ABSTRACT
Candida guilliermondii shows intrinsic reduced echinocandin susceptibility. It harbors two polymorphisms (L633M and T634A) in the Fks1p hot spot 1 region. Our objective was to confirm that the reduced echinocandin susceptibility of C. guilliermondii is due to those naturally occurring substitutions. We constructed a Saccharomyces cerevisiae mutant in which a region of the FKS1 gene (including hot spot 1) was replaced with that from C. guilliermondii. The chimeric mutants showed 32-fold increases in echinocandin MIC values, confirming the hypothesis.
KEYWORDS: Candida guilliermondii, FKS mutations, echinocandin resistance, molecular mechanism
TEXT
Echinocandins are the treatment of choice for candidemia, since most Candida spp. are susceptible to these drugs (1–4). However, some highly prevalent Candida spp., such as species of the Candida parapsilosis complex and Candida guilliermondii, show natural reduced echinocandin susceptibility (RES) (5–7). Echinocandin drugs interact with the Fksp subunits of the β-1,3-glucan synthase complex, inhibiting the biosynthesis of the principal cell wall glucan. Clinical resistance was linked to hot spot mutations in Fksp (5). It was molecularly confirmed that the C. parapsilosis sensu lato RES phenotype is due to a naturally occurring substitution (P660A) in the hot spot 1 region of Fks1p (8). C. guilliermondii harbors two polymorphisms (L633M and T634A) in the same Fks1p region; it was assumed that the RES in this species is attributable to these amino acid substitutions, but the hypothesis was never confirmed (5).
The objective of this work was to verify that the intrinsic RES of C. guilliermondii is due to the naturally occurring amino acid changes in the hot spot 1 region of Fks1p. To reach this goal, we constructed a Saccharomyces cerevisiae strain with a hybrid FKS1 gene. A region of the FKS1 gene that includes hot spot 1 was replaced with that from C. guilliermondii. We then evaluated the echinocandin susceptibility.
S. cerevisiae BY4742 was used as the parental strain to obtain S. cerevisiae LMDM 537 (BY4742 FKS1 deletant in which codons 453 to 649 were replaced with URA3, i.e., fks1Δ453–649::URA3) (8). C. guilliermondii ATCC 6260 (Meyerozyma guilliermondii) was used to obtain the 695-nucleotide (nt) region of the FKS1 gene, which was fused to generate the chimeric FKS1 gene (S. cerevisiae-C. guilliermondii). DNA was extracted by phenol-based extraction (9). Caspofungin (CSF) and anidulafungin (ANF) susceptibility tests were performed following CLSI guidelines (10–12), with modifications. For S. cerevisiae susceptibility evaluations, RPMI 1640 broth and Mueller-Hinton 2% glucose-methyl blue agar were replaced by YPD broth (1% yeast extract, 2% peptone, 2% glucose) and YPD agar (YPD broth plus 1.5% agar), respectively. Also, the incubation temperature was changed to 30°C. These modifications were necessary since S. cerevisiae strains did not grow properly with the CLSI-proposed culture media and temperature (8).
A two-step method for PCR-based FKS1 mutagenesis was employed to generate a S. cerevisiae FKS1 chimeric mutant (8, 13, 14). Briefly, the first step consisted of partial deletion of the FKS1 gene of S. cerevisiae BY4742 using an URA3 cassette. The deletion included the Fks1p hot spot 1 region (amino acid residues 453 to 649). This disruption leads to FK506 (tacrolimus) and echinocandin hypersensitivity (8, 15). The transformation was performed and the deletion was confirmed using methodologies described previously (8, 16). The second step of the method was the replacement of the partially deleted