Abstract
The gibberellin (GA) phytohormones are produced not only by plants but also by fungi and bacteria. Previous characterization of a cytochrome P450 (CYP)-rich GA biosynthetic operon found in many symbiotic, nitrogen-fixing rhizobia led to the elucidation of bacterial GA biosynthesis and implicated GA9 as the final product. However, GA9 does not exhibit hormonal/biological activity and presumably requires further transformation to elicit an effect in the legume host plant. Some rhizobia that contain the GA operon also possess an additional CYP (CYP115), and here we show that this acts as a GA 3-oxidase to produce bioactive GA4 from GA9. This is the first GA 3-oxidase identified for rhizobia, and provides a more complete scheme for biosynthesis of bioactive GAs in bacteria. Furthermore, phylogenetic analyses suggest that rhizobia acquired CYP115 independently of the core GA operon, adding further complexity to the horizontal gene transfer of GA biosynthetic enzymes among bacteria.
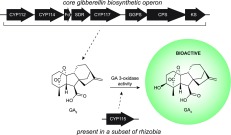
The gibberellin (GA) phytohormones are critical endogenous regulators of plant growth and development1 but are also produced by some plant-associated fungi and bacteria.2 The plant and fungal GA biosynthetic pathways have been extensively studied,1 but only more recently has GA biosynthesis in bacteria been fully elucidated.3,4 Bacterial GA biosynthetic genes are found in an operon (GA operon) that exhibits a scattered distribution, although it seems to be exclusively found in plant-associated species (personal communication, Dr. Asaf Levy, US-DOE Joint Genome Institute). The core GA operon (Figure 1a), encompassing the genes found in essentially all instances, encodes three cytochrome P450 monooxygenases (CYPs; CYP112, CYP114, and CYP117), a ferredoxin (FdGA), a short-chain dehydrogenase/reductase (SDRGA), an (E,E,E)-geranylgeranyl diphosphate synthase (GGPS), an ent-copalyl diphosphate synthase (CPS), and an ent-kaurene synthase (KS). Previous work with the operons from the rhizobia Bradyrhizobium japonicum USDA110, Sinorhizobium fredii NGR234, and Mesorhizobium loti MAFF303099 has led to the characterization of each of these genes in GA biosynthesis (Figure 1b), which collectively act to produce GA9 (1).3−7 The functional characterization of this pathway further established that bacteria evolved GA biosynthesis independently of plants and fungi, at least for the downstream oxidative steps.4
Figure 1.
GA biosynthesis in bacteria. (a) GA biosynthetic operon. The core operon (CYP112 → KS) is mostly conserved, while other genes such as CYP115 and IDI (isopentenyl diphosphate δ-isomerase) are only present in certain strains. Additionally, the core GA operon found in most rhizobia contains a 5′ CYP115 fragment, and most full-length CYP115 in rhizobia, when present, are followed by a 3′ CYP112 fragment. (b) GA biosynthesis in core operon containing rhizobia ends at GA9 (1) with C-3β hydroxylation of GA9 by a GA3ox required to produce the bioactive GA4 (2).
While these studies elucidated the core bacterial GA pathway, it was surprising that biosynthesis ceased at 1, as this GA (of which there are >130) does not seem to function as a phytohormone1—e.g., 1 does not elicit characteristic plant growth phenotypes8 nor does it bind effectively to the appropriate plant receptor (GID1).9,10 Conversion of 1 to a bioactive GA only requires hydroxylation at the C-3β position, which would produce bioactive GA4 (2). This 3β-hydroxyl group is crucial for bioactivity in plants, as 2 binds GID1 roughly 3 orders of magnitude more effectively than 1.9,10 It has been hypothesized that the legume host plants, which necessarily contain GA biosynthetic enzymes, can convert 1 secreted by the symbiotic rhizobial bacteroids to bioactive GAs through the action of endogenous GA 3-oxidases (GA3ox).3 Indeed, incubation of 1 in root nodule extracts from Lotus japonicus plants nodulated with M. loti MAFF303099 led to its conversion into bioactive GAs.3 Given the absence of any additional uncharacterized genes within the operon, this transformation was assumed to be a result of plant enzymatic activity.
