Abstract
The innate immune system represents the first line of defense during infection and is initiated by the detection of conserved microbial products by germline-encoded pattern recognition receptors (PRRs). Sensing through PRRs induces broad transcriptional changes that elicit powerful inflammatory responses. Tight regulation of these processes depends on multiple regulatory checkpoints including noncoding RNA species such as microRNAs. In addition, long noncoding RNAs (lncRNAs) have recently gained attention as important regulators of gene expression acting through versatile interactions with DNA, RNA, or proteins. As such, these RNAs have a multitude of mechanisms to modulate gene expression. Here, we summarize recent advances in this rapidly moving and evolving field, highlighting the contribution of lncRNAs to both the development and activation of innate immune cells. In addition, we discuss experimental approaches required to comprehensively investigate the function of a candidate noncoding RNA locus.
Keywords: (long) non-coding RNA, innate immunity, inflammation, gene expression, immune regulation
Introduction
The innate immune system is a multicomponent system that involves extensive gene regulation in order to elicit antimicrobial defenses and establish immune homeostasis [1]. Rapid changes in gene expression are a major component of this dynamic response that lead to the production of cytokines, chemokines and additional immune mediators to establish an acute inflammatory response [2]. Immune gene expression must be tightly coordinated, regulated and scaled in order to prevent autoinflammatory and autoimmune diseases and immune mediated damage of the host. A multitude of transcriptional and post-transcriptional regulators have been identified allowing both the magnitude and duration of these events to be carefully measured (reviewed in [3]).
In recent years, the complexity of eukaryotic transcriptomes has become the subject of intense scrutiny and curiosity. The finding that the vast majority of transcription in mammalian cells originates from noncoding, rather than protein-coding DNA elements, is one of the biggest surprises of the post-genomic era [4]. RNA-sequencing from diverse tissues and in diverse biological contexts has revealed that at least 75% of the genome is transcribed, whereas only about 2% encodes for proteins [5]. Most of these non-coding transcripts are larger than 200 nucleotides and are thus referred to as long noncoding RNAs (lncRNAs). lncRNAs are expressed in cell-type specific manners, and many are differentially expressed during cell differentiation or activation [4, 6]. It is becoming increasingly evident that immune cells express specific lncRNAs that execute important regulatory functions in the development or activation status of these cells.
lncRNAs are the largest class of noncoding transcripts
Non protein-coding RNAs (ncRNAs) comprise many different species that can broadly be categorized into small noncoding RNAs and lncRNAs, using 200 nucleotides as an arbitrary cutoff value. The concept of RNAs as regulatory molecules is well established for many years, especially for small RNAs. Classic regulatory functions include splicing (small nuclear RNAs), nuclear organization (small nuclear and small nucleolar RNAs), transposon silencing (piwi-interacting RNAs) or inhibition of mRNA translation (microRNAs). The latest release of the GENECODE project (www.genegodegenes.org) indicates that lncRNAs are the largest class of ncRNAs with approximately 16,000 identified in humans and 9,000 in mice. How many of these transcripts have RNA-mediated physiological however is currently unknown. lncRNAs are usually classified based on their genomic localization and orientation relative to protein coding genes and can be either long intergenic noncoding RNAs (lincRNAs) (between protein coding genes), [6], intronic (within introns of protein-coding genes), antisense to protein coding genes (natural antisense transcripts, NATs [7]), or transcribed from divergent bidirectional promoters [8, 9].
Most lncRNAs are transcribed by RNA polymerase II (RNAPII), spliced and modified with a 5′cap and a poly-A tail, all of which makes them largely indistinguishable from mRNAs [10, 11]. In contrast to mRNAs, lncRNAs however lack protein-coding potential. A combination of computational approaches (open reading frame size, codon substitution frequencies) and experimental techniques (in vitro translation, association with polysomes, ribosome footprinting in combination with RNA-Seq) are applied to interrogate the protein-coding capacity of these transcripts. The cellular compartment of a specific transcript can also be a helpful indication of protein coding capacity, since a predominantly cytoplasmic RNA has a higher likelihood of protein coding capacity than a strictly nuclear molecule. Even for genes annotated as lncRNAs, it is critical that their protein coding capacity is tested experimentally, since it has been shown that some lncRNAs encode small peptides as their functional elements [12]. This critical assessment of the coding capacity of novel transcripts is equally important for uncharacterized transcripts from pathogens. As an example, a recent study found novel translated open reading frames in the human cytomegalovirus (hCMV) genome outside of annotated protein-coding genes using ribosome footprinting. As a convincing measure for the “real-life” presence of those proteins in the context of a human infection in vivo, the authors could detect a T-cell response directed against one of these proteins which was previously annotated as a lncRNA. This immune response was only present in CMV-experienced T cells, but absent when testing T cells from CMV-negative donors [13].