S. cerevisiae FKS1 gene (fks1Δ453–649::URA3) by a construction that included a 695-nt portion of the C. guilliermondii FKS1 gene (nt 1591 to nt 2286 of the sequence reported in GenBank under accession number XM_001487741.1) surrounded by S. cerevisiae FKS1 regions from nt 1123 to nt 1611 and from nt 2308 to nt 2548 (S. cerevisiae FKS1 sequence reported in GenBank under accession number U12893.1). This construction was designed to yield, after homologous recombination, a functional FKS1 gene encoding a chimeric Fks1p (S. cerevisiae fks1Δ537–762::C. guilliermondii fks1 531–762) (Fig. 1). The transformation vector was obtained by four PCRs, including amplification of the C. guilliermondii FKS1 fragment (695 nt), the 488-nt portion of the 5′ region of S. cerevisiae FKS1, and the 240 nt of the 3′ fragment of S. cerevisiae FKS1 using the primer pairs Fus5-FKS1-Sc-CguiF/Fus3-FKS1-Sc-CguiR, Fus5-FKS1-Sc-CguiR/FKS1-375, and Fus3-FKS1-Sc-CguiF/LMDM85, respectively. The last PCR was designed to fuse the three fragments using FKS1-375 and LMDM85 primers (see Table 1 for primer details). PCRs were performed in a 25-μl volume, following the manufacturer's instructions for the Pegasus DNA polymerase (PBL, Buenos Aires, Argentina), in an Applied Biosystems thermocycler (Tecnolab-AB, Buenos Aires, Argentina). The construction was cloned into a PGEM-T Easy vector (Promega; Biodynamics, Buenos Aires, Argentina) to yield the plasmid LMDM-P113. S. cerevisiae LMDM 537 (BY4742 fks1Δ453–649::URA3) was transformed with 1 μg of the LMDM-P113 plasmid, following the protocol (lithium acetate) described by Geitz et al. (17). Transformants were selected on FK506 plates and screened for the loss of URA3 as described previously (8). The RES phenotype was screened for using YPD agar with 0.25 μg/ml CSF. Seven chimeric mutants showing the expected phenotype, i.e., uracil auxotrophic, FK506 resistant, and able to grow in the presence of 0.25 μg/ml CSF, were obtained. The FKS1 reconstitution was assessed in all seven mutants by PCR using the primers FKS1-305F and Cg/h-R. These primers hybridize to the S. cerevisiae FKS1 gene (around 900 nt upstream of the start codon) and C. guilliermondii FKS (near hot spot 1). The expected PCR band of 872 nt would be obtained only if the chimeric FKS1 gene was present (Fig. 1). The FKS1 reconstitution was confirmed by sequencing. A revertant control strain (LMDM 537R) was obtained by transforming a 1,000-bp PCR fragment into the S. cerevisiae LMDM 537 strain (17). This fragment was amplified using S. cerevisiae BY4742 DNA and the primers FKS1-375 and FKS1-707R (Table 1). The amplified fragment included the S. cerevisiae FKS1 hot spot 1 region. Homologous recombination produced a strain with a functional FKS1 that was FK506 resistant.
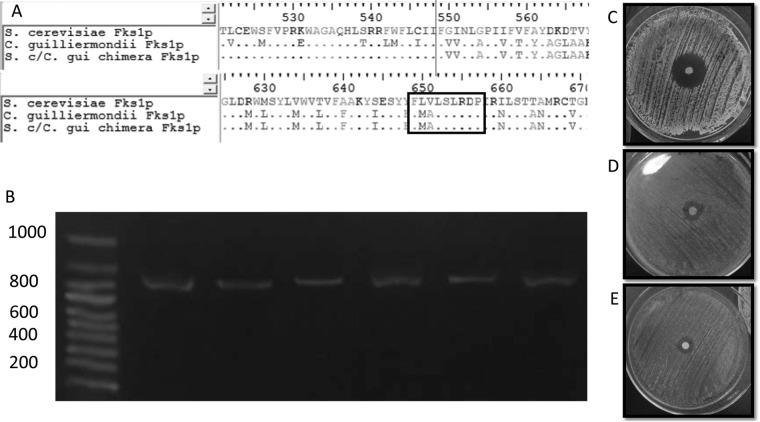
FIG 1.
(A) Clustal alignment of the deduced Fks1p sequences of S. cerevisiae BY4742, S. cerevisiae harboring the chimeric FKS1, and C. guilliermondii ATCC 6260. Black lines show where the C. guilliermondii FKS1 fragment was fused with the S. cerevisiae gene. The black box shows the FKS1 hot spot 1 region. (B) PCR confirmation of the FKS1 reconstitution. Lane 1, markers; lanes 2 to 7, 872-nt PCR fragment of the chimeric FKS1 gene obtained by using the FKS1-305F and Cg/h-R primers and DNA isolated from uracil-auxotrophic and CSF- and FK506-resistant S. cerevisiae transformants. (C, D, and E) Diffusion susceptibility testing using caspofungin disks, following CLSI guidelines (11), for S. cerevisiae BY4742 (C), S. cerevisiae harboring the chimeric FKS1 (D), and C. guilliermondii ATCC 6260 (E).
TABLE 1.