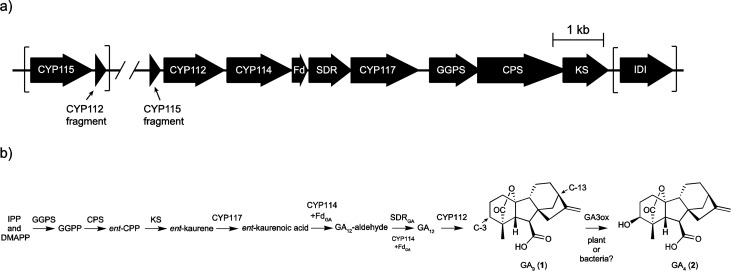
For rhizobia with fully assembled genomes, the GA operon seems to be invariably located within the symbiotic island or plasmid,11−15 which establishes the symbiotic, nitrogen-fixing lifestyle of rhizobia.16 It has previously been determined that these symbiotic modules can be transferred between species of rhizobia,16 and it further seems that the GA operon undergoes independent horizontal gene transfer (HGT) between the symbiotic island/plasmid of various rhizobia.6 This operon also is found in several plant pathogens, particularly Xanthomonas species.17 Interestingly, the operons present in the phytopathogens all contain an additional CYP (CYP115) tightly appended to the 5′ end of the core operon (Figure 1a), which suggests a potential role for this gene in bacterial GA metabolism.
Initially, it appeared that rhizobia either did not contain CYP115 or contained only a presumably nonfunctional fragment of this gene.18 To re-explore the possibility that rhizobia might carry out further metabolism of GA beyond the recently demonstrated production of 1, BLAST searches were carried out with CYP115 from the GA operon found in Xanthomonas oryzae pv oryzicola BLS256. This revealed the presence of nominally full-length CYP115 family members (∼410 amino acids) in 20 unique strains of rhizobia (Supporting Information Table S1), including M. loti MAFF303099 and Sinorhizobium meliloti WSM4191 (a.k.a. Ensifer medicae WSM4191), with each of these strains also containing the core GA operon. Surprisingly, the CYP115 genes were often not found in close proximity to the GA operon, and almost all differ in exact arrangement from the tight 5′ integration found in the phytopathogens (Supporting Information Table S2). For example, while CYP115 in S. meliloti WSM4191 is found close to the core GA operon, this is located proximal to the 3′ rather than 5′ end. However, in M. loti MAFF303099, which has already been shown to produce at least 1,3 CYP115 is found >200 kb away from the GA operon, although it is still located within the symbiotic island.12
To determine if rhizobial CYP115 genes are involved in GA metabolism, those from S. meliloti WSM4191 (SmCYP115) and M. loti MAFF303099 (MlCYP115) were heterologously expressed in S. meliloti 1021 (which does not contain GA biosynthetic enzymes), much as previously described,4 and the resulting recombinant strains were incubated with 1. SmCYP115 was also coexpressed with FdGA from S. fredii NGR234, as this has been shown to be necessary for full activity of CYP114,4 and might be needed to supply electrons to CYP115 as well.
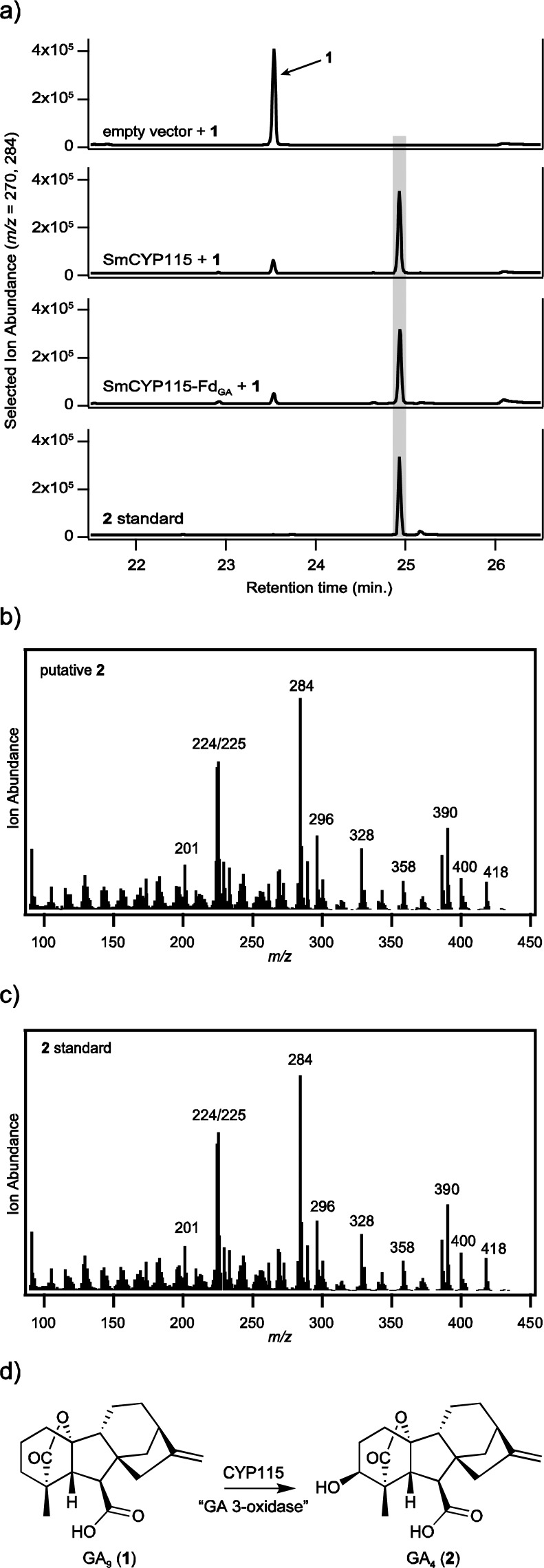
Recombinant S. meliloti 1021 cells expressing SmCYP115 efficiently convert 1 to 2 (Figure 2), and thus, CYP115 acts as a GA3ox. This activity was independent of FdGA, as statistically similar turnover of 1 to 2 was observed either with or without coexpression of FdGA (88 ± 2% and 87 ± 2%, respectively). Accordingly, FdGA seems to only be necessary for CYP114 activity.
Figure 2.