lncRNAs regulate gene expression via a multitude of mechanisms
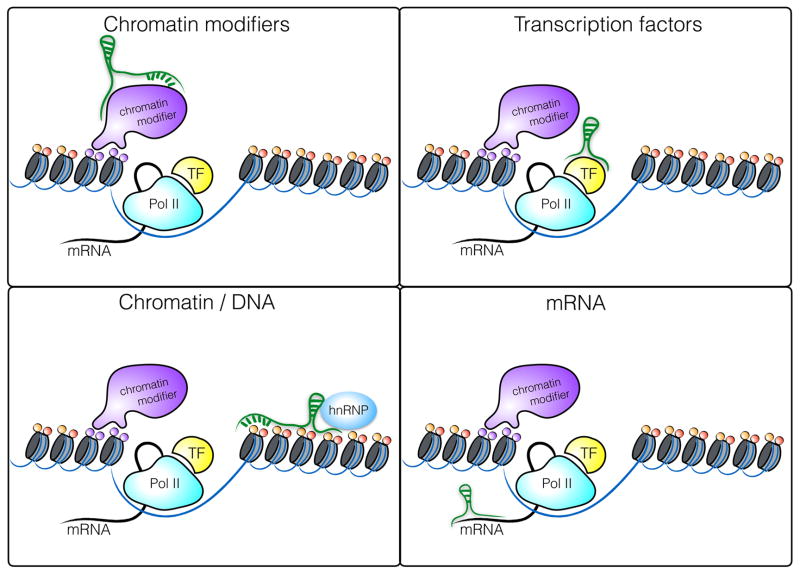
The molecular mechanisms by which lncRNAs modulate gene expression are diverse and have been reviewed extensively [14, 15]. lncRNAs can act in cis to regulate the expression of nearby genes on the same allele or in trans to regulate genes at other genomic locations across the genome. In general, lncRNAs function through interactions with DNA, RNA or proteins, where the formation of ribonucleoproteins is by far the most important interaction for mediating functional roles (Figure 1) lncRNAs can regulate gene expression at the level of transcription, RNA processing or translation. As mentioned above, the subcellular localization of a given lncRNA provides important clues to its potential mode of action. A large percentage of lncRNAs are localized in the nucleus where they alter the transcription of target genes through interaction with transcription factors, chromatin modifying complexes or heterogenous ribonucleoprotein complexes (hnRNPs), a class of nuclear RNA-binding proteins interacting with precursor mRNAs. One of the best studied nuclear lincRNAs, Xist, is a 17 kb lincRNA transcribed from the inactive X chromosome involved in the silencing of the inactive X chromosome in female cells. Xist recruits Polycomb Repressive Complex 2 (PRC2) through interaction with several nuclear proteins [16] to mark chromatin with repressive histone modifications that block transcription of target genes by exclusion of RNA Polymerase II across the inactivated X chromosome [17, 18]. HnRNPs are important functional partners for numerous lincRNAs including hnRNPK, which associates with lincRNA-p21 [19, 20], hnRNPU which binds Xist and Firre [21, 22], hnRNPL which binds THRIL [23] and hnRNPA2/B1 which binds lincRNA-Cox2 [24].
Figure 1.
Molecular mechanisms of gene regulation by lncRNAs in the nucleus. lncRNAs can interact with chromatin modifying complexes, transcription factors, the chromatin itself or mRNA. With respect to protein interaction, heterogenous ribonucleoprotein complexes (hnRNPs) have emerged as one of the most important protein families interacting with lncRNAs.
In the cytosol, lncRNAs have been shown to control the stability of target mRNAs. An example of such a mechanism in the immune system is iNOS-AS, an antisense RNA, which is encoded on the antisense strand and corresponding to the 3′UTR of the iNOS mRNA. iNOS and iNOS-AS are both regulated in IL1β-treated hepatocytes. Gene perturbation experiments with overexpression and knockdown approaches revealed that the AS RNA molecule stabilized iNOS mRNA by binding its 3′UTR [25]. Conversely, cytoplasmic antisense lncRNAs can have the opposite effect when direct complementarity to their target mRNA leads to gene silencing in both Dicer-dependent and independent manners [4, 26, 27].
