Abstract
CRISPR-Cas immune systems defend prokaryotes against viruses and plasmids. CRISPR RNAs (crRNAs) associate with various CRISPR-associated (Cas) protein modules to form structurally and functionally diverse (Type I–VI) crRNP immune effector complexes. Previously, we identified three, co-existing effector complexes in Pyrococcus furiosus -Type I-A (Csa), Type I-G (Cst), and Type III-B (Cmr)—and demonstrated that each complex functions in vivo to eliminate invader DNA. Here, we reconstitute functional Cst crRNP complexes in vitro from recombinant Cas proteins and synthetic crRNAs and investigate mechanisms of crRNP assembly and invader DNA recognition and destruction. All four known Cst-affiliated Cas proteins (Cas5t, Cst1, Cst2, and Cas3) are required for activity, but each subunit plays a distinct role. Cas5t and Cst2 comprise a minimal set of proteins that selectively interact with crRNA. Further addition of Cst1, enables the four subunit crRNP (Cas5t, Cst1, Cst2, crRNA) to specifically bind complementary, double-stranded DNA targets and to recruit the Cas3 effector nuclease, which catalyzes cleavages at specific sites within the displaced, non-target DNA strand. Our results indicate that Type I-G crRNPs selectively bind target DNA in a crRNA and, protospacer adjacent motif dependent manner to recruit a dedicated Cas3 nuclease for invader DNA destruction.
Keywords: CRISPR, Cas, Cst, Cas3, Type I-G, Pyrococcus furiosus
Introduction
CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR-associated) systems are RNA-based adaptive immune systems that protect bacteria and archaea from viruses, plasmids, and other foreign genetic elements. These systems capture fragments of foreign DNA, called protospacers, and integrate them at host CRISPR genomic loci. The CRISPR locus is made up of arrays of direct repeats (typically 30–40 base pairs in length) interspersed by similar-sized, viral- and plasmid-derived spacer sequences, together with an upstream leader element (Grissa et al. 2007). CRISPR loci are transcribed and processed into CRISPR RNAs (crRNAs) that each contains invader-derived (guide) sequences flanked on their 5′ and/or 3′ ends by CRISPR repeat-derived (tag) sequences. crRNAs assemble with Cas proteins to recognize and degrade complementary nucleic acids (reviewed in Jackson and Wiedenheft 2015; Jiang and Doudna 2015; Makarova et al. 2015; Sorek et al. 2013; Terns and Terns 2011; van der Oost et al. 2014).
Due to tremendous diversity in Cas proteins and molecular mechanisms, CRISPR-Cas immune systems are currently grouped into two classes, six distinct types (I–VI), and at least 16 specific subtypes (Haft et al. 2005; Makarova et al. 2006, 2011, 2015; Vestergaard et al. 2014). A host organism can harbor more than one CRISPR-Cas system wherein crRNAs associate with Cas subtype proteins to form crRNA-Cas effector complexes (crRNPs) and guide homology-dependent destruction of foreign nucleic acids. Type I and II systems directly recognize and target destruction of DNA targets (Brouns et al. 2008; Cady and O’Toole 2011; Elmore et al. 2015; Garneau et al. 2010; Gasiunas et al. 2012; Jinek et al. 2012; Magadan et al. 2012; Maier et al. 2013; Mulepati et al. 2014; Nam et al. 2012; Plagens et al. 2014; Sinkunas et al. 2013; Westra et al. 2012; Wiedenheft et al. 2011b). In contrast, Type III systems degrade target RNA and use the bound target RNA to also destroy the source target DNA (Deng et al. 2013; Elmore et al. 2016; Estrella et al. 2016; Hale et al. 2009; Jiang et al. 2016b; Marraffini and Sontheimer 2008; Samai et al. 2015; Staals et al. 2013; Tamulaitis et al. 2014; Zebec et al. 2014; Zhang et al. 2012). The function of Type I and II systems requires that their crRNP effector complexes recognize short (typically 3–5 base) sequence elements in the foreign DNA called protospacer adjacent motifs (PAMs) (Deveau et al. 2008; Mojica et al. 2009; Shah et al. 2013). Following PAM recognition by a Cas protein component (Anders et al. 2014; Hayes et al. 2016; Hochstrasser et al. 2014; Nishimasu et al. 2014; Sashital et al. 2012), Type I and II crRNP effector complexes then disrupt local DNA pairing to form R-loop structures in which the crRNA guide elements base-pair with the complementary target strand to cause displacement of the non-target strand of the DNA (Hayes et al. 2016; Szczelkun et al. 2014). The binding and unwinding of double-stranded DNA targets by crRNP complexes is required for DNA cleavage and destruction by Type-specific Cas effector nucleases (Cas3 and Cas9, respectively, for Type I and II systems) (Hochstrasser et al. 2014; Huo et al. 2014; Jiang and Doudna 2015; Jiang et al. 2016a; Mulepati and Bailey 2013; Mulepati et al. 2014; Westra et al. 2012).
Characterized Type I crRNP effector complexes contain members of Cas5, Cas7, Cas8 (large subunit) and Cas11 (small subunit; sometimes fused to the large subunit) superfamily Cas proteins as structural components (Brendel et al. 2014; Brouns et al. 2008; Jackson et al. 2014; Lintner et al. 2011; Majumdar et al. 2015; Makarova et al. 2011; Mulepati et al. 2014; Nam et al. 2012; Plagens et al. 2012; van Duijn et al. 2012; Wiedenheft et al. 2011a; Zhao et al. 2014). Cas3 superfamily proteins contain nuclease (HD) and helicase (DExH) domains and are the effector nucleases of DNA silencing, Type I effector complexes (Cady and O’Toole 2011; Elmore et al. 2015; Huo et al. 2014; Mulepati and Bailey 2013; Plagens et al. 2014; Sinkunas et al. 2013; Westra et al. 2012). Cas3 is a stable component of Type I-A crRNPs (Majumdar et al. 2015; Plagens et al. 2014) but is recruited in trans to the crRNP-bound DNA targets by other Type I systems (e.g., I-E and I-F) (Cady and O’Toole 2011; Mulepati and Bailey 2013; Sinkunas et al. 2013). Cas3 nicks the displaced, non-target strand of the R-loop structure through its HD nuclease active site (Huo et al. 2014; Mulepati and Bailey 2013; Plagens et al. 2014; Westra et al. 2012). An intrinsic ATP-dependent helicase activity of Cas3 can further promote DNA destruction through progressive DNA unwinding and further DNA cleavages, at least in some systems (Huo et al. 2014; Mulepati and Bailey 2013; Sinkunas et al. 2013; Westra et al. 2012). Bioinformatic analyses and our previous biochemical and molecular genetic studies revealed that the function of Type I-G (Cst) systems requires four Cas proteins: Cst1 (Cas8a1), Cst2 (Cas7), Cas5t (Cas5) and Cas3 (Elmore et al. 2015; Haft et al. 2005; Majumdar et al. 2015; Makarova et al. 2015; Vestergaard et al. 2014).
