lane 2 B. licheniformis BL10T; Lane 3 B. licheniformis BL10S; Lane 4 B. licheniformis BL10GT; Lane 5 B. licheniformis BL10GS. Data are represented as the means of three replicates and bars represent the standard deviations, *P < 0.05; and **P < 0.01 indicate the significance levels between recombinant strains and control strain
Effects of signal peptide peptidase deficiency on the target protein production
In this study, nattokinase and α-amylase were selected as the target proteins to evaluate the function of sppA and tepA. Nattokinase expression vector pP43SacCNK was made in a previous study [7], and the α-amylase expression vector pHY-amyL harboring P43 promoter (K02174.1), amyL (FJ556804.1) coding for α-amylase (containing its own signal peptide and terminator) was verified by DNA sequence. The pP43SacCNK and pHY-amyL plasmids were individually electro-transferred into the parental strain B. licheniformis BL10, as well as the SPP gene deficient strains BL10S and BL10T. PCR verification and plasmid extraction confirmed that the recombinant strains were constructed successfully.
Furthermore, the extracellular activities of α-amylase and nattokinase produced by different strains were measured. As shown in Fig. 3, the extracellular activities of α-amylase and nattokinase were decreased by 51 and 48%, and the maximum biomass was decreased by 18 and 15% respectively due to the deficiency of sppA. Furthermore, the specific activities of α-amylase and nattokinase in BL10S were 17011.86 U/gDCW and 3436.71 FU/gDCW (1 OD600 = 0.363 gDCW/L), which were 40 and 38% lower compared with those of BL10 (28238.63 U/gDCW and 5585.39 FU/gDCW), respectively. There were no differences in the yields of the target proteins and specific enzyme activities between the tepA deficient strain BL10T and BL10. These results suggest that SppA and not TepA, plays an important role for protein secretion. Additionally, the deficiency of sppA might not only affect the target protein yields in B. licheniformis, but also the cell biomass was decreased in the sppA deficient strains (Fig. 4a, c).
Fig. 3.
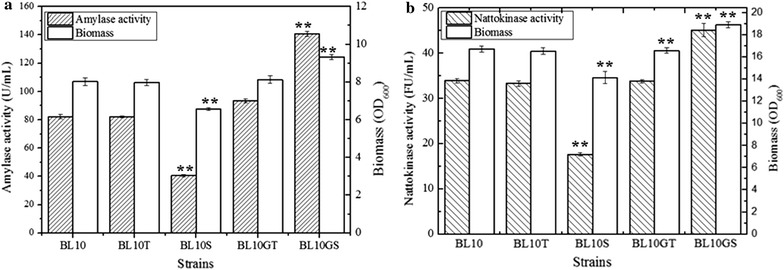
The activities of target proteins and biomass of different host strains harboring pHY-amyL or pP43SacCNK. a α-Amylase activities and biomass of different host strains harboring pHY-amyL; b nattokinase activities and biomass of different host strains harboring pP43SacCNK. Data are represented as the means of three replicates and bars represent the standard deviations, *P < 0.05; and **P < 0.01 indicate the significance levels between recombinant strains and control strain
Fig. 4.