Role of lncRNAs during innate immune cell development
lncRNAs have initially been studied in the context of development and cancer, but accumulating evidence also supports important roles for these non-coding RNAs in both the innate and adaptive immune system. Deep sequencing approaches have identified lncRNAs in virtually all immune cells including macrophages [24], dendritic cells [6], T-cells [28, 29] and B-cells [30]. While the expression of these lncRNAs are differentially regulated during the differentiation and/or activation of these cells, examples of lncRNAs with defined biological roles in immune cells are only beginning to emerge [31]. Several lncRNAs have been identified which control the differentiation of hematopoietic cells. For example, the differentiation of granulocytes is partly mediated by HOX antisense intergenic RNA myeloid 1 (HOTAIRM1), an antisense lncRNA within the HOXA gene locus. Knockdown of HOTAIRM1 abrogated retinoic acid-dependent activation of HOXA1/A2 and CD11b and CD18 (Mac-1), two beta2 integrin transcripts that are hallmark myeloid maturation-associated genes [32, 33]. Human dendritic cell (DC) differentiation has also been shown to depend on lncRNA activity. An RNA sequencing study of DC maturation in humans and mice identified a cluster of lncRNAs whose expression correlated with DC maturation [34]. One of these lncRNAs, lnc-DC was upregulated during human DC development and highly expressed in Lin−MHCII+CD11c+ conventional DCs, but absent from plasmacytoid DCs, monocytes or other leukocyte subsets. Lentiviral knockdown of lnc-DC during human DC development had broad functional consequences including impaired expression of surface receptors critical for T cell activation (CD80/86, HLA-DR, CD40), impairment in antigen uptake by monocyte-derived DCs and decreased IL-12 production after stimulation with LPS. Mechanistically, lnc-DC was shown to regulate STAT3 activity, a critical regulator of DC maturation. lnc-DC bound directly to STAT3 in the cytoplasm, preventing STAT3 binding to and dephosphorylation by SHP1. This resulted in maintenance of STAT3 phosphorylation in the presence of lnc-DC. It is worth mentioning that a later study suggested that the murine lnc-DC actually encodes a small secreted protein called Wdnm1-like and therefore further studies are required to better understand the role of lnc-DC in murine DC differentiation [35]. To understand the dynamic expression of lncRNAs during lymphoid development, a recent study performed RNA-Seq with de-novo assembly of the lncRNA transcriptome during human B and T cell development and could identify several thousand novel lncRNAs. Interestingly, the lncRNA transcriptome had a higher specificity for a given lymphocyte lineage and its differentiation stage than the protein-coding transcriptome [30]. Together, these studies suggest that lncRNAs impact immune cell differentiation.
Regulation of immune effector functions through lncRNAs
Besides lncRNAs regulating the differentiation of immune cells, there is growing evidence that lncRNAs also impact gene expression programs in activated immune cells. For example, several lncRNAs have been identified which alter the activity of transcription factors crucial for innate and adaptive immune cell activation. Lethe (Rps15a-ps4) was identified in mouse embryonic fibroblasts (MEF) treated with TNFα and other stimuli, and it;s induction was shown to depend on NF-κB signaling [36]. Lethe in turn binds the NF-κB subunit p65 (RelA) preventing it from directing transcription of IL-6, IL-8 and SOD2. Lethe therefore functions as a negative feedback regulator of the NF-κB signaling pathway to limit proinflammatory signaling. lncRNAs also interact with transcription factors important in T-cells. The lncRNA NRON negatively regulates T cell activation through its interaction with nuclear factor of activated T-cells (NFAT). NFAT is localized to the cytoplasm and imported into the nucleus in response to calcium-dependent signaling. NRON inhibits the nuclear accumulation of NFAT either by binding to nuclear transport factors, or by sequestering inactive NFAT in the cytosol [6, 37].