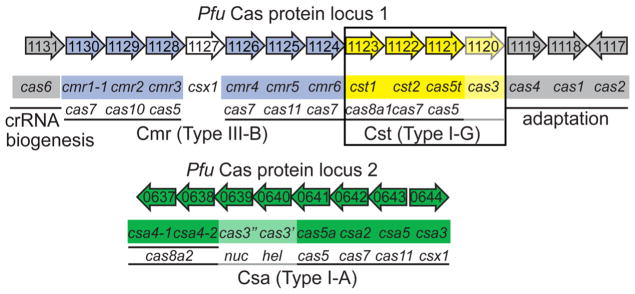
The cas genes of Pyrococcus furiosus (Pfu) are organized in two separate gene clusters that encode Cas proteins involved in DNA uptake into CRISPR loci (Cas1, Cas2, Cas4), crRNA biogenesis (Cas6), and formation of three effector modules (Csa or Type I-A, Cst or Type I-G, and Cmr or Type III-B) (Fig. 1). Additionally, Pfu contains seven CRISPR loci that encode approximately 200 crRNAs (Terns and Terns 2013). We have previously identified and characterized three distinct native crRNP complexes in Pfu [Csa (Type I-A), Cst (Type I-G) and Cmr (Type III-B)] (Hale et al. 2009, 2012; Majumdar et al. 2015). All three effector complexes associate with mature crRNAs derived from all seven CRISPR loci. Pfu Type III-B (Cmr) complex mediates crRNA-guided cleavage of invader RNA and transcript-dependent DNA destruction (Elmore et al. 2016; Hale et al. 2009, 2012, 2014; Spilman et al. 2013). Furthermore, our in vivo analysis demonstrated plasmid DNA silencing by either the Csa (Type I-A) or Cst (Type I-G) systems in Pfu (Elmore et al. 2015). In this study, we have investigated the assembly and function of Cst (Type I-G) crRNPs by reconstituting active complexes from synthetic crRNA and purified recombinant Cst Cas proteins. crRNA features and Cas protein components that are essential for formation of Cst crRNPs were delineated. Target DNA elements important for Cst crRNP-mediated DNA identification and destruction were also identified. Like some other characterized Type I systems, our results indicate that Cst crRNPs function as surveillance complexes and specifically bind non-self target DNA sequences in a PAM- and crRNA-dependent manner. Furthermore, we show that the target-bound Cst crRNPs recruit Cas3 to cleave the strand of the target DNA that is displaced following crRNA/target DNA strand pairing.
Fig. 1.
Pyrococcus furiosus cas gene organization and annotation. Genes encoding the crRNA biogenesis protein [(cas6), grey], predicted CRISPR DNA uptake proteins [(cas 1, cas2, and cas4), grey], and three effector modules—Type III-B [(Cmr), blue], Type I-G [(Cst), yellow (boxed), and Type I-A (Csa), green] are shown. The annotated superfamily designations (e.g., cas5, cas7, etc.) are denoted below each gene name
Materials and methods
Expression and purification of recombinant Cst and Cas3 proteins
The genes encoding N-terminal, 6 × His-tagged Pfu Cst proteins (Cas5t, Cst2, Cst1) and Cas3 (Pfu and Tko) were subcloned into expression vectors (pET-24) and transformed into a BL21 RIPL E. coli expression strain. 1–2 L LB cultures were grown at 37 °C under antibiotic selection to an OD600 between 0.4 and 0.7. Protein expression was induced with 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) overnight at room temperature. Cell pellets were resuspended in buffer A [25 mM Tris (pH 7.6), 250 mM NaCl] with 10 mM imidazole and 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and lysed by sonication (Misonix Sonicator 3000). The lysate was incubated at 70 °C for 30 min with frequent mixing to precipitate heat-labile proteins, then centrifuged at 14,000 rpm at 4 °C, and the soluble fraction was filtered (0.8 μm filter pore size Millex filter unit, Millipore). His-tagged proteins were purified via Ni2+-affinity chromatography, washed with buffer A and 20 mM imidazole, and eluted in buffer A at increasing concentrations of imidazole (50, 100, 200, and 500 mM). Peak elution fractions were dialyzed in buffer A using Slide-A-Lyzer MINI Dialysis Cassettes to remove imidazole (Thermo Fisher). Cst2 and Pfu Cas3 were concentrated with Vivaspin 500 Centrifugal Filters (GE Health-care). Proteins were quantified using a Qubit 2.0 Fluorometer (Life Technologies) and stored at 4 °C prior to use for functional assays.
Preparation of RNA and DNA substrates
Synthetic RNA and DNA oligonucleotide sequences are listed in Table S1. 7.01 crRNA sequences (wild-type tag and mutant tag) were previously described (Hale et al. 2009, 2012). crRNAs were 3′-labeled [Figs. 2b, 3a, b (5′-OH, 3′-P)] with T4 RNA ligase (New England Biolabs) and [α-32P] pCp (3000 Ci/mmol, Perkin Elmer) as described (Carte et al. 2010). Internally labeled 7.01 crRNA (Fig. 3b, 5′-OH/3′-OH) was prepared by in vitro transcription of repeat-spacer unit with [α-32P]-GTP (800 Ci/mmol, 10 mCi/mL, Perkin Elmer) followed by Cas6 cleavage as described (Spilman et al. 2013). crRNA (Fig. 3b, 5′-P/3′-OH) and DNA oligos (MWG Eurofins) were 5′-labeled with T4 polynucleotide kinase (PNK) and [γ-32P] ATP as described (Carte et al. 2008). Radiolabeled, double-stranded DNA substrates (Table S2) were generated by annealing complementary single-stranded DNA oligos, using a 2:1 molar ratio of unlabeled to radiolabeled DNA, in 10 mM Tris (pH 7.5–8.0), 50 mM NaCl, 1 mM EDTA at 95 °C for 5 min followed by slow cooling to 23 °C (at the rate of 1 °C per minute). Annealing was confirmed and substrates were extracted from 8 % non-denaturing 0.5× TBE polyacrylamide gels. The annealed DNA duplex was extracted with PCI [phenol/chloroform/isoamyl alcohol (pH 8.0)], precipitated with ethanol, resuspended in buffer A and stored at −20 °C for future use in functional assays.
Fig. 2.
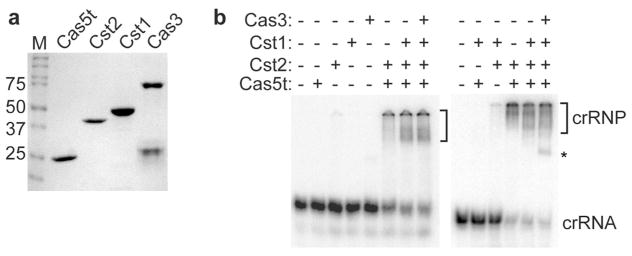
Cas protein requirements for crRNA binding. a SDS-PAGE showing purified recombinant Cas5t, Cst2, Cst1, and Cas3 and molecular mass marker proteins (M). b Native gel electrophoresis of 3′-radiolabeled crRNA incubated in the absence (−) or presence (+) of Cas5t, Cst2, Cst1, and/or Cas3 proteins as indicated. The positions of unbound crRNA (crRNA), non-specific binding associated with Cas3 (asterisk) and Cst complexes (crRNP, bracket) are highlighted
Fig. 3.