CYP115 is a GA 3-oxidase. (a) Incubating GA9 (1) in cultures expressing SmCYP115 results in efficient transformation to a compound corresponding to GA4, as shown here in gas chromatograms. (b) Mass spectrum of GA4 product. (c) Mass spectrum of GA4 authentic standard. (d) GA 3-oxidase reaction catalyzed by CYP115. Chromatograms and mass spectra correspond to compounds derivatized as methyl esters and/or trimethylsilyl ethers.
CYP115 seems to act specifically as a GA3ox, as cells expressing SmCYP115 do not react with other bacterial GA biosynthetic intermediates (Figure 1b) to any significant extent. Interestingly, SmCYP115-expressing cells carried out 3β-hydroxylation with GA20 (which differs from 1 in containing a C-13 hydroxyl group distal to the targeted C-3β position) to form bioactive GA1, but this transformation did not appear to be as efficient as that of 1 to 2 (Supporting Information Figure S1). This is similar to the activity of plant GA3ox, as these also typically react with both 1 and GA20 within the parallel non-13-hydroxylation and 13-hydroxylation GA biosynthetic pathways found in plants1 (although not in bacteria4,7). This specificity further supports the role of CYP115 as a GA3ox.
GA3ox activity (conversion of 1 to 2) was also observed with cells expressing MlCYP115, though this enzyme was less active (Supporting Information Figure S2). It is possible that this Mesorhizobium-adapted CYP115 family member is not expressed as well in S. meliloti 1021, and/or that this does not pair as well with the endogenous redox partners (e.g., ferredoxins). Regardless, the GA3ox activity of CYP115 appears to be conserved across rhizobia, indicating that that CYP115-containing species are at least capable of producing bioactive 2, rather than just the penultimate phytohormone precursor 1. This is the first example of a GA3ox in rhizobia and provides evidence that bacteria are capable of synthesizing bioactive GAs without the aid of plant metabolism. As such, this result affords a more complete understanding of GA biosynthesis in bacteria. Also, much like the previously characterized bacterial GA biosynthetic enzymes,4 it seems clear that GA3ox activity in bacteria has evolved independently of that in plants or fungi. Specifically, plants make use of 2-oxogluturate dependent dioxygenases for GA3ox activity instead of CYPs,1 and while fungi use a CYP for this reaction, this belongs to the distinct CYP68A subfamily.19
The 20 unique strains of rhizobia with CYP115 represent ∼16% of the total number of strains found to contain the core GA operon (>120 strains of rhizobia total). Of the four major rhizobia genera, Mesorhizobium, Sinorhizobium/Ensifer, and Rhizobium are each represented with at least two strains containing CYP115, while a full-length copy of this gene is notably absent within Bradyrhizobium species. Thus, CYP115 seems to exhibit somewhat restricted distribution within the broader Rhizobiales order. More specifically, CYP115 appears to be particularly enriched within Mesorhizobium species, as over half (13/20) of the strains containing CYP115 are in this genus, which also represents over one-third (13/34) of Mesorhizobium strains with the core GA operon.