While lncRNAs that bind transcription factors have the capacity to alter the expression of multiple target genes in immune cells, lncRNAs also modulate the expression of individual immune genes such as cytokines by a multitude of additional mechanisms. Several distinct lncRNAs modulate the levels of the interleukin-1 family of pro-inflammatory cytokines. One example is a NAT that is antisense to IL-1β [38]. In response to LPS signaling this IL-1β NAT is upregulated. Using ectopic expression approaches the authors showed that overexpression of this NAT decreased H3K4me3 histone marks at the IL-1β promoter, resulting in decreased RNA polymerase II (RNAPII) occupancy and reduced transcription of IL-1β. Additional lncRNAs have also been described which regulate the IL-1 β locus. LPS-induced mRNA transcription of IL-1β and CXCL8 were attenuated upon knockdown of two lncRNAs derived from the enhancer region surrounding the IL-1β (IL1β-eRNA) locus [38]. Beyond IL-1 β, IL-1α expression is also regulated by lncRNAs: Antisense IL-1α AS- IL1α RNA shows similar expression patterns with the IL-1α protein coding gene, with which it partially overlaps. Using loss of function shRNA approaches AS- IL-1α was shown to be essential to facilitate IL-1α gene transcription. Knockdown of AS- IL-1α by RNA interference compromised the recruitment of RNAPII to the promoter of IL-1α and as a result decreased levels of IL-1α mRNA in macrophages exposed to LPS [39].
Just like many inflammatory genes, many lncRNAs are only expressed in innate immune cells following their activation, while other lncRNAs that are abundantly expressed are downregulated when cells are exposed to inflammatory stimuli [24]. The LPS-TLR4 axis, which leads to activation of NF-κB and interferon regulatory factors (IRFs), induces and represses large numbers of lncRNAs in macrophages [6, 24]. Amongst the top regulated lncRNA genes in macrophages exposed to LPS or TLR2 ligands was lincRNA-Cox2 (also known as Ptgs2os2). This RNA was named due to its genomic organization proximal to the protein-coding gene Ptgs2, which encodes for cyclooxygenase-2 (Cox-2) [6]. lincRNA-Cox2 is induced via a canonical TLR signaling pathway involving the adaptor protein MyD88 and NF-κB [24]. Loss (shRNA) and gain (retroviral overexpression) of function studies revealed that lincRNA-Cox2 broadly regulated the expression of inflammatory response genes. lincRNA-Cox2 was shown to repress expression of a large number of immune genes through its interactions with hnRNPA2/B1 and hnRNP-A/B(Figure 2). In addition, this lincRNA-Cox2 facilitated the inducible expression of a distinct cluster of immune response genes including pro-inflammatory cytokines and other inflammatory mediators. In addition to its role in macrophages, lincRNA-Cox2 is also regulated downstream of NF-kB in epithelial cells [40]. Similar to what was observed in macrophages, knockdown of lincRNA-Cox2 resulted in reprogramming of the gene expression profile in intestinal epithelial cells exposed to TNF-α. In particular lincRNA-Cox2 appears to repress the transcription of IL12b, lincRNA-Cox2 mediates these effects via its interactions with the Mi-2/nucleosome remodeling and deacetylase (Mi- 2/NuRD) repressor complex, which this lincRNA appears to guide to the Il12b promoter region. These data provide mechanistic insight into the role of lincRNA-Cox2 in promoting epigenetic modulation of cytokine genes and identify lincRNA-Cox2 as a novel regulator of both macrophage and intestinal epithelial inflammatory responses.
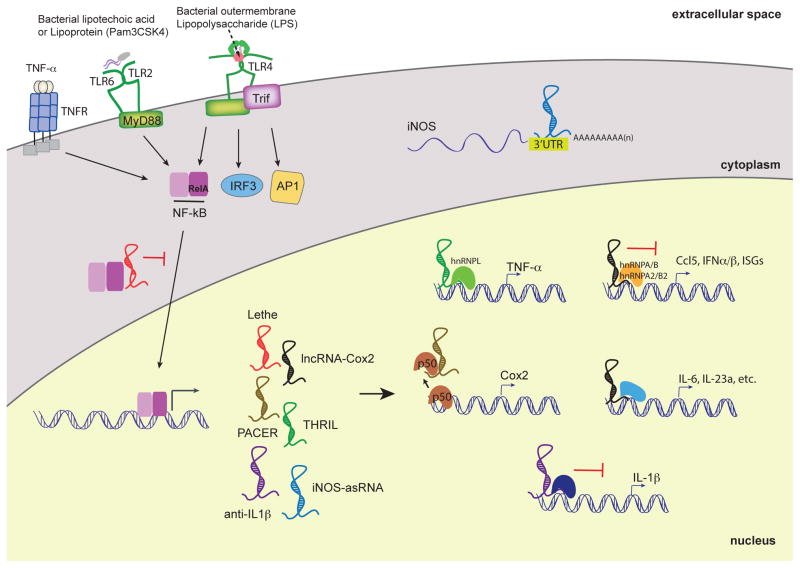
Figure 2.