SDS-PAGE analysis of the extracellular proteins from different host strains harboring pHY-amyL or pP43SacCNK. a SDS-PAGE analysis of extracellular proteins from tepA and sppA deficient strains harboring pHY-amyL, Lane 1 BL10/pHY-amyL; Lane 2 BL10T/pHY-amyL; Lane 3 BL10S/pHY-amyL. b SDS-PAGE analysis of extracellular proteins from tepA and sppA overexpression strains harboring pHY-amyL, Lane 1 BL10/pHY-amyL; Lane 2 BL10GT/pHY-amyL; Lane 3 BL10GS/pHY-amyL; c SDS-PAGE analysis of extracellular proteins from the tepA and sppA deficient strains harboring pP43SacCNK, Lane 1 BL10/pP43SacCNK; Lane 2 BL10T/pP43SacCNK; Lane 3 BL10S/pP43SacCNK; d SDS-PAGE analysis of extracellular proteins from tepA and sppA overexpression strains harboring pP43SacCNK, Lane 1 BL10/pP43SacCNK; Lane 2 BL10GT/pP43SacCNK; Lane 3 BL10GS/pP43SacCNK. M Protein marker (120, 100, 95, 72, 55, 43, 34, 26, 14 kDa)
Effects of overexpression of signal peptide peptidase on the protein secretion
To verify the vital role of SPP in extracellular protein production, the genes sppA and tepA mediated by P43 promoter were further overexpressed in B. licheniformis BL10 by inserting the relevant expression cassettes into the chromosome by homologous recombination, respectively [8]. The sppA insert was verified by PCR and the bands amplified from the mutant strain exhibited the same length exactly as the length combination of the homologous arms and the sppA gene plus P43 promoter. Furthermore, DNA sequencing results confirmed the successful integration of sppA into B. licheniformis BL10, resulting in the new strain B. licheniformis BL10GS. The tepA overexpression strain was constructed and verified using the similar methods. The resulting new strain was named B. licheniformis BL10GT.
Both SPP overexpression strains BL10GS and BL10GT were cultivated in the ME medium for 24 h. As shown in Fig. 2a, the yield of total extracellular proteins increased by 37% in the sppA overexpression strain BL10GS, compared with that of the parental strain BL10. These results positively correlated with the SDS-PAGE results in Fig. 2b. Meanwhile, overexpression of tepA had no effect on the concentration of the total extracellular proteins. Furthermore, in order to analyze the influence of sppA overexpression on cell lysis, the green fluorescent protein (GFP) was expressed in the parental strain BL10, the sppA mutant strain BL10S and the sppA overexpression strain BL10GS (Additional file 1: Figure S2). As shown in Additional file 1: Figure S3, GFP was expressed successfully in these strains, and no GFP was detected in ME medium after cultivation for 24 h. These results suggest that overexpression of sppA has no effect on the cell lysis. Furthermore, these results indicate that SppA could be used to improve the concentration of extracellular proteins produced by B. licheniformis.
The nattokinase and α-amylase expression vectors were electro-transferred into the sppA and tepA overexpression strains, and the recombinant strains were cultivated in the nattokinase and amylase production media for 48 h to evaluate the effects of overexpression of SPP genes on the target protein production, respectively. As shown in Fig. 3, the activities of target enzymes and cell yields both increased in the sppA overexpression strain, which was consistent with the improvement of target protein concentrations (Table 2). Based on our results, the cell biomass of sppA deficiency strains were decreased by 16 and 13% during α-amylase and nattokinase production, and the decreases in cell biomass observed were much lower than decreases in protein yields (67 and 30%, respectively). Also, the specific activities of α-amylase and nattokinase were increased by 43 and 18% respectively in the sppA overexpression strain, compared with those produced by B. licheniformis BL10. These results indicate that overexpression of sppA improves both cell biomass and protein yields. Meanwhile, in the tepA overexpression strain, only a 12% improvement of specific α-amylase activity was obtained and overexpression of tepA had no effect on the concentrations of nattokinase (Fig. 4b, d). These results were consistent with the enzymes activities (Fig. 3). Moreover, the concentrations of intracellular protein were measured in these strains duringα-amylase and nattokinase production, the SPPs deficiency and overexpression have no effects on the concentrations of intracellular protein (Additional file 1: Table S3).
Table 2.