While CYP115 is tightly coupled to the GA operon in plant pathogens (within 100 bp of the 5′ end of CYP112), only one rhizobium species (Mesorhizobium sp. AA22) exhibits a similar arrangement. Even when nominally located proximal to the 5′ end of the core GA operon, such as it is in S. fredii CCBAU83666 and S. arboris LMG 14919, CYP115 is located at least 700 bp away. In two other strains, S. meliloti WSM4191 and S. medicae WSM1369, CYP115 is located proximal to the 3′ end of the GA operon. This presumably reflects reincorporation of CYP115 into the operon, which in theory could occur on either end of the pre-existing gene cluster. However, in most of the strains (15/20), CYP115 is located elsewhere within the symbiotic island or plasmid, ranging from ∼3 kb away in Mesorhizobium sp. WSM3626 to the >200 kb distance in M. loti MAFF303099. This distal location relative to the core GA operon presumably underlies the omission of CYP115 in the initial characterization of GA biosynthesis in M. loti MAFF303099.3 Overall, the varying location of CYP115 relative to the core GA operon suggests that this gene is in the process of either being gained or lost from the operon.
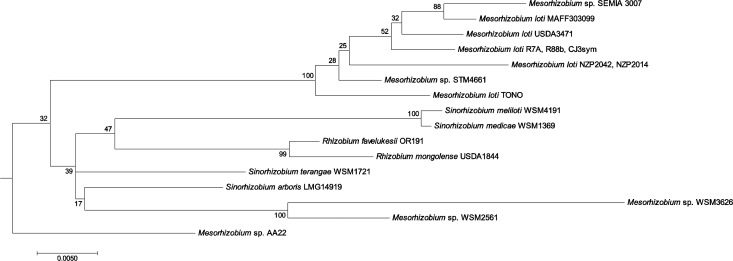
Closer examination of the region surrounding the distal, full-length copy of CYP115 in M. loti MAFF303099 revealed that this is followed by a fragment of CYP112, similar to the arrangement of CYP115 (i.e., 5′ to CYP112) in the phytopathogen operon. This suggests that a full-length CYP115, along with at least a portion of the CYP112 gene, was excised from an operon structured like that found in the phytopathogens prior to its incorporation into, or mobilization within, the M. loti MAFF303099 genome. Notably, similar CYP112 fragments follow the full-length CYP115 in most other strains identified here (Supporting Information Table S2). Indeed, even in S. fredii CCBAU83666 and S. arboris LMG 14919, where the full-length CYP115 is located near the 5′ of the operon, there is still an intervening fragment of CYP112 between CYP115 and the full-length CYP112 that marks the 5′ border of the core GA operon. Thus, it appears that CYP115 is undergoing separate mobilization relative to the core GA operon within these two strains, as well as most other rhizobia where this is found. Consistent with this hypothesis, many of the CYP115 genes outside of the operon are flanked by transposases and other insertion sequence (IS) elements (Supporting Information Table S3). Phylogenetic analysis hints at differences between the relationships of rhizobial full-length CYP115 relative to the core GA operon from the same species, but the low bootstrap values of the relevant trees prevent any strong conclusion (Supporting Information Figures S3 and S4). This is presumably due to the low number of available sequences, in addition to their high level of overall identity to one another (>91% at the nucleotide level). However, it is notable that CYP115 from Mesorhizobium sp. AA22, which is tightly integrated 5′ to the GA biosynthetic operon (i.e., resembling that in the more distant phytopathogens), appears to be that most closely related to the ancestral rhizobial CYP115 (Figure 3 and Supporting Information Figures S3 and S4).