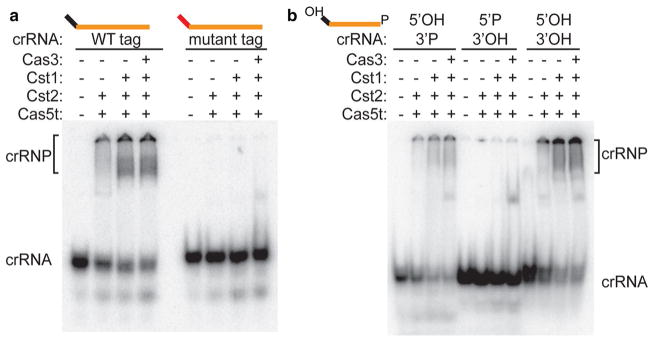
Key crRNA features for assembly of Cst crRNP complexes. a Gel mobility shift assay showing Cst crRNP formation with 2, 3, or 4 Cst Cas proteins as indicated and 3′-radiolabeled (32P) crRNA containing either a wild-type 5′ tag (black) or mutated 5′ tag (red). b Gel mobility shift assay showing Cst crRNP formation with 2,3, or 4 Cst Cas proteins and crRNA bearing either 5′ hydroxyl and 3′ phosphate [(5′ OH/3′ P), wild type shown in schematic], 5′ phosphate and 3′ hydroxyl (5′ P/3′ OH), or 5′ hydroxyl and 3′ hydroxyl (5′ OH/3′ OH). The guide sequence (orange) is wild type in all cases. Unbound crRNAs (crRNA) and Cst complexes (crRNP, bracket) are indicated on right
Native gel mobility shift assays to study crRNP formation
Cst crRNP formation (Figs. 2, 3) was assayed as follows: 1 μM each of recombinant Cst proteins (as indicated) were incubated with 0.01–0.05 pmol of 32P-radiolabeled crRNA (end-labeled or internally labeled as specified) in 25 mM Tris (pH 7.6), 250 mM NaCl, 1.5 mM MgCl2, 1 unit of SUPERase-In ribonuclease inhibitor (Applied Biosystems) and 5 μg of E. coli tRNA at 70 °C for 30 min. Reactions were mixed with DNA loading dye (4 % glycerol, 0.05 % bromophenol blue, 0.05 % xylene cyanol in 1× TBE) and separated on a 6 % native PAGE (containing 0.5X TBE and 8 % glycerol). The gel was run at 110 V for 70 min, dried, and exposed on a phosphor-imaging screen.
DNA binding and cleavage assays
crRNPs were formed as specified above except with 0.5 pmol unlabeled crRNA. After 30 min incubation at 70 °C to promote crRNP formation, 0.01–0.05 pmol of 32P-radiolabeled double-stranded DNA and 100 μM of NiCl2 were added to the reactions and incubated at 70 °C for 3 h. To assess DNA binding, half of each reaction was resolved by electrophoresis through a 6 % native polyacrylamide gel at 110 V for 110 min, dried, and exposed on a phosphor-imaging screen. For assaying DNA cleavage, the remaining half of the reactions were treating with 1 μg of proteinase K (New England Biolabs) and incubated at 37 °C for 15 min, followed by PCI extraction and ethanol precipitation and resolved on 15 % polyacrylamide (7 M urea) sequencing gels. Gels were dried and exposed on phosphor-imaging screen. 5′ end-labeled (32P) low molecular weight single-stranded DNA markers (10–100 nucleotides, Affymetrix) were used to determine the sizes of the observed target DNA cleavage products.
RNA–protein UV crosslinking
UV light-induced, crRNA-Cst protein crosslinking using photo-activable 4-thiouridine (4SU) and internally (32P) radiolabeled crRNAs was performed as described (Spilman et al. 2013) with the following modifications: crRNP reconstitution reactions were doubled relative to standard Cst crRNP reconstitution experiments and RNase inhibitor was omitted. A quarter of each reaction was assessed for crRNA binding by native gel electrophoresis (Fig. 4a). Following crosslinking with 312 nm UV light, complexes were disrupted in 1 % SDS and boiling for 10 min, followed by RNase treatment at 37 °C for 1 h. Reactions containing crRNA and 8-nt 5′-tag RNA were treated with RNase T1 (1,000 U, Thermo Fisher), and guide RNA was treated with RNase A (10 μg, Thermo Fisher). The samples were separated by 12.5 % SDS-PAGE. Proteins were visualized by G-250 Coomassie Blue staining, dried, and subjected to phosphor-imaging and autoradiography to allow for alignment of protein bands with autoradiographs (Fig. 4b).
Fig. 4.
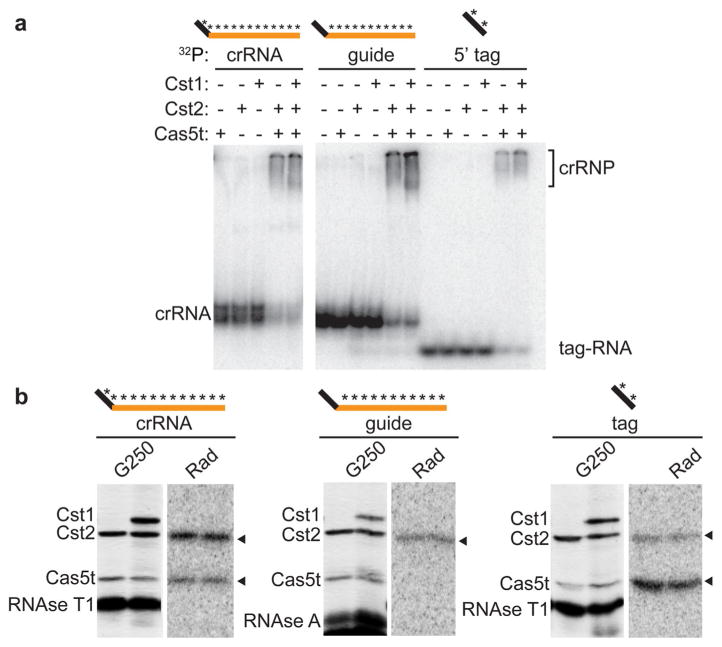
Identification of direct interactions between crRNA and Cst proteins via UV-crosslinking. a Gel mobility shift assay showing crRNPs formed under crosslinking conditions in the presence (+) or absence (−) of individual Cst proteins, Cas5t + Cst2, or Cas5t + Cst2 + Cst1, as indicated. b SDS polyacrylamide gel (G250) of Cst Cas proteins, RNAse T1, and/or RNase A in each reaction plus corresponding autoradiographic images (Rad) identifying crRNA-interacting Cst Cas proteins (arrowheads). Schematic above each planel indicates the 32P- and 4-thiouridine-labeled RNA used in each experiment [5′ tag sequence (black), guide sequence (orange), radiolabeled region (asterisks)]
Site-directed mutagenesis
To make HD-AA mutants of Pfu Cas 3 (His 53, Asp 54) and Tko Cas3 (His 53, Asp 54), QuikChange site-directed mutagenesis was performed as per manufacture’s guidelines (Agilent Technologies) using primers (MWG Eurofins) listed in Table S1. The mutants were generated by PCR amplification of wild-type Pfu cas3 (PF1120) and Tko cas3 (PF0460) genes followed by DpnI digestion and transformation into an E. coli BL21 RIPL strain. Mutations were confirmed by sequencing. The proteins were expressed and purified alongside wild-type Pfu and Tko Cas3 (Fig. S1).
Protein–protein interaction
N-terminal, 6x His-tagged Cas5t (pET-24) and untagged (native) Cst2 (modified pET-21) were purified as described above. The supernatant obtained after cell-lysis, thermal purification, and centrifugation was used. Approximately 30 μg of 6x His-tagged Cas5t, 100 μg of Cst2 and 30 μg of BSA (used as internal control) were incubated at 70 °C for 30 min with frequent mixing. One half of the samples were precipitated with 7.6 % trichloroacetic acid, 0.18 % sodium deoxycholate and 100 % ice cold acetone, centrifuged, resuspended in SDS loading buffer and used as ‘input’. The other half of the sample was affinity-purified with Ni–NTA (Qiagen) by incubating with 15 μl of nickel beads at room temperature for 20 min with end-over-end rotation, followed by four washes in buffer A with 25 mM imidazole and elution by boiling in SDS loading dye. The proteins were separated by electrophoresis on 12.5 % SDS–polyacrylamide gels at 200 V and visualized by Coomassie Blue R-250 staining.