Effects of deletion or overexpression of sppA and tepA on the concentrations of target proteins
| Strains | BL10 | BL10T | BL10S | BL10GT | BL10GS | |
|---|---|---|---|---|---|---|
| Concentrations (mg/L) | pHY-amyL | 89.04 (± 5.43) | 86.93 (± 6.25) | 46.78 (± 2.43) | 104.29 (± 9.54) | 147.95 (± 11.32) |
| pP43SacCNK | 215.30 (± 10.19) | 205.41 (± 21.64) | 108.94 (± 8.11) | 212.62 (± 14.46) | 281.19 (± 12.65) |
Effect of SppA on the α-amylase secretion and cell growth during the fermentation process
Figure 5 shows the time courses of the cell growth and α-amylase yield of B. licheniformis BL10/pHY-amyL, B. licheniformis BL10S/pHY-amyL, B. licheniformis BL10T/pHY-amyL, B. licheniformis BL10GT/pHY-amyL, and B. licheniformis BL10GS/pHY-amyL in the α-amylase production medium. The culture was sampled every 4 h, and α-amylase activity and biomass were measured at the defined time. Based on our results, the α-amylase activities of BL10GS/pHY-amyL were higher than those of BL10/pHY-amyL and BL10S/pHY-amyL throughout the whole fermentation process, and the α-amylase activity was the highest in B. licheniformis BL10GS/pHY-amyL and the lowest in BL10S/pHY-amyL. Also, the biomass of BL10GS/pHY-amyL was higher than those of other strains throughout the fermentation process. Meanwhile, deletion or overexpression of tepA had no effect on the cell growth and target production during the production of α-amylase (Fig. 5). These results indicated that overexpression of sppA could not only improve the protein secretion level, but also enhance the biomass yield.
Fig. 5.
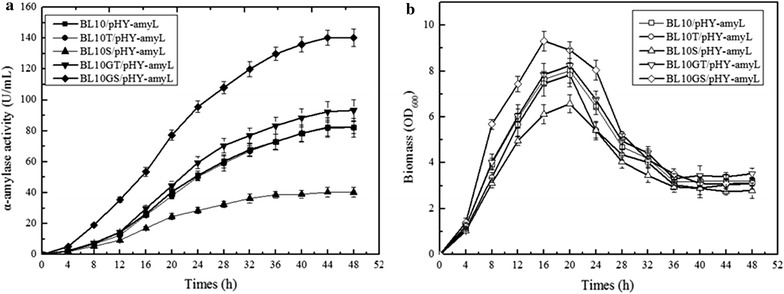
Fermentation process curves of α-amylase production by different host strains harboring pHY-amyL. a α-Amylase activities; b OD600
The relative gene expression levels of BL10/pHY-amyL, BL10T/pHY-amyL, BL10S/pHY-amyL, BL10GT/pHY-amyL and BL10GS/pHY-amyL were also evaluated at the mid-log phase during the α-amylase production process (8 h). As shown in Fig. 6, the transcriptional expression levels of sppA and tepA could not be detected in the sppA and tepA deficient strains, and the sppA and tepA transcriptional expression levels increased by 11.43- and 11.47-fold in the BL10GS and BL10GT strains, respectively compared to the parental strain.
Fig. 6.
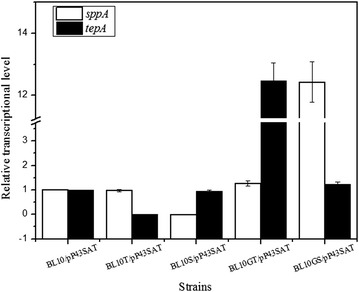
Transcriptional levels of sppA and tepA in the different strains during α-amylase production
Discussion
Heterologous expression is an effective strategy to improve the target protein production, and many strategies have been carried out to improve the protein yield [3, 9, 29–31]. It has been previously shown that SPP can play an important role during protein secretion [32], However, prior to the current study, no research has been reported on the improvement of protein secretion by overexpression of SPP. In this study, we have demonstrated that the SppA SPP plays an important role for protein secretion in B. licheniformis, and overexpression of sppA could improve the yields of the total extracellular proteins and target proteins.