Figure 3.
Phylogeny of CYP115 in rhizobia. Maximum likelihood tree analysis (1000 bootstraps) of rhizobial full-length CYP115 nucleotide sequences from a codon-based alignment (MUSCLE). The generation of this phylogenetic tree is described in the Supporting Information Methods. See Supporting Information Figure S4 for trees generated with different algorithms.
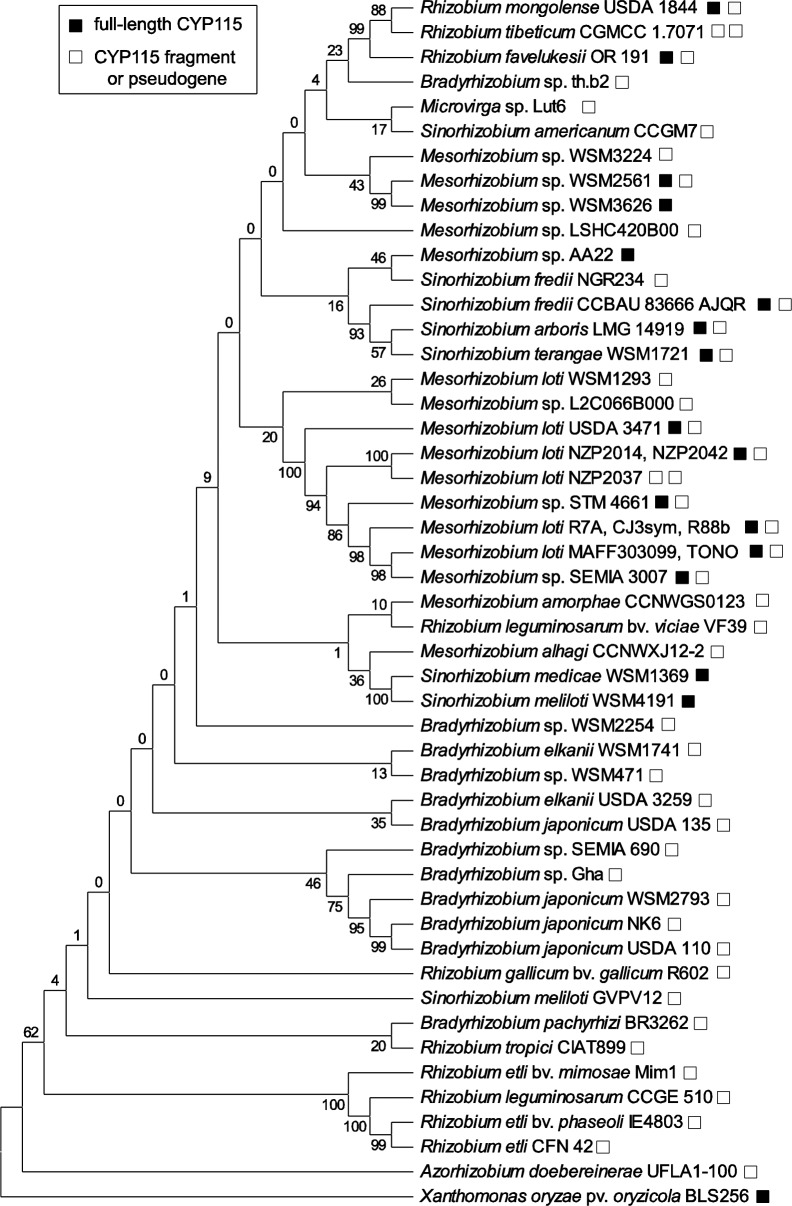
To further evaluate the scattered distribution of CYP115 among rhizobia, the phylogeny of the core GA operon was examined via alignments of individual operon genes (e.g., CPS and CYP114) from all (123 total) of the currently identified GA operon-containing rhizobia (Figure 4 and Supporting Information Figure S5). This revealed that the full-length CYP115-containing rhizobia fall into several distinct clades that also include strains without a full-length CYP115, suggesting that these lineages gained CYP115 independently of each another. In addition, all of these strains were evaluated for the presence of a CYP115 pseudogene or gene fragment, as described for the originally reported copy of the operon from B. japonicum USDA110.18 This analysis revealed that nearly every strain of rhizobia in which the core GA operon is found also contains a CYP115 fragment tightly appended 5′ to CYP112 (Figure 4 and Supporting Information Figure S5). Strikingly, this also is true for most rhizobia that contain a full-length CYP115, with one notable exception being Mesorhizobium sp. AA22, where the full-length CYP115 is tightly appended to the core operon.
Figure 4.
Scattered distribution of CYP115 full-length genes and gene fragments within GA operon-containing rhizobia. Shown is a representative cladogram (maximum likelihood algorithm, 1000 bootstraps) from codon-based alignment (MUSCLE) of CPS from the GA operon with the CPS from the phytopathogen X. oryzae pv. oryzicola BLS236 included as an outgroup (see Supporting Information Figure S6 for a comprehensive phylogenetic tree depicting all GA operon-containing rhizobia with nonfixed branch lengths). The presence of multiple open boxes indicates multiple distinct CYP115 fragments within a strain’s genome.
Collectively, our results suggest that most rhizobia with the extant, full-length CYP115 acquired this gene in HGT events independent from their acquisition of the core GA operon. Such separate acquisition of CYP115 would represent an additional layer of HGT underlying GA biosynthesis in rhizobia. It is known that symbiotic plasmids and genomic islands are exchanged via HGT among species within the Rhizobiales order.16 Furthermore, the GA operon itself seems to undergo separate HGT within these symbiotic plasmids/islands, as indicated by both their distinct GC content and phylogenetic relationships.6 Therefore, additional HGT of CYP115 within rhizobia that also contain the core GA operon adds a third layer of HGT within the Rhizobiales order and provides a means by which rhizobia can acquire the final biosynthetic step to convert 1, the penultimate, nonbioactive product of the core GA operon, into 2, a bioactive GA.
Consistent with a role in production of GA, CYP115 appears to be coexpressed with the core GA operon in rhizobia, regardless of relative genomic location. In particular, numerous studies have shown selective expression of the core operon during rhizobial symbiosis.3,12,20−26 Additionally, at least in M. loti MAFF303099, CYP115 transcripts and proteins have also been demonstrated to be upregulated during symbiosis.12,25 Thus, rhizobia containing CYP115 may directly produce bioactive 2 rather than just 1. Notably, this includes M. loti MAFF303099, where the core GA operon was shown to lead to production of 1.3 While this was suggested to be further transformed by GA3ox from the host plant,3 CYP115 is clearly expressed during symbiosis,25 and based upon our characterization of this enzyme as a GA3ox, this strain is capable of direct production of 2.
The presence of a CYP115 fragment at the 5′ end of the GA core operon in almost all rhizobia suggests that this gene was lost soon after rhizobial acquisition of the operon. Given the apparent ability to reacquire a functional full-length CYP115 via independent HGT, the limited distribution of CYP115 suggests that GA3ox activity by rhizobia may be deleterious in most rhizobial-legume symbiotic relationships. It has recently been reported that GA production by M. loti MAFF303099 affects nodulation, presumably to limit nodule numbers and, thus, competition within individual host plants.3 Additionally, it has been shown that X. oryzae pv. oryzicola, which contains CYP115, can produce 2(27) and that this production contributes to the virulence of this rice pathogen via reduction of the jasmonic acid-mediated defense response.17 Given that bioactive GA production is advantageous to phytopathogens, it may be that production of 2 by rhizobia is deleterious to host legume defense. To ameliorate this effect, it is possible that rhizobia are under selective pressure to lose CYP115 and leave control of the final step in bioactive GA production to the host plant. The continued presence of CYP115 in some rhizobia may then reflect a lack of such GA3ox expression in nodules by their host legume species. Although CYP115 seems to be enriched in Mesorhizobium species, there is no obvious correlation between the rhizobial strains that contain a full-length CYP115 and their host plants, leaving this hypothesis on a speculative basis.
Methods
Heterologous expression of CYP115 was performed much as previously described for the other CYPs from the GA operon.4 Phylogenetic analyses were carried out using MEGA7.28 Detailed materials and methods are available in the Supporting Information.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM109773) and National Science Foundation (CHE-1609917) to R.J.P.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.6b01038.