Results
Proteins required for Cst crRNP assembly
Toward understanding the role of individual Cst Cas protein subunits in assembly of functional Cst complexes, we initially investigated which Cst proteins are required for recognition and incorporation of crRNAs into the complex by native gel electrophoresis with 3′-radiolabeled crRNAs and various combinations of Cst proteins (Fig. 2a, b). None of the individual Cst proteins (Cas5t, Cst2, Cst1) or Cas3 stably associated with crRNA (Fig. 2b, left panel). In contrast, stable crRNP complexes formed when both Cas5t and Cst2 (but no other combination of two Cst proteins) were incubated with crRNA (Fig. 2b, left and right panels). A reproducible but modest apparent increase in crRNA affinity was observed when all three Cst proteins (Cas5t, Cst2 and Cst1) were present versus the two Cst proteins (Cas5t and Cst2) (Fig. 2b, left and right panels and see Figs. 3, 4). Further addition of Cas3 did not observably influence the binding affinity of the three Cst proteins with crRNA (Fig. 2b, left and right panels) suggesting that Cas3 does not stably associate with the crRNP even in the presence of the three crRNA-associated Cst Cas proteins (Fig. 2b). These results indicate that Cas5t and Cst2 are minimally required for crRNA recognition in vitro and promote the recruitment of Cst1 to the crRNP complex.
Cst crRNP formation depends on the 5′ tag and 5′ hydroxyl group of the crRNA
We previously determined that each of the ~200 crRNA species associated with native Pfu Cst crRNPs contained an 8-nt (AUUGAAAG) CRISPR repeat-derived sequence (or 5′ tag) at their 5′ termini (Majumdar et al. 2015). This common 5′ tag sequence results from endonucleolytic cleavage of larger precursor crRNA transcripts by the Cas6 crRNA biogenesis enzyme and leads to mature crRNAs containing 5′-OH ends (Carte et al. 2008, 2010). The 3′ ends of Cst-associated native crRNAs are heterogenous in length and likely harbor 3′ phosphate end groups (Majumdar et al. 2015). Native gel mobility shift analysis indicates that both the 5′ tag sequence and terminal 5′ hydroxyl group of the crRNA are essential for Cst crRNP assembly (Fig. 3). Cst crRNP formation was abolished by substitution of all 8 nucleotides of the 5′ tag sequence (Fig. 3a) or by conversion of the 5′ hydroxyl group of the wild-type 5′ tag sequence to a 5′ phosphate (Fig. 3b). In contrast, alteration of the 3′ end group (from a phosphate to a hydroxyl group) did not significantly affect binding by Cst proteins (Fig. 3b). The effects of 5′ tag and 5′ OH alteration in preventing crRNA binding were observed when two (Cas5t + Cst2), three (Cas5t + Cst2 + Cst1) or four (Cas5t + Cst2 + Cst1 + Cas3) proteins were assayed, indicating that Cas5t and/or Cst2 specifically recognize the crRNA 5′ tag (Fig. 3b). Taken together, the results indicate that the Cas6-generated, crRNA 5′ tag is a critical component for recognition and assembly of Cst crRNPs.
Cas5t and Cst2 directly interact with crRNA
To investigate which Cst protein(s) directly contact the crRNA 5′ tag and guide regions, we performed UV light-induced protein crosslinking of Cas5t + Cst2 and Cas5t + Cst2 + Cst1 crRNP complexes assembled with 32P-labeled, 4-thiouridine containing crRNA or 8-nt 5′ tag RNA (Fig. 4). By this approach, proteins that interact directly with crRNA become covalently associated with 32P-labeled crRNA binding sites upon exposure to UV light and can be identified by RNase digestion, SDS gel electrophoresis, and autoradiography (Spilman et al. 2013). We confirmed crRNP assembly by analyzing a fraction of the UV-crosslinked samples by native gel electrophoresis (Fig. 4a). The Cst proteins associated with both full-length crRNA and the 5′ tag sequence alone (Fig. 4a). The presence of each protein in the UV crosslinking reactions was verified by SDS-PAGE and Coomassie Blue (G250) staining, and proteins that directly contact each crRNA substrate were identified by autoradiography (Rad) (Fig. 4b). Both Cst2 and Cas5t crosslinked to crRNAs that were uniformly radiolabeled, with signals for Cst2 being higher than those for Cas5t (Fig. 4b, left panel). Cst2 was the only protein observed to crosslink to crRNAs in which the radiolabel was incorporated selectively within the guide region, suggesting that Cst2 (but not Cas5t) interacts with the guide RNA element (Fig. 4b, middle panel). Cas5t is the primary protein that interacts with 5′ tag RNA (weak signals are also observed for Cst2) (Fig. 4b, right panel). There was no evidence of Cst1 crosslinking to any region of the crRNA or 5′ tag RNA, as all samples containing two Cst proteins had identical crosslinking patterns to samples containing all three Cst proteins. These results indicate that Cst2 contacts the crRNA guide region, while Cas5t primarily contacts the 5′ tag.
All three Cst proteins are required for crRNA-guided, target DNA binding
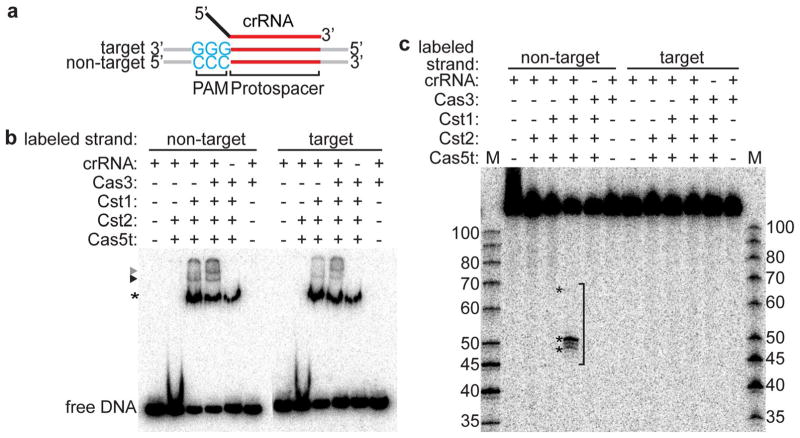
Pfu Cst crRNPs recognize and destroy target DNA in vivo with a requirement that the target DNA contain both a region of complementarity to the crRNA guide element (protospacer) as well as a three base protospacer adjacent motif (PAM) (Elmore et al. 2015). To investigate target DNA recognition in greater detail, we examined binding of various combinations of Cst proteins to target DNA by native gel electrophoresis (Fig. 5). The target DNA substrate used is a 32P 5′ end-labeled double-stranded (ds) DNA that contains the 7.01 protospacer sequence (complementary to first crRNA of Pfu CRISPR locus seven) and including a 5′-GGG-3′ PAM sequence on the target strand and 5′-CCC-3′ on the non-target strand (Fig. 5a). The 5′ GGG/CCC 3′ PAM sequence and 7.01 protospacer sequence (in the context of a plasmid) was previously shown to be targeted by Cst crRNPs in vivo (Elmore et al. 2015).
Fig. 5.
Components of Cst crRNP essential for DNA targeting. a Schematics of double-stranded DNA substrates used in the assay. Wild-type protospacer (red), wild-type PAM (blue), target strand (complementary to crRNA) and non-target strand of DNA, and 5′ tag (black) and guide (red) of crRNA (assembled in Cst crRNP) are labeled. b Gel mobility shift assay with either no protein (−) or Cst crRNP formed with either Cas5t + Cst2, Cas5t + Cst2 + Cst1 (black triangle), Cas5t + Cst2 + Cst1 + Cas3 (grey triangle), all 4 Cst proteins without crRNA, and Cas3 + crRNA in the absence of Cst proteins. Asterisk indicates non-specific binding of DNA by Cst1. c Corresponding DNA cleavages (asterisk, bracket) of samples in b. 5′-radiolabeled DNA size-standard (Affymetrix, low molecular weight marker, 10–100 bases) is labeled (M). The top line of b, c indicates the location of the 32P-label in either the non-target or target strand
To determine the minimal protein components important to selectively recognize target DNA, we performed DNA binding assays with preformed crRNPs comprised of different combinations of Cst proteins and dsDNA containing either a radiolabeled target or non-target DNA strand (Fig. 5b). The minimal crRNP formed with Cas5t and Cst2 (Figs. 2, 3, 4) did not associate with the target DNA. In contrast, target DNA binding was observed upon further addition of the Cst1 protein (Fig. 5b, DNA binding by the crRNP formed when all three Cst proteins (Cas5t, Cst2, Cst1) plus crRNA were added is indicated by a black triangle). The addition of Cas3 to the DNA binding Cst crRNP triggered formation of an additional lower mobility complex indicating that Cas3 is capable of joining the crRNP in the presence of Cst crRNP-bound DNA (Fig. 5b, complexes denoted by a grey triangle). Cst1 alone exhibits apparent non-specific DNA binding that is independent of the presence of crRNA (asterisk, Fig. 5b). In contrast, the Cst crRNP-target DNA complexes formed with Cas5t, Cst2, Cst1 (−/+ Cas3, black and grey triangles) are crRNA-dependent (Fig. 5b). The results suggest that all three Cst proteins (Cas5t, Cst2, and Cst1) are required to specifically associate with target DNA in a crRNA-dependent manner and that the resultant target DNA-bound Cst crRNP is capable of recruiting Cas3 to the target DNA.
Cst crRNP recruits Cas3 for DNA cleavage
Next, we tested whether the in vitro assembled Cst crRNPs were capable of guiding cleavage of the target dsDNA (Fig. 5c). We found that while the crRNP that contains three Cst proteins (Cas5t, Cst2, and Cst1 plus crRNA) is sufficient for specific recognition of target DNA (Fig. 5b), this complex is not capable of cleaving the DNA (Fig. 5c). However, addition of Cas3 to the Cst crRNP led to specific cleavages of the non-target DNA strand, but no cleavages within the (crRNA-interacting) target strand (Fig. 5c). Cas3 alone did not cleave double-stranded DNA and DNA cleavage is dependent on the presence of crRNA (Fig. 5c). The results indicate that the Cst crRNP (containing Cas5t, Cst2, Cst1 and crRNA) targets DNA by binding and recruiting Cas3 to cleave at the non-target strand of the protospacer DNA.
Role of the PAM in target DNA recognition and cleavage
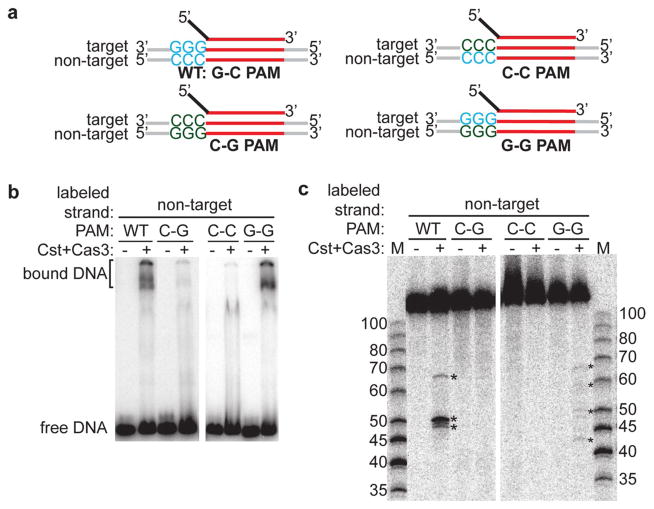
To test for PAM specificity of Cst crRNP-Cas3 mediated DNA binding and cleavage, we made different mutations in the wild-type (5′ GGG/CCC 3′) PAM element of the target DNA (Fig. 6a). We mutated the PAM on target strand from 5′-GGG-3′ to 5′-CCC-3′ and made compensatory mutations on non-target strand to maintain complementarity (5′ CCC/GGG 3′ or C-G PAM, Fig. 6a). Both Cst crRNP-mediated DNA binding and Cst crRNP and Cas3-mediated cleavage of non-target strand of DNA was abolished by this PAM mutation (compare WT versus C-G PAM, Fig. 6b, c, respectively). Moreover, DNA binding and cleavage did not occur for substrate DNA containing a wild-type PAM but containing a protospacer sequence that is not complementary to the crRNA (data not shown). Therefore, Cst crRNP and Cas3-mediated target DNA binding and cleavage is dependent upon recognition of a proper PAM sequence and is guided by crRNA homology.
Fig. 6.
PAM sequence requirements for DNA recognition and destruction. a Schematics of double-stranded DNA substrates used in the assay: crRNA (top) containing 5′ tag (black) and guide (red), target and non-target strands of double-stranded DNA with wild-type protospacer (red) plus either wild-type PAM (blue), C-G PAM (double-stranded but mutant in both strands), C-C PAM (single-stranded wild-type PAM in non-target strand), or G-G PAM (single-stranded wild-type PAM in target strand). b DNA binding and c DNA cleavages (asterisk) by either no protein (−) or Cst crRNP formed with Cas5t + Cst2 + Cst1 + Cas3 and the specified substrates, as labeled. Non-target strand of DNA is 5′-radiolabeled (32P) in all cases. Mobility of 32P-labeled DNA size-standards (in basepairs) are indicated (M)
Cst crRNP recognizes a single-stranded PAM sequence for DNA targeting
We next sought to delineate whether the Cst crRNP recognizes the 5′ GGG/CCC 3′ PAM element as a single-stranded entity or if there is a requirement for the motif to be in a base-paired, double-stranded form during DNA binding and/or Cas3-mediated cleavage. To address this question, we made mutations on one strand of the PAM and did not make compensatory mutations on the other strand, leaving a region of non-complementarity in the PAM region [5′ GGG/GGG 3′ (G-G) PAM or 5′ CCC/CCC 3′ (C-C) PAM, Fig. 6a]. We found that Cst crRNP-Cas3 did not bind or cleave the DNA substrate when the PAM mutation was made on the target strand (5′ CCC/CCC 3′ or C-C PAM, Fig. 6b, c, respectively). In contrast, maintaining the native sequence of the target strand but preventing base-pairing of this sequence (5′ GGG/GGG 3′ or G-G PAM) supported Cst crRNP DNA binding at levels comparable to that observed with the wild-type PAM (compare WT PAM vs. G-G PAM, Fig. 6b). Furthermore, target DNA cleavage occurred with the G-G PAM substrate, albeit at reduced efficiency and producing similar but apparently non-identical cleavage patterns relative to those observed for substrates with a wild-type PAM (compare G-G PAM vs. WT PAM, Fig. 6c). Our data suggest that the Cst crRNP primarily recognizes the PAM sequence on the target DNA strand for both DNA binding and Cas3-mediated cleavage of the non-target DNA strand.