In this study, the yields and specific activities of target proteins were decreased significantly in the sppA deficient strain, and they were increased in the sppA overexpression strain, compared with those of the parental strain BL10. However, different expression levels of an alternative SPP, TepA, had no effect on the concentrations of heterologously produced extracellular target proteins. These results suggest that SppA is very important for protein secretion in B. licheniformis, and are consistent with previously reported results in B. subtilis [18, 19]. Previously, several researches have implied that the TepA of B. subtilis is a cytoplasmic, ClpP-like germination protease involved in spore outgrowth, and it is expressed almost exclusively during sporulation, and researches proposed that the TepA of B. subtilis is not the SPP [33, 34]. Since B. licheniformis have high homology with B. subtilis, it was suggested that the TepA of B. licheniformis BL10 might also not be the SPP by our results. Meanwhile, SPPs are required for degradation of signal peptides that are inhibitory to protein translocation [35, 36], and the deficiency of sppA might influence the ability to remove remnant signal peptides in the cellular membrane. It has been proposed that the accumulation of these remnant signal peptides might block the membrane channel for protein translocation and hinder protein secretion [18]. Therefore, we postulated that overexpression of sppA would increase the removal of the remnant signal peptides, therefore improving protein secretion. The extracellular activities of α-amylase produced by B. licheniformis BL10GS/pHY-amyL were higher than those of BL10/pHY-amyL throughout the whole fermentation process. These results indicate that overexpression of sppA could improve the α-amylase yield from the beginning of the fermentation, and SppA is necessary for highly efficient protein secretion. Meanwhile, the concentrations of intracellular protein showed no differences among these recombinant strains, which indicated that the higher levels of extracellular proteins might be due to the increase level of recombinant proteins production. Thus, in-depth research on remnant signal peptides in the cell membrane should be carried out to further understand this mechanism. In addition, our results indicate that overexpression of sppA could improve the cell growth (Fig. 3), which partially contributed to the increase of the α-amylase activity. However, since the increase in cell biomass observed was much lower than the increase in the α-amylase activity, we concluded that the enhanced α-amylase activity was mainly due to the increased protein secretion.
In order to improve extracellular protein production, several strategies have been developed for signal peptide processing by manipulating the signal peptidase and signal peptidase [37]. Malten et al. [29] have overexpressed the type I signal peptidase SipM in Bacillus megaterium MS941, and the yield of target protein Dsrs, mediated by its own signal peptide, was increased by 3.7-fold. Also, the signal peptidase I SipV was confirmed to play a vital role for nattokinase (mediated by the signal peptide of AprE) production in our previous study [8]. Previous studies have shown that either signal peptidase I (SipS or SipT) was sufficient for protein secretion [38]. Signal peptidases serve as the “scissors” to cut off the signal peptides from the pre-proteins [39], and multiple signal peptidases were responsible for the cleavage of different signal peptides [40]. Therefore, overexpression of a single signal peptidase could not be used as a universal strategy for enhancing protein secretion. In this research, the yield of total extracellular proteins was improved markedly in the sppA overexpression strain, and the concentrations of nattokinase and α-amylase mediated by different signal peptides were increased by 30 and 67%, respectively. These results implied that overexpression of sppA might act as an efficient and useful strategy for protein production.
In conclusion, this study implied that SppA is the main functional SPP for protein secretion in B. licheniformis, and overexpression of sppA indeed improved the concentrations of target proteins, biomass and specific activities. In this study, overexpression of sppA might act as a useful and efficient strategy to improve protein secretion. The host strain B. licheniformis BL10GS demonstrated high efficiency for protein secretion and could be employed as a promising industrial strain for extracellular production of valuable proteins.
Authors’ contributions
DC, XW and SC designed the study. DC and HW carried out the molecular biology studies and construction of engineering strains. DC, HW and PH carried out the fermentation studies. DC, HW, CZ, QW, CTN and SC analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its Additional file 1].
Funding
This work was supported by the National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (2013AA102801-52), the Science and Technology Program of Wuhan (20160201010086).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1. All the sequences of the primers and concentrations of total intracellular proteins were listed in the Additional file (Table S1, Table S2 and Table S3). The agarose gel electrophoresis analysis of the SPPs deficient strains and the fluorescence detection of the cell and the fermentation supernatant of the BL10/pHY-GFP, BL10S/pHY-GFP, BL10GS/pHY-GFP were also contained in the Additional file (Fig. S1, Fig. S2 and Fig. S3). This information is available free of charge via the Internet http://microbialcellfactories.biomedcentral.com/.