LncRNAs functions in TLR-dependent inflammatory responses in myeloid cells. lncRNAs are induced by PRRs such as TLRs and TNFR. Upon PRR activation, the transcription factor NF-κB translocates to the nucleus and induces the expression of various lncRNAs. Their proposed functions are either nuclear, where they can activate or repress the transcription of pro-inflammatory cytokines and chemokines. Lethe binds to RelA, a subunit of NF-kB, to sequester and prevent binding to NF-kB-target promoters. PACER binds to the p50 subunit of NF-kB and modulates the basal levels of Cox2. Anti-IL1β inhibits IL-1β transcription via chromatin remodeling. LincRNA-Cox2 possesses suppressive functions by interacting with hnRNP-A/B and A2/B1 and activating functions through yet-to-be discovered mechanisms. THRIL regulates TNFα expression through its interaction with hnRNP-L. iNOS-asRNA, mediates its cis-activity is in the cytoplasm and stabilizes the iNOS mRNA transcript. In order to more thoroughly understand lncRNA-dependent innate immunoregulatory functions, the interferon pathway mediated by IRFs and the MAP kinase pathway will need to be more deeply explored.
The genomic locus encoding Ptgs2 (Cox2) in mice encodes a second lncRNA called Cox2-divergent (Ptgs2 opposite strand; Ptgs2os) [24, 36]. The Cox2-divergent lncRNA is located at the 5′-end of Ptgs2 (non-overlapping), and is transcribed on the opposite (negative) DNA strand [36]. Although functions of Cox2-divergent are as yet unclear, it is highly inducible in mouse embryonic fibroblasts (MEF) exposed to TNFα and LPS [36]. Additionally, another lncRNA PACER also exists in this genomic region in humans [41]. PACER (p50-associated COX-2 extragenic RNA) is exclusively involved in controlling the expression of COX-2 in cis in human monocyte-macrophage cell lines, and primary human mammary epithelial cells [41]. PACER expression is regulated by the chromatin-boundary/insulator factor CTCF, which establishes an open chromatin domain in the upstream region of COX-2 to promote PACER expression. In turn, PACER binds the NF-κB homodimer p50/p50 (a transcriptional repressor complex) and titrates it away from the COX-2 promoter. These events then favor the recruitment of the active NF-κB heterodimer p65/p50, which promotes the assembly of the transcription pre-initiation complex containing the histone acetyltransferase p300, and RNA polymerase II at the COX-2 promoter. Therefore, PACER appears to function by occluding the repressor complex (p50/p50) to facilitate the expression of COX-2.
Like lincRNA-Cox2, THRIL (TNFα and hnRNPL related immunoregulatory lncRNA) is another inducible lncRNA shown to function in part via its interaction with hnRNPL [23]. This lncRNA was identified in the THP-1 human monocyte cell line and was amongst 159 lncRNAs that were differentially expressed upon activation with Pam3CSK4 treatment. THRIL loss-of- function (shRNA) studies revealed that THRIL contributes to the inducible expression of the pro-inflammatory cytokine mediators TNF-α and IL-6 upon Pam3CSK4 stimulation. Further supporting a role for THRIL in immune gene regulation, chromatin immunoprecipitation (ChIP) experiments indicated that heterogenous ribonucleoprotein (hnRNP)-L localized to the TNF-α promoter upon Pam3CSK4 stimulation (Figure 2).
A very different mechanism by which lncRNAs can induce inflammatory responses seems to be a direct inflammatory response directed against the ssRNA. This has recently been demonstrated by transfection of in vitro transcribed lncRNAs into myeloid cells, which led to a strong induction of proinflammatory cytokines[42]. Interestingly, this response showed some specificity since lncRNAs which are specifically upregulated in tumor cells induced a strong cytokine response, whereas other physiologically expressed lncRNAs did not. A bioinformatic motif anaylsis showed that tumor-associated lncRNAs had a higher frequency of immunostimulatory motifs such as CpG dinucleotides. It is tempting to speculate if tumor-associated lncRNAs can represent a physiological DAMP which can evoke an antitumor immune response. The finding that lncRNAs might evoke an RNA-driven inflammatory response in myeloid cells is also important from an experimental perspective, especially, when observing proinflammatory responses in myeloid cells after ectopic overexpression of a lncRNA.