Cas3 cleaves at multiple sites within the non-target DNA strand
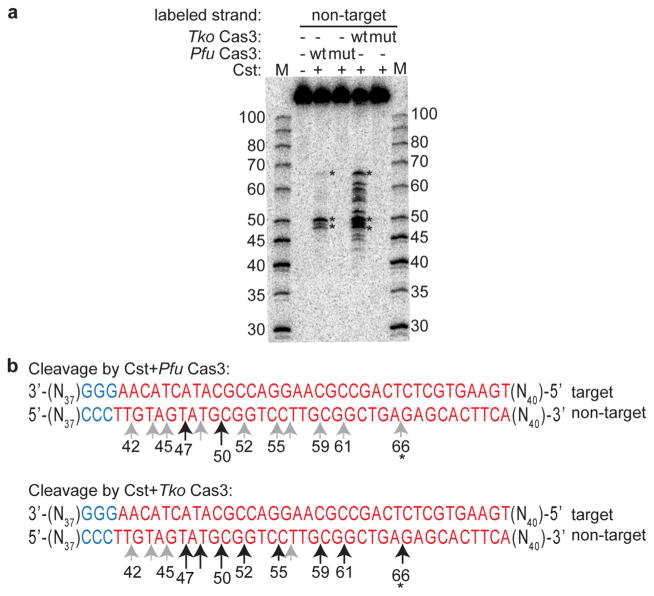
Our data indicate that interaction of the Cst crRNP with target DNA subsequently recruits Cas3 for DNA cleavage (Fig. 5). We found that target DNA-bound, Pfu Cst crRNPs can recruit Cas3 from either Pfu or a related species, Tko (Thermococcus kodakarensis) (Fig. 7). Tko Cas3 shares 58 % sequence identity with Pfu Cas3, and we previously determined that Tko also has a functional Type I-G (Cst) CRISPR-Cas system (Elmore et al. 2013). To gain a more detailed understanding of the Cas3-mediated target DNA cleavage mechanism, we mapped and compared the sites of target DNA cleavages by both Pfu Cas3 and Tko Cas3 following recruitment by Pfu Cst crRNP (Fig. 7). While the relative abundance of cleavage products generated by Pfu and Tko Cas3 nucleases differs somewhat [denoted by black arrows (major products) or grey arrows (minor products) in Fig. 7b], the cleavages map to mostly identical positions. All cleavages map to the protospacer region of the non-target strand and span from 10 nts from the 3′ end to 2 nts from 5′ PAM side of the protospacer (Fig. 7b). The cleavage activity of both Pfu and Tko Cas3 was abolished by site-directed mutation of the conserved HD nuclease active sites (Fig. 7a). The loss of Cas3 cleavage activity was not due to target DNA binding defects as both HD mutant Cas3 proteins bind target DNA in the presence of Pfu Cst crRNPs with efficiencies comparable to those of the wild-type Cas3 protein counterparts (Fig. S1b). These results suggest that Cst crRNPs can recruit related Type I-G associated Cas3 effector nucleases to cleave target DNA utilizing their conserved HD nuclease active sites.
Fig. 7.
DNA cleavages by Cas3 (HD) nuclease map to the protospacer of the non-target strand. a DNA cleavage by no protein (−) or Cst crRNP (Cas5t + Cst2 + Cst1) assembled with either wild-type Cas3 (wt) or mutated Cas3 (mut, HD-AA) from either Pfu or Tko. Non-target strand of DNA is 5′-radiolabeled (32P). Mobility of 32P-labeled DNA size-standards (in basepairs) are indicated (M). b Comparison of Pfu Cas3- and Tko Cas3-catalyzed major cleavages (long black arrows) and minor cleavages (short grey arrows) mapped to DNA sequence [PAM (blue), protospacer (red)]. Numbers accompanying arrows indicate size of products. Prominent products present in both digests are marked by an asterisk on the gel a and in the sequence b
Discussion
Of the numerous and diverse CRISPR-Cas adaptive immune systems identified to date, Type I systems are by far the most prevalent and diverse. Type I systems represent ~60–64 % of the total systems found in sequenced bacteria and archaea, respectively, and are currently classified into 7–8 distinct subtypes (I-A through I-G and I-U) (Haft et al. 2005; Makarova et al. 2015; Vestergaard et al. 2014). All Type I systems share in common a Cas3 effector nuclease/helicase responsible for target DNA cleavage and immunity against viruses and other foreign genetic elements (Brouns et al. 2008; Cady and O’Toole 2011; Elmore et al. 2015; Hochstrasser et al. 2014; Huo et al. 2014; Makarova et al. 2015; Mulepati and Bailey 2013; Plagens et al. 2014; Westra et al. 2012). Otherwise, there is considerable variability in the number and forms of Cas protein subunits that interact with crRNAs to form the distinct, Type I subtype-specific crRNP effector complexes. In this study, we have, for the first time, reconstituted functional Type I-G Cst crRNPs from purified RNA and protein components and obtained important insight into mechanisms governing crRNP assembly and target DNA interaction and destruction. Our study has shed light on the roles of the individual Cst subunits (Cas5t, Cst1, Cst2) as well as affiliated Cas3 protein and defined key features of the crRNA essential for assembly of functional Cst crRNP complexes.
Roles of the Cst Cas proteins in assembly of functional crRNP effector complexes
The results of our crRNA/protein interaction assays (Figs. 2, 4) and protein–protein interaction study (Fig. S2) indicate that Cas5 and Cst2 physically associate with one another and directly and specifically interact with crRNA to make up a minimal core of the Cst crRNP (Fig. 8, upper panel). Cas5t and Cst2 are members of the broader Cas5 and Cas7 superfamily of Cas proteins, respectively, that are common to both Type I and III systems (Makarova et al. 2015). Our work shows that Cas5t (Cas5) directly contacts the CRISPR repeat-derived 5′ tag element and that both nucleotide identity of the tag sequence and the RNA 5′ terminal hydroxyl group are key determinants for binding of both the Cas5t/Cst2 core and full (Cas5t, Cst2, Cst1) Cst crRNP complex (Figs. 3, 4). Type III-B (Cmr) effector crRNPs also exhibit the same requirement for 5′ tag sequence and 5-OH end group for their formation and function (Hale et al. 2014; Spilman et al. 2013), highlighting the general importance of the short common tag sequence in guiding formation of Type I and III effector crRNPs. Like Cas5t, Cst2 (Cas7) also directly contacts the crRNA but preferentially recognizes the ~37 nt guide sequence (Fig. 4). Consistent with structural studies of related Type I-E and Type I-F systems (Jackson et al. 2014; Mulepati et al. 2014; van Duijn et al. 2012; Wiedenheft et al. 2011b; Zhao et al. 2014) and Type III systems (Rouillon et al. 2013; Spilman et al. 2013; Staals et al. 2013), it is predicted that several copies of Cst2 (e.g., 4–6) multimerize and bind along the guide element and make up a helical backbone of the Cst crRNP particle (Fig. 8, upper panel). In contrast, a single copy of Cas5t in the crRNP particle is predicted.
Fig. 8.
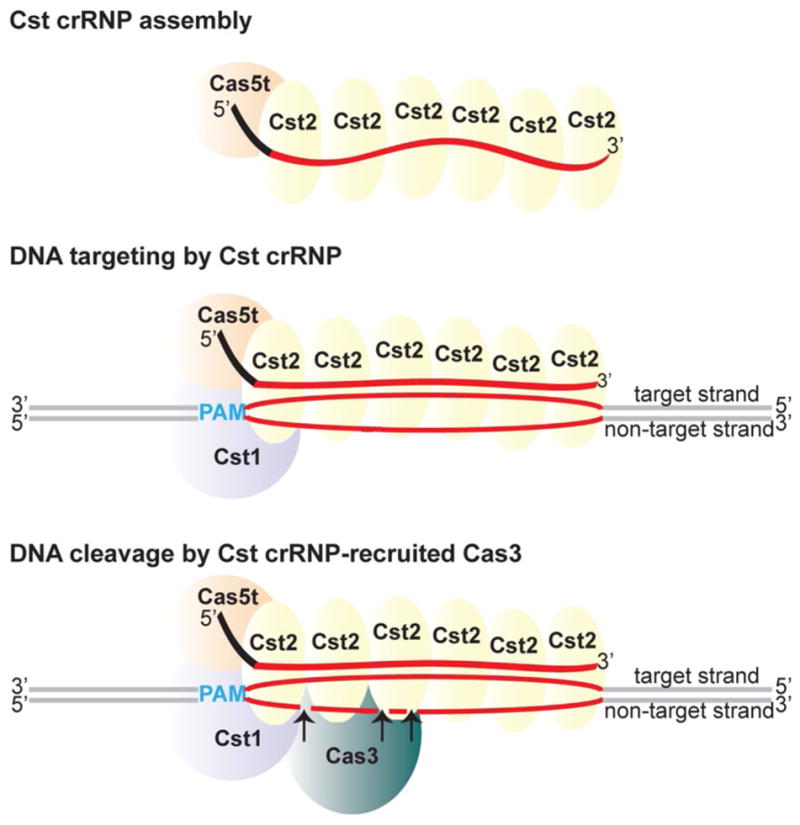
Mechanistic model of Cst crRNP complex assembly, DNA recognition, and cleavage in Pfu. Cas5t (orange) and Cst2 (yellow) interact with 5′ tag (black) and Cst2 interacts with guide (red) of crRNA to form minimal crRNP. Guided by crRNA, Cas5t + Cst2 + Cst1 forms a surveillance complex. Cst1 (purple) scans and interacts with the PAM (blue), enabling the guide sequence of crRNA to base-pair with the target strand of the cognate protospacer (red), displacing the non-target strand and forming an R-loop. The Cas3 nuclease (teal) is recruited to the Cst crRNP/R-loop complex, and the Cas3 nuclease cuts the non-target strand of the protospacer at multiple sites within the R-loop (indicated by arrows)
We show that the Cst1 subunit enables the Cst crRNP complex to specifically recognize and bind PAM-containing, double-stranded DNA targets that are complementary to the crRNA (Figs. 5, 6 and see Fig. 8, middle panel). Cst1 corresponds to the Cas8 or “large subunit” superfamily of Cas proteins that have been implicated in target DNA binding through recognition of the PAM sequence in other Type I systems (Cass et al. 2015; Hochstrasser et al. 2014; Mulepati et al. 2014; Sashital et al. 2012). Accordingly, we predict a role for Cst1 in recognizing the PAM sequences on the target DNA that then facilitates interaction of the crRNA with the target strand. Our results indicate that prior binding of Cas5t and Cst2 to the crRNA, leads to recruitment of Cst1 to the Cst crRNP complex (Fig. 2) likely through protein–protein interactions with Cas5 and/or Cst2 [we did not observe a direct interaction of Cst1 with crRNA in UV crosslinking analyses (Fig. 4)].
Cas3 is the effector DNA nuclease
Cas3 is a known effector DNA nuclease common to all Type I systems, and genetic studies have revealed that the Cst-associated cas3 gene product (Fig. 1) is critical for Cst crRNP function in vivo (Elmore et al. 2015). Not surprisingly, we found that Cas3 is also critical in vitro for DNA cleavage activity and utilizes its HD nuclease domain for this function (Figs. 5c, 7 and see Fig. 8, lower panel). Our observation, that Cas3 is required for crRNA-guided and PAM-dependent cleavage of target DNA (Figs. 5, 6) but appears not to be a stable component of the crRNP complexes unless they are bound to DNA (Figs. 2, 5) is consistent with findings from prior biochemical studies indicating that native Cst crRNP complexes consist of crRNAs in stable association with three Cst protein subunits (Cas5t, Cst1, Cst2) but not Cas3 (Majumdar et al. 2015; Menon et al. 2009). Taken together, the in vivo and in vitro results suggest that the Cst crRNP (crRNA, Cas5t, Cst2, Cst1) acts as a surveillance complex that recognizes and binds target DNA, which subsequently informs Cas3 where to cut DNA.
PAM recognition by the Cst crRNP complex
Consistent with our previous in vivo studies that showed that a target DNA PAM is critical for Cst crRNP function (Elmore et al. 2015), we found that a PAM sequence is essential in vitro for both Cst crRNP binding and subsequent cleavage of the target DNA by Cas3 (Figs. 5, 6). Furthermore, our results indicate that the PAM sequence on the target strand (same strand as the crRNA-DNA interaction) is being specifically recognized during Cst crRNP association but that maximal DNA cleavage efficiency may require that the PAM be in a base-paired configuration (Fig. 6). Similar conclusions were obtained for a Type I-E system where a single-stranded PAM (on target strand) was found to be sufficient for binding (Westra et al. 2012) while base-pairing at the PAM region appeared to be important for conferring optimal DNA cleavage (Hochstrasser et al. 2014; Westra et al. 2012). Insight into this phenomenon comes from a recent crystal structure of Type I-E crRNP complex bound to target dsDNA (Hayes et al. 2016). The structure revealed that the PAM is recognized by the Cse1 (Cas8) subunit in a double-stranded form but with a bias for recognition of bases in the minor groove such that recognition of a single-stranded PAM on the target strand is expected to be tolerated. In contrast, biochemical studies suggest that a Type I-F complex recognizes double-stranded PAM through both major and minor groove interactions (Rollins et al. 2015), highlighting likely subtle differences in the way that PAM-interacting components of diverse Type I crRNP complexes discern PAM DNA elements. Future studies should be focused on understanding PAM recognition by the Cst crRNP.
Model for Cst crRNP function
Based on our collective findings and those reported for other Type I crRNP subtypes, we propose a mechanistic model of Cst crRNP assembly and function (Fig. 8). The three Cst Cas proteins—Cas5t (Cas5), Cst2 (Cas7), and Cst1 (Cas8a1 or large subunit)—assemble with crRNA to form a surveillance complex that binds double-stranded DNA targets in a PAM-dependent fashion involving recognition of the bases on the target strand. PAM recognition leads to crRNA strand invasion and base-pairing of the crRNA guide element with the target strand of the protospacer DNA triggering directional R-loop formation and displacement of the non-target strand. Cas3 nuclease is recruited to the Cst crRNP/R-loop structure and nicks the displaced non-target DNA strand at multiple sites within the protospacer region (Fig. 7). In other studied Type I systems, ATP-dependent Cas3 helicase-mediated unwinding of DNA leads to progressive 3′–5′ cleavage by Cas3 outside of the protospacer region of the non-target strand as well as at the target strand (Huo et al. 2014; Mulepati and Bailey 2013; Sinkunas et al. 2013; Westra et al. 2012). In contrast, in our work, the Cst crRNP cleavages were confined to the non-target strand within the protospacer region whether or not ATP was present (Figs. 5, 7 and data not shown). It is unclear whether our reaction conditions are simply not appropriate for supporting the Cas3 helicase activity of both the tested Pfu and Tko Cas3 proteins or if the observed restricted cleavage of the non-target DNA strand is an intrinsic property of Cst crRNP-Cas3 system.