Contributor Information
Dongbo Cai, Email: hzauskycai@126.com.
Hao Wang, Email: 18071729546@163.com.
Penghui He, Email: penghuihehubu@163.com.
Chengjun Zhu, Email: zhuchengjun@gmail.com.
Qin Wang, Email: qinwang327@gmail.com.
Xuetuan Wei, Email: weixuetuan@mail.hzau.edu.cn.
Christopher T. Nomura, Email: ctnomura@esf.edu
Shouwen Chen, Phone: +86 027-88666081, Email: mel212@126.com.
References
- 1.Yang H, Liu L, Shin HD, Chen RR, Li J, Du G, Chen J. Comparative analysis of heterologous expression, biochemical characterization optimal production of an alkaline alpha-amylase from alkaliphilic Alkalimonas amylolytica in Escherichia coli and Pichia pastoris. Biotechnol Prog. 2013;29:39–47. doi: 10.1002/btpr.1657. [DOI] [PubMed] [Google Scholar]
- 2.Lin S, Zhang M, Liu J, Jones GS. Construction and application of recombinant strain for the production of an alkaline protease from Bacillus licheniformis. J Biosci Bioeng. 2015;119:284–288. doi: 10.1016/j.jbiosc.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Wang P, Wang P, Tian J, Yu X, Chang M, Chu X, Wu N. A new strategy to express the extracellular alpha-amylase from Pyrococcus furiosus in Bacillus amyloliquefaciens. Sci Rep. 2016;6:22229. doi: 10.1038/srep22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westers L, Westers H, Quax WJ. Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Biochim Biophys Acta. 2004;1694:299–310. doi: 10.1016/j.bbamcr.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 5.van Dijl JM, Hecker M. Bacillus subtilis: from soil bacterium to super-secreting cell factory. Microb Cell Fact. 2013;12:3. doi: 10.1186/1475-2859-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pohl S, Harwood CR. Heterologous protein secretion by Bacillus species from the cradle to the grave. Adv Appl Microbiol. 2010;73:1–25. doi: 10.1016/S0065-2164(10)73001-X. [DOI] [PubMed] [Google Scholar]
- 7.Wei X, Zhou Y, Chen J, Cai D, Wang D, Qi G, Chen S. Efficient expression of nattokinase in Bacillus licheniformis: host strain construction and signal peptide optimization. J Ind Microbiol Biotechnol. 2015;42:287–295. doi: 10.1007/s10295-014-1559-4. [DOI] [PubMed] [Google Scholar]
- 8.Cai D, Wei X, Qiu Y, Chen Y, Chen J, Wen Z, Chen S. High-level expression of nattokinase in Bacillus licheniformis by manipulating signal peptide and signal peptidase. J Appl Microbiol. 2016;121:704–712. doi: 10.1111/jam.13175. [DOI] [PubMed] [Google Scholar]
- 9.Kang Z, Yang S, Du G, Chen J. Molecular engineering of secretory machinery components for high-level secretion of proteins in Bacillus species. J Ind Microbiol Biotechnol. 2014;41:1599–1607. doi: 10.1007/s10295-014-1506-4. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Liu Y, Wang Z, Lu F. Influence of promoter and signal peptide on the expression of pullulanase in Bacillus subtilis. Biotechnol Lett. 2014;36:1783–1789. doi: 10.1007/s10529-014-1538-x. [DOI] [PubMed] [Google Scholar]
- 11.Bolhuis A, Broekhuizen CP, Sorokin A, van Roosmalen ML, Venema G, Bron S, Quax WJ, van Dijl JM. SecDF of Bacillus subtilis, a molecular Siamese twin required for the efficient secretion of proteins. J Biol Chem. 1998;273:21217–21224. doi: 10.1074/jbc.273.33.21217. [DOI] [PubMed] [Google Scholar]