lncRNAs as novel regulators of antimicrobial defense
Several reports recently revealed new roles for lncRNAs as regulators of antimicrobial functions. As an example, the lncRNA nuclear enriched abundant transcript 1 (NEAT1), is induced by herpes simplex virus and influenza virus infection. Mechanistically, NEAT1 was shown to mediate remodeling of specialized heterochromatin structures called paraspeckles. By relocating a repressor protein called SFPQ from the IL-8 promoter to these paraspeckles NEAT1 promoted IL-8 synthesis. Moreover, NEAT1 seems to not only regulate the immune response of the host, but also directly affect viral replication, as knockdown of NEAT1 in HIV-infected T cells increased HIV replication, possibly through export of unspliced HIV-1 mRNA from the nucleus to the cytoplasm [43].
lncRNAs with antiviral functions also play a role in antimicrobial defenses in epithelial cells. Microarray profiling from human alveolar epithelial cells (A549) infected with influenza virus showed that the lncRNA negative regulator of antiviral response (NRAV) was amongst the most significantly downregulated lncRNAs following infection with various viruses, including influenza virus [44]. Ectopic overexpression of NRAV in vivo worsened the disease course as shown by increased weight loss and decreased overall survival. In line with these observations, NRAV gain-of-function in vivo led to increased viral replication and relatively reduced expression levels of MxA, IFITM3 and other ISGs. Similar observations were made in mice infected with Sendai Virus (SeV), Muscovy Duck Revirus, and Herpes Simplex Virus. Although located in both nuclear and cytoplasmic compartments, NRAV was shown to have limited effects on ISG mRNA stability in the cytosol, and most likely regulated ISG expression through histone modifications - ChIP analysis at target gene loci showed a decreased H3K4me3 signature, a marker of active transcription, and increased H3K27me3, which reflects transcriptional repression.
Utilizing comparative genomics to map disease susceptibility loci in two mouse strains that differed in their resistance to Theiler’s murine encephalomyelitis virus infection also showed the importance of lncRNAs as regulators of antiviral defense in vivo. Comparing polymorphisms between the SJL/J and B10.S mouse strains led to the identification of a susceptibility locus that was mapped to a conserved lncRNA called NeST (nettoie Salmonella pas Theiler’s). This locus conferred enhanced clearance of Theiler’s virus but increased susceptibility to Salmonella infection [45]. NeST, originally described as Tmevpg1, was shown to control the expression of the IFN-γ gene through histone modifications. NeST acts in cis as an enhancer RNA by binding to WDR5, a component of the histone 3 lysine 4 (H3K4) methyltransferase complex, that in turn alters histone 3 methylation at the IFN-γ locus.
Dissecting a lncRNA locus – a multifaceted approach
Comprehensive investigation of a genomic noncoding RNA locus requires multiple complimentary approaches, since the biological function can be attributed to the genomic DNA, the act of transcription, the RNA molecule itself – or a combination of these mechanisms. It is therefore crucial to design experimental strategies that allow one to distinguish these possibilities, and to interpret observed phenotypes in the context of the chosen perturbation strategy. Ablation of lncRNA function by genetic deletion of a genomic locus has been used to demonstrate that deletion of noncoding loci can have in vivo consequences. Sauvaugeau et al. deleted 18 individual lncRNA genes, by replacing the genomic locus of each lncRNA with a LacZ reporter cassette so that the locus remained transcriptionally active. This led to peri- and postnatal lethality in three of these animals (Fendrr, Peril, Mdgt), and developmental abnormalities in two others (lnc-Brn1b and lnc-Pint) [46]. While these approaches are useful and likely to yield novel insights into physiological roles for lncRNAs, it is critical to perform rescue experiments to test whether the observed phenotypes are due to deficiency of the RNA molecule itself or rather a consequence of the deletion of a regulatory DNA element such as an enhancer [47]. A caveat to this complementation approach is that for lncRNAs which function strictly in cis, it is thought that it may be impossible to rescue biological effects by ectopic expression since these lncRNAs may need to be induced from a specific genomic location. In comparison to large deletions, insertion of transcriptional terminators such as polyadenylation termination signals directly after the transcription start site have the advantage of more limited impact on the DNA locus despite a complete loss of the transcript.