Supporting Methods, Figures S1–S6, and Tables S1–S5 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Hedden P.; Thomas S. G. (2012) Gibberellin biosynthesis and its regulation. Biochem. J. 444, 11–25. 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- MacMillan J. (2002) Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 20, 387–442. 10.1007/s003440010038. [DOI] [PubMed] [Google Scholar]
- Tatsukami Y.; Ueda M. (2016) Rhizobial gibberellin negatively regulates host nodule number. Sci. Rep. 6, 27998. 10.1038/srep27998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett R. S.; Montanares M.; Marcassa A.; Lu X.; Nagel R.; Charles T. C.; Hedden P.; Rojas M. C.; Peters R. J. (2017) Elucidation of gibberellin biosynthesis in symbiotic rhizobia reveals convergent evolution. Nat. Chem. Biol. 13, 69–74. 10.1038/nchembio.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone D.; Chambers J.; Lowry L.; Kim G.; Anterola A.; Bender K.; Peters R. J. (2009) Gibberellin biosynthesis in bacteria: Separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett. 583, 475–480. 10.1016/j.febslet.2008.12.052. [DOI] [PubMed] [Google Scholar]
- Hershey D. M.; Lu X.; Zi J.; Peters R. J. (2014) Functional conservation of the capacity for ent-kaurene biosynthesis and an associated operon in certain rhizobia. J. Bacteriol. 196, 100–106. 10.1128/JB.01031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez C.; Baginsky C.; Hedden P.; Gong F.; Carú M.; Rojas M. C. (2014) Gibberellin oxidase activities in Bradyrhizobium japonicum bacteroids. Phytochemistry 98, 101–109. 10.1016/j.phytochem.2013.11.013. [DOI] [PubMed] [Google Scholar]
- Nishijima T.; Koshioka M.; Yamazaki H. (1994) Use of several gibberellin biosynthesis inhibitors in sensitized rice seedling bioassays. Biosci., Biotechnol., Biochem. 58, 572–573. 10.1271/bbb.58.572. [DOI] [Google Scholar]
- Ueguchi-Tanaka M.; Ashikari M.; Nakajima M.; Itoh H.; Katoh E.; Kobayashi M.; Chow T. Y.; Hsing Y. I.; Kitano H.; Yamaguchi I.; Matsuoka M. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- Nakajima M.; Shimada A.; Takashi Y.; Kim Y. C.; Park S. H.; Ueguchi-Tanaka M.; Suzuki H.; Katoh E.; Iuchi S.; Kobayashi M.; Maeda T.; Matsuoka M.; Yamaguchi I. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46, 880–889. 10.1111/j.1365-313X.2006.02748.x. [DOI] [PubMed] [Google Scholar]
- Göttfert M.; Röthlisberger S.; Kündig C.; Beck C.; Marty R.; Hennecke H. (2001) Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 183, 1405–1412. 10.1128/JB.183.4.1405-1412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiumi T.; Ohwada T.; Itakura M.; Mitsui H.; Nukui N.; Dawadi P.; Kaneko T.; Tabata S.; Yokoyama T.; Tejima K.; Saeki K.; Omori H.; Hayashi M.; Maekawa T.; Sriprang R.; Murooka Y.; Tajima S.; Simomura K.; Nomura M.; Suzuki A.; Shimoda Y.; Sioya K.; Abe M.; Minamisawa K. (2004) Expression islands clustered on the symbiosis island of the Mesorhizobium loti genome. J. Bacteriol. 186, 2439–2448. 10.1128/JB.186.8.2439-2448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret X.; Freiberg C.; Rosenthal A.; Broughton W. J.; Fellay R. (1999) High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mol. Microbiol. 32, 415–25. 10.1046/j.1365-2958.1999.01361.x. [DOI] [PubMed] [Google Scholar]