- 12.Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Fu G, Gai Y, Zheng P, Zhang D, Wen J. Combinatorial Sec pathway analysis for improved heterologous protein secretion in Bacillus subtilis: identification of bottlenecks by systematic gene overexpression. Microb Cell Fact. 2015;14:92. doi: 10.1186/s12934-015-0282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Gai Y, Fu G, Zhou W, Zhang D, Wen J. Enhanced extracellular production of alpha-amylase in Bacillus subtilis by optimization of regulatory elements and over-expression of PrsA lipoprotein. Biotechnol Lett. 2015;37:899–906. doi: 10.1007/s10529-014-1755-3. [DOI] [PubMed] [Google Scholar]
- 15.Fu LL, Xu ZR, Li FW, Shuai JB, Lu P, Hu CX. Protein secretion pathways in Bacillus subtilis: implication for optimization of heterologous protein secretion. Biotechnol Adv. 2007;25:1–12. doi: 10.1016/j.biotechadv.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Harwood CR, Cranenburgh R. Bacillus protein secretion: an unfolding story. Trends Microbiol. 2008;16:73–79. doi: 10.1016/j.tim.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 17.van Roosmalen ML, Geukens N, Jongbloed JD, Tjalsma H, Dubois JY, Bron S, van Dijl JM, Anne J. Type I signal peptidases of Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:279–297. doi: 10.1016/j.bbamcr.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Bolhuis A, Matzen A, Hyyrylainen HL, Kontinen VP, Meima R, Chapuis J, Venema G, Bron S, Freudl R, MaartenvanDijl J. Signal peptide peptidase- and ClpP-like proteins of Bacillus subtilis required for efficient translocation and processing of secretory proteins. J Bio Chem. 1999;274:24585–24592. doi: 10.1074/jbc.274.35.24585. [DOI] [PubMed] [Google Scholar]
- 19.Nam SE, Paetzel M. Structure of signal peptide peptidase A with C-termini bound in the active sites: insights into specificity, self-processing, and regulation. Biochemistry. 2013;52:8811–8822. doi: 10.1021/bi4011489. [DOI] [PubMed] [Google Scholar]
- 20.Qiu Y, Xiao F, Wei X, Wen Z, Chen S. Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Appl Microbiol Biotechnol. 2014;98:8895–8903. doi: 10.1007/s00253-014-5978-y. [DOI] [PubMed] [Google Scholar]
- 21.Qi G, Kang Y, Li L, Xiao A, Zhang S, Wen Z, Xu D, Chen S. Deletion of meso-2,3-butanediol dehydrogenase gene budC for enhanced D-2,3-butanediol production in Bacillus licheniformis. Biotechnol Biofuels. 2014;7:16. doi: 10.1186/1754-6834-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue GP, Johnson BP, Dalrymple BP. High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis. J Microbiol Meth. 1999;34:183–191. doi: 10.1016/S0167-7012(98)00087-6. [DOI] [Google Scholar]
- 23.Qiu Y, Zhang J, Li L, Wen Z, Nomura CT, Wu S, Chen S. Engineering Bacillus licheniformis for the production of meso-2,3-butanediol. Biotechnol Biofuels. 2016;9:117. doi: 10.1186/s13068-016-0522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Zhou Y, Zhao X, Chen S, Wei X. Comparative analysis of different Bacillus licheniformis host strains on the secretion expression of α-amylase. Food Sci. 2015;9:275–284. [Google Scholar]
- 25.Grintzalis K, Georgiou CD, Schneider YJ. An accurate and sensitive Coomassie Brilliant Blue G-250-based assay for protein determination. Anal Biochem. 2015;480:28–30. doi: 10.1016/j.ab.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Voigt B, Schweder T, Sibbald MJ, Albrecht D, Ehrenreich A, Bernhardt J, Feesche J, Maurer KH, Gottschalk G, van Dijl JM, Hecker M. The extracellular proteome of Bacillus licheniformis grown in different media and under different nutrient starvation conditions. Proteomics. 2006;6:268–281. doi: 10.1002/pmic.200500091. [DOI] [PubMed] [Google Scholar]