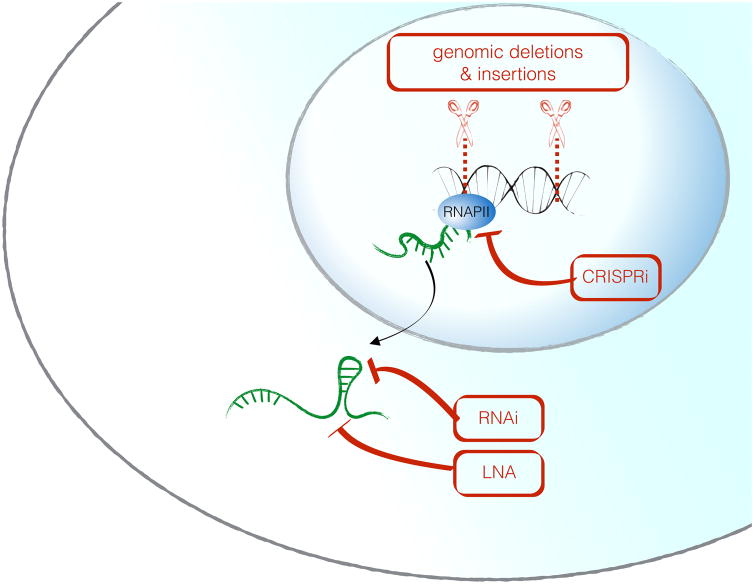
Loss-of-function approaches to directly test the functional role of the RNA molecule itself include RNA interference [48] or the use of chemically modified antisense oligos with higher stability called locked nucleic acids (LNA) [49]. Adaptions of the CRISPR/Cas9 technology called CRISPRi and CRISPRa can also be useful to manipulate lncRNA expression from their endogenous loci: a catalytically inactive Cas9 protein (dCas9) coupled either to a transcriptional repressor (CRISPRi), or a transcriptional activator protein (CRISPRa) can be used to specifically inhibit or activate transcription of lncRNAs from their endogenous loci [50, 51]. In summary, none of the experimental perturbation techniques alone are sufficient to fully characterize a lncRNA, and combinatorial approaches are strongly recommended. An overview of these approaches is shown in Figure 3 and Table 1.
Figure 3. Perturbation approaches for the functional characterization of a candidate lncRNA locus.
Whereas RNA-centered methods (RNAi, LNA) specifically perturb the RNA molecule without affecting the DNA locus, genomic deletions are powerful loss-of function approaches, but can not differentiate between DNA-mediated and RNA-mediated functions of a given gene. CRISPRi is a technique where a nuclease-dead Cas9 protein is coupled to a transcriptional repressor and thereby mediates gene silencing.
Table 1.
Different loss-of function strategies for the dissection of a lncRNA locus. Affected elements of the gene by a specific perturbation method are indicated by (+)
| perturbation | DNA | transcription | RNA | protein |
|---|---|---|---|---|
| whole gene ablation | + | + | + | + |
| reporter gene insertion | + | − | + | + |
| CRISPRi | (+) | + | + | + |
| RNAi | − | − | + | + |
| LNA | − | − | + | + |
Concluding Remarks
Advances in deep sequencing have revealed the enormous complexity of the mammalian genome. Despite clear evidence that lncRNAs are expressed and regulated during immunity our knowledge of the mechanisms and scope of lncRNA-mediated regulation of immune gene expression is still in its infancy. There have been several elegant mechanistic studies providing new insights into immune gene regulation by lncRNAs. However, the vast majority of lncRNAs in in the immune system remain largely uncharacterized. The challenge going forward will be to identify functional lncRNAs regulating innate immune responses and to define their molecular mode of action. More globally, it will be important to identify specific sequences and domains that execute the biological functions of lncRNAs. Ideally, this could lead to a classification of lncRNAs based on function, rather than their genomic locations relative to protein-coding genes. The multifarious toolbox of CRISPR-Cas9 mediated manipulation of lncRNAs and their genomic loci in immune cells will certainly facilitate efforts to test the functional relevance of candidate lncRNA loci, and to differentiate RNA-mediated from DNA-mediated functions [52].
Finally, the vast majority (>90%) of disease-associated single nucleotide polymorphisms (SNPs) are located in noncoding DNA elements, and a functional understanding of these loci to disease contribution is urgently needed [38, 53]. It is likely, that genetic variation could affect the expression or function of non-coding RNAs, which can have far-reaching implications for a variety of inflammatory diseases [38]. We believe that the functional and molecular characterization of lncRNA loci that regulate the immune response has enormous potential for the development of novel diagnostic and therapeutic approaches to treat immune diseases.
Acknowledgments
This work was supported by a postdoctoral fellowship the German Research Foundation (DFG) to R.E. (790/1-1), as well as by grants from the NIH (A1095213, a T32 Training Grant) to J.C and (AI067497 and AI115448) to K.A.F.
Footnotes
Conflict of interest
The authors declare no financial or commercial conflict of interest.
References
- 1.Janeway CA, Jr, Medzhitov R. Introduction: the role of innate immunity in the adaptive immune response. Semin Immunol. 1998;10:349–350. doi: 10.1006/smim.1998.0142. [DOI] [PubMed] [Google Scholar]
- 2.Smale ST, Tarakhovsky A, Natoli G. Chromatin contributions to the regulation of innate immunity. Annu Rev Immunol. 2014;32:489–511. doi: 10.1146/annurev-immunol-031210-101303. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 4.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinger ME, Amaral PP, Mercer TR, Mattick JS. Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Brief Funct Genomic Proteomic. 2009;8:407–423. doi: 10.1093/bfgp/elp038. [DOI] [PubMed] [Google Scholar]
- 6.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li K, Ramchandran R. Natural antisense transcript: a concomitant engagement with protein-coding transcript. Oncotarget. 2010;1:447–452. doi: 10.18632/oncotarget.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 9.Wu X, Sharp PA. Divergent transcription: a driving force for new gene origination? Cell. 2013;155:990–996. doi: 10.1016/j.cell.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009;10:973–982. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, Olson EN. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McHugh CA, Chen C-K, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015 doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, Jacks T. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19:469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, Rinn JL. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O’Neill LA, Moore MJ, Caffrey DR, Fitzgerald KA. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsui K, Nishizawa M, Ozaki T, Kimura T, Hashimoto I, Yamada M, Kaibori M, Kamiyama Y, Ito S, Okumura T. Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology. 2008;47:686–697. doi: 10.1002/hep.22036. [DOI] [PubMed] [Google Scholar]
- 26.Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 27.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, Zhu J, Zhao K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013;14:1190–1198. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S, Gruarin P, Provasi E, Sugliano E, Marconi M, De Francesco R, Geginat J, Bodega B, Abrignani S, Pagani M. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol. 2015;16:318–325. doi: 10.1038/ni.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casero D, Sandoval S, Seet CS, Scholes J, Zhu Y, Ha VL, Luong A, Parekh C, Crooks GM. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nature immunology. 2015 doi: 10.1038/ni.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol Med. 2014;20:623–631. doi: 10.1016/j.molmed.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayadas TN, Cullere X. Neutrophil beta2 integrins: moderators of life or death decisions. Trends Immunol. 2005;26:388–395. doi: 10.1016/j.it.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 35.Dijkstra JM, Ballingall KT. Non-human lnc-DC orthologs encode Wdnm1-like protein. F1000Res. 2014;3:160. doi: 10.12688/f1000research.4711.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 38.Kumar V, Westra HJ, Karjalainen J, Zhernakova DV, Esko T, Hrdlickova B, Almeida R, Zhernakova A, Reinmaa E, Vosa U, Hofker MH, Fehrmann RS, Fu J, Withoff S, Metspalu A, Franke L, Wijmenga C. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet. 2013;9:e1003201. doi: 10.1371/journal.pgen.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan J, Atianand M, Jiang Z, Carpenter S, Aiello D, Elling R, Fitzgerald KA, Caffrey DR. Cutting Edge: A Natural Antisense Transcript, AS-IL1alpha, Controls Inducible Transcription of the Proinflammatory Cytokine IL-1alpha. J Immunol. 2015;195:1359–1363. doi: 10.4049/jimmunol.1500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong Q, Gong A-Y, Zhang X, Lin C, Ma S, Chen J, Hu G, Chen X-M. LincRNA-Cox2 modulates TNF-α–induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi-2/NuRD-mediated epigenetic histone modifications. The FASEB Journal. 2015 doi: 10.1096/fj.15-279166. fj. 15-279166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. Elife. 2014;3:e01776. doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanne A, Muniz LR, Puzio-Kuter A, Leonova KI, Gudkov AV, Ting DT, Monasson R, Cocco S, Levine AJ, Bhardwaj N. Distinguishing the immunostimulatory properties of noncoding RNAs expressed in cancer cells. Proceedings of the National Academy of Sciences. 2015;112:15154–15159. doi: 10.1073/pnas.1517584112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio. 2013;4:e00596–00512. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao GF, Wang G, Chen JL. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe. 2014;16:616–626. doi: 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, Liapis SC, Mallard W, Morse M, Swerdel MR, D’Ecclessis MF, Moore JC, Lai V, Gong G, Yancopoulos GD, Frendewey D, Kellis M, Hart RP, Valenzuela DM, Arlotta P, Rinn JL. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, Herrmann BG. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci U S A. 2010;107:22196–22201. doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]