- González V.; Bustos P.; Ramírez-Romero M. a; Medrano-Soto A.; Salgado H.; Hernández-González I.; Hernández-Celis J. C.; Quintero V.; Moreno-Hagelsieb G.; Girard L.; Rodríguez O.; Flores M.; Cevallos M. a; Collado-Vides J.; Romero D.; Dávila G. (2003) The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments. Genome Biol. 4, R36. 10.1186/gb-2003-4-6-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J. T.; Trzebiatowski J. R.; Brown S. D.; Elliot R. M.; Fleetwood D. J.; Mccallum N. G.; Stuart G. S.; Weaver J. E.; Webby R. J.; de Bruijn F. J.; Ronson C. W.; Cruickshank R. W.; Gouzy J.; Rossbach U. (2002) Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184, 3086–3095. 10.1128/JB.184.11.3086-3095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U.; Hochhut B.; Hentschel U.; Hacker J. (2004) Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424. 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- Lu X.; Hershey D. M.; Wang L.; Bogdanove A. J.; Peters R. J. (2015) An ent-kaurene-derived diterpenoid virulence factor from Xanthomonas oryzae pv. oryzicola. New Phytol. 206, 295–302. 10.1111/nph.13187. [DOI] [PubMed] [Google Scholar]
- Tully R. E.; van Berkum P.; Lovins K. W.; Keister D. L. (1998) Identification and sequencing of a cytochrome P450 gene cluster from Bradyrhizobium japonicum. Biochim. Biophys. Acta, Gene Struct. Expression 1398, 243–255. 10.1016/S0167-4781(98)00069-4. [DOI] [PubMed] [Google Scholar]
- Hedden P.; Phillips A. L.; Rojas M. C.; Carrera E.; Tudzynski B. (2001) Gibberellin biosynthesis in plants and fungi: A case of convergent evolution?. J. Plant Growth Regul. 20, 319–331. 10.1007/s003440010037. [DOI] [PubMed] [Google Scholar]
- Pessi G.; Ahrens C. H.; Rehrauer H.; Lindemann A.; Hauser F.; Fischer H.-M.; Hennecke H. (2007) Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol. Plant-Microbe Interact. 20, 1353–1363. 10.1094/MPMI-20-11-1353. [DOI] [PubMed] [Google Scholar]
- Chang W.-S.; Franck W. L.; Cytryn E.; Jeong S.; Joshi T.; Emerich D. W.; Sadowsky M. J.; Xu D.; Stacey G. (2007) An oligonucleotide microarray resource for transcriptional profiling of Bradyrhizobium japonicum. Mol. Plant-Microbe Interact. 20, 1298–1307. 10.1094/MPMI-20-10-1298. [DOI] [PubMed] [Google Scholar]
- Li Y.; Tian C. F.; Chen W. F.; Wang L.; Sui X. H.; Chen W. X. (2013) High-resolution transcriptomic analyses of Sinorhizobium sp. NGR234 bacteroids in determinate nodules of Vigna unguiculata and indeterminate nodules of Leucaena leucocephala. PLoS One 8, e70531. 10.1371/journal.pone.0070531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar E.; Díaz-Mejía J. J.; Moreno-Hagelsieb G.; Martínez-Batallar G.; Mora Y.; Mora J.; Encarnación S. (2010) Characterization of the NifA-RpoN regulon in Rhizobium etli in free life and in symbiosis with Phaseolus vulgaris. Appl. Environ. Microbiol. 76, 4510–4520. 10.1128/AEM.02007-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J. T.; Brown S. D.; Ronson C. W. (2013) The NifA-RpoN regulon of Mesorhizobium loti strain R7A and its symbiotic activation by a novel LacI/GalR-family regulator. PLoS One 8, e53762. 10.1371/journal.pone.0053762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsukami Y.; Nambu M.; Morisaka H.; Kuroda K.; Ueda M. (2013) Disclosure of the differences of Mesorhizobium loti under the free-living and symbiotic conditions by comparative proteome analysis without bacteroid isolation. BMC Microbiol. 13, 180. 10.1186/1471-2180-13-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma A. D.; Emerich D. W. (2005) Global protein expression pattern of Bradyrhizobium japonicum bacteroids: A prelude to functional proteomics. Proteomics 5, 4170–4184. 10.1002/pmic.200401296. [DOI] [PubMed] [Google Scholar]
- Nagel R., Turrini P. C. G., Nett R. S., Leach J. E., Verdier V., Van Sluys M.-A., and Peters R. J. (2017) An operon for production of bioactive gibberellin A4 phytohormone with wide distribution in the bacterial rice leaf streak pathogen Xanthomonas oryzae pv. New Phytol. Published Online January 30, 2017, DOI: 10.1111/nph.14441 (accessed January 30, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.; Stecher G.; Tamura K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.