- 27.Tian G, Fu J, Wei X, Ji Z, Qi G, Chen S. Enhanced expression of pgdS, gene for high production of poly-γ-glutamic aicd with lower molecular weight in Bacillus licheniformis WX-02. J Chem Technol Biotechnol. 2013;89:1825–1832. doi: 10.1002/jctb.4261. [DOI] [Google Scholar]
- 28.Yangtse W, Zhou Y, Lei Y, Qiu Y, Wei X, Ji Z, Qi G, Yong Y, Chen L, Chen S. Genome sequence of Bacillus licheniformis WX-02. J Bacteriol. 2012;194:3561–3562. doi: 10.1128/JB.00572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malten M, Nahrstedt H, Meinhardt F, Jahn D. Coexpression of the type I signal peptidase gene sipM increases recombinant protein production and export in Bacillus megaterium MS941. Biotechnol Bioeng. 2005;91:616–621. doi: 10.1002/bit.20523. [DOI] [PubMed] [Google Scholar]
- 30.Degering C, Eggert T, Puls M, Bongaerts J, Evers S, Maurer KH, Jaeger KE. Optimization of protease secretion in Bacillus subtilis and Bacillus licheniformis by screening of homologous and heterologous signal peptides. Appl Environ Microbiol. 2010;76:6370–6376. doi: 10.1128/AEM.01146-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang JK, Kang Z, Ling Z, Cao W, Liu L, Wang M, Du G, Chen J. High-level extracellular production of alkaline polygalacturonate lyase in Bacillus subtilis with optimized regulatory elements. Bioresour Technol. 2013;146:543–548. doi: 10.1016/j.biortech.2013.07.129. [DOI] [PubMed] [Google Scholar]
- 32.Nam SE, Kim AC, Paetzel M. Crystal structure of Bacillus subtilis signal peptide peptidase A. J Mol Biol. 2012;419:347–358. doi: 10.1016/j.jmb.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 33.Westers H, Darmon E, Zanen G, Veening JW, Kuipers OP, Bron S, Quax WJ, Van Dijl JM. The Bacillus secretion stress response is an indicator for alpha-amylase production levels. Lett Appl Microbiol. 2004;39:65–73. doi: 10.1111/j.1472-765X.2004.01539.x. [DOI] [PubMed] [Google Scholar]
- 34.Traag BA, Pugliese A, Setlow B, Setlow P, Losick R. A conserved ClpP-like protease involved in spore outgrowth in Bacillus subtilis. Mol Microbiol. 2013;90:160–166. doi: 10.1111/mmi.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CY, Malchus NS, Hehn B, Stelzer W, Avci D, Langosch D, Lemberg MK. Signal peptide peptidase functions in ERAD to cleave the unfolded protein response regulator XBP1u. EMBO J. 2014;33:2492–2506. doi: 10.15252/embj.201488208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voss M, Schroder B, Fluhrer R. Mechanism, specificity, and physiology of signal peptide peptidase (SPP) and SPP-like proteases. Biochim Biophys Acta. 2013;1828:2828–2839. doi: 10.1016/j.bbamem.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 37.Dalbey RE, Wang P, van Dijl JM. Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol Mol Biol Rev. 2012;76:311–330. doi: 10.1128/MMBR.05019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tjalsma H, Bolhuis A, van Roosmalen ML, Wiegert T, Schumann W, Broekhuizen CP, Quax WJ, Venema G, Bron S, van Dijl JM. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 1998;12:2318–2331. doi: 10.1101/gad.12.15.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Auclair SM, Bhanu MK, Kendall DA. Signal peptidase I: cleaving the way to mature proteins. Protein Sci. 2012;21:13–25. doi: 10.1002/pro.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song Y, Nikoloff JM, Zhang D. Improving protein production on the level of regulation of both expression and secretion pathways in Bacillus subtilis. J Microbiol Biotechnol. 2015;25:963–977. doi: 10.4014/jmb.1501.01028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its Additional file 1].