In summary, we have defined requirements for the assembly of Type I-G Cst effector crRNPs from their Cas protein and crRNA components and illuminated the mechanism by which the assembled crRNPs interact with and destroy target DNA. This work sets the stage for more detailed understanding of the molecular mechanism of DNA recognition and destruction by Cst crRNP complexes. High resolution structures of the crRNP in the presence and absence of target DNA and Cas3 effector nuclease would offer exciting functional insights including mechanisms of PAM recognition and Cas3 recruitment. In general, a detailed understanding of Type I-G Cst crRNP structure and function would contribute to a global understanding of how evolutionarily distinct Type I crRNPs act to defend prokaryotes from viral attack.
Supplementary Material
Acknowledgments
We thank Lesha Spencer and Julie Grainy for their contribution in protein–protein interaction studies. We are grateful to Caryn Hale for intellectual input and technical assistance and Claiborne V. C. Glover III for critical reading of this manuscript. This work was supported by National Institutes of Health (NIH) Grant 1R35GM118160.
Footnotes
Communicated by L. Huang.
Electronic supplementary material The online version of this article (doi:10.1007/s00792-016-0871-5) contains supplementary material, which is available to authorized users.
This article is part of a special feature based on the 11th International Congress on Extremophiles held in Kyoto, Japan, September 12–16, 2016.
References
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel J, et al. A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crRNAs) in Haloferax volcanii. J Biol Chem. 2014;289:7164–7177. doi: 10.1074/jbc.M113.508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady KC, O’Toole GA. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol. 2011;193:3433–3445. doi: 10.1128/JB.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. doi: 10.1261/RNA.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass SD, et al. The role of Cas8 in type I CRISPR interference. Biosci Rep. 2015 doi: 10.1042/BSR20150043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Garrett RA, Shah SA, Peng X, She Q. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol. 2013;87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JR, et al. Programmable plasmid interference by the CRISPR-Cas system in Thermococcus kodakarensis. RNA Biol. 2013;10:828–840. doi: 10.4161/rna.24084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore J, Deighan T, Westpheling J, Terns RM, Terns MP. DNA targeting by the type I-G and type I-A CRISPR-Cas systems of Pyrococcus furiosus. Nucleic Acids Res. 2015;43:10353–10363. doi: 10.1093/nar/gkv1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JR, Sheppard NF, Ramia N, Deighan T, Li H, Terns RM, Terns MP. Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR-Cas system. Genes Dev. 2016;30:447–459. doi: 10.1101/gad.272153.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrella MA, Kuo FT, Bailey S. RNA-activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Genes Dev. 2016 doi: 10.1101/gad.273722.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. doi:10.1016/j. cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. doi:10.1016/j. molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Cocozaki A, Li H, Terns RM, Terns MP. Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex. Genes Dev. 2014;28:2432–2443. doi: 10.1101/gad.250712.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes RP, et al. Structural basis for promiscuous PAM recognition in Type I-E Cascade from E. coli. Nature. 2016;530:499–503. doi: 10.1038/nature16995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci USA. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, et al. Structures of CRISPR Cas3 offer mechanistic insights into cascade-activated DNA unwinding and degradation. Nat Struct Mol Biol. 2014 doi: 10.1038/nsmb.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Wiedenheft B. A conserved structural chassis for mounting versatile CRISPR RNA-guided immune responses. Mol Cell. 2015;58:722–728. doi: 10.1016/j.molcel.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, et al. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014 doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Doudna JA. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol. 2015;30C:100–111. doi: 10.1016/j.sbi.2015.02.002. doi:10.1016/j. sbi.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016a;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Samai P, Marraffini LA. Degradation of phage transcripts by CRISPR-associated RNases enables Type III CRISPR-Cas immunity. Cell. 2016b;164:710–721. doi: 10.1016/j.cell.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintner NG, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286:21643–21656. doi: 10.1074/jbc.M111.238485. doi:10.1074/jbc. M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magadan AH, Dupuis ME, Villion M, Moineau S. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS One. 2012;7:e40913. doi: 10.1371/journal.pone.0040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier LK, et al. Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system I-B. RNA Biol. 2013;10:865–874. doi: 10.4161/rna.24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S, et al. Three CRISPR-Cas immune effector complexes coexist in Pyrococcus furiosus. RNA. 2015;21:1147–1158. doi: 10.1261/rna.049130.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon AL, et al. Novel multiprotein complexes identified in the hyperthermophilic archaeon Pyrococcus furiosus by non-denaturing fractionation of the native proteome. Mol Cell Proteomics: MCP. 2009;8:735–751. doi: 10.1074/mcp.M800246-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Mulepati S, Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J Biol Chem. 2013;288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Heroux A, Bailey S. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014 doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Haitjema C, Liu X, Ding F, Wang H, DeLisa MP, Ke A. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. doi:10.1016/j. str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. doi:10.1016/j. cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A, Tjaden B, Hagemann A, Randau L, Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J Bacteriol. 2012;194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A, et al. In vitro assembly and activity of an archaeal CRISPR-Cas type I-A Cascade interference complex. Nucleic Acids Res. 2014;42:5125–5138. doi: 10.1093/nar/gku120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins MF, Schuman JT, Paulus K, Bukhari HS, Wiedenheft B. Mechanism of foreign DNA recognition by a CRISPR RNA-guided surveillance complex from Pseudomonas aeruginosa. Nucleic Acids Res. 2015;43:2216–2222. doi: 10.1093/nar/gkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon C, et al. Structure of the CRISPR interference complex CSM reveals key similarities with Cascade. Mol Cell. 2013;52:124–134. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. Co-transcriptional DNA and RNA cleavage during Type III CRISPR-Cas immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SA, Erdmann S, Mojica FJ, Garrett RA. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. The EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- Spilman M, et al. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell. 2013;52:146–152. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun MD, et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, et al. Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol Cell. 2014;56:506–517. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. doi:10.1016/j. mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns RM, Terns MP. The RNA- and DNA-targeting CRISPR-Cas immune systems of Pyrococcus furiosus. Biochem Soc Trans. 2013;41:1416–1421. doi: 10.1042/BST20130056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijn E, et al. Native tandem and ion mobility mass spectrometry highlight structural and modular similarities in clustered-regularly-interspaced shot-palindromic-repeats (CRISPR)-associated protein complexes from Escherichia coli and Pseudomonas aeruginosa. Mol Cell Proteomics: MCP. 2012;11:1430–1441. doi: 10.1074/mcp.M112.020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard G, Garrett RA, Shah SA. CRISPR adaptive immune systems of Archaea. RNA Biol. 2014;11:156–167. doi: 10.4161/rna.27990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. doi:10.1016/j. molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011a;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. 2011b;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebec Z, Manica A, Zhang J, White MF, Schleper C. CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2014;42:5280–5288. doi: 10.1093/nar/gku161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, et al. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014 doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.