Primers used in this study
| Primer | Target organism | Orientation (5′→3′) | Sequence (5′→3′) | Purpose | Reference |
|---|---|---|---|---|---|
| FKS1-453-URA3F | S. cerevisiae | Sense | AAAGAGACCCGTACTTGGTTACATTTGGTCACCAACTTCAGAGTGCACCATACCACAGCT | Generation of S. cerevisiae FKS1-deleted mutant | 8 |
| FKS1-649-URA3R | S. cerevisiae | Antisense | GTATTCACCTGTACACCTCATTGCAGTGGTGGACAAAATTGGTATTTCACACCGCATAGG | Generation of S. cerevisiae FKS1-deleted mutant | 8 |
| URA3iR | S. cerevisiae | Antisense | TGCCTTTAGCGGCTTAACTG | Confirmation of S. cerevisiae FKS1 deletion | 8 |
| FKS1-375 | S. cerevisiae | Sense | GGTCGTTTTGTCAAGCGTGA | Confirmation of S. cerevisiae FKS1 deletion, chimeric FKS1 construction, and S. cerevisiae FKS1 reconstruction | 8 |
| FKS1-707R | S. cerevisiae | Antisense | ATTTCCCAACAGAGAAAATGG | S. cerevisiae FKS1 reconstruction | 8 |
| LMDM85 | S. cerevisiae | Antisense | CGTAGTGAGGAGTCAATACTGTG | Chimeric FKS1 construction | This study |
| Fus5-FKS1-Sc-CguiF | S. cerevisiae and C. guilliermondii | Sense | CGTAGATTCTGGTTTTTATGCATCATCTTCGTGGTTAACTTGGCCCC | Chimeric FKS1 construction | This study |
| Fus5-FKS1-Sc-CguiR | S. cerevisiae and C. guilliermondii | Antisense | GGGGCCAAGTTAACCACGAAGATGATGCATAAAAACCAGAATCTACG | Chimeric FKS1 construction | This study |
| Fus3-FKS1-Sc-CguiF | S. cerevisiae and C. guilliermondii | Sense | GTACAGAGAACATTTGTTGGCTATTGACCATGTACAAAAATTACTATATC | Chimeric FKS1 construction | This study |
| Fus3-FKS1-Sc-CguiR | S. cerevisiae and C. guilliermondii | Antisense | GATATAGTAATTTTTGTACATGGTCAATAGCCAACAAATGTTCTCTGTAC | Chimeric FKS1 construction | This study |
| FKS1-305F | S. cerevisiae | Sense | CCCTGGAAAGAGTTCGTCATATC | Confirmation of FKS1 chimeric reconstitution | This study |
| Cg/h-R | C. guilliermondii | Antisense | CAAACCACCCAAAGGCATAAC | Confirmation of FKS1 chimeric reconstitution | This study |
Parental and revertant S. cerevisiae strains (BY4742 and LMDM 537R, respectively) showed very low CSF and ANF MIC values (0.008 μg/ml), while the seven chimeric strains showed 32-fold higher MICs for both echinocandins (0.25 μg/ml). These values mimicked the C. guilliermondii ATCC 6260 RES phenotype (MICs of 0.25 μg/ml).
C. guilliermondii causes 1.4% of all Candida infections worldwide but is more prevalent in South America (>4%) (6, 18–20). Despite the RES phenotype, most of these infections are treated with echinocandins, since this yeast is also less susceptible to fluconazole than other Candida spp. (21). Most of these infections respond well to echinocandin therapy. However, C. guilliermondii breakthrough infections during caspofungin therapy have been reported (22). Under such circumstances, there is a concern that the use of echinocandins would contribute to an increased prevalence of this species.
It is well known that echinocandin clinical resistance is linked to FKS hot spot substitutions (5). The 32-fold increases in echinocandins MICs observed for the seven chimeric S. cerevisiae mutants support the assumption that L633M and T634A polymorphisms at C. guilliermondii Fks1p hot spot 1 are responsible for the observed RES in this species.
ACKNOWLEDGMENTS
This study was supported in part by a CONICET grant (grant PIP 2011-331) and an Universidad Nacional del Litoral CAI+D grant to G.G.-E. C.D., M.S.C., and F.L. have fellowships from CONICET, and D.M. has a fellowship from FONCYT.
REFERENCES
- 1.Cornely OA, Vazquez J, De Waele J, Betts R, Rotstein C, Nucci M, Pappas PG, Ullmann AJ. 2014. Efficacy of micafungin in invasive candidiasis caused by common Candida species with special emphasis on non-albicans Candida species. Mycoses 57:79–89. doi: 10.1111/myc.12104. [DOI] [PubMed] [Google Scholar]
- 2.Reboli AC, Rotstein C, Pappas PG, Chapman SW, Kett DH, Kumar D, Betts R, Wible M, Goldstein BP, Schranz J, Krause DS, Walsh TJ. 2007. Anidulafungin versus fluconazole for invasive candidiasis. N Engl J Med 356:2472–2482. doi: 10.1056/NEJMoa066906. [DOI] [PubMed] [Google Scholar]
- 3.Reboli AC, Shorr AF, Rotstein C, Pappas PG, Kett DH, Schlamm HT, Reisman AL, Biswas P, Walsh TJ. 2011. Anidulafungin compared with fluconazole for treatment of candidemia and other forms of invasive candidiasis caused by Candida albicans: a multivariate analysis of factors associated with improved outcome. BMC Infect Dis 11:261. doi: 10.1186/1471-2334-11-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vazquez J, Reboli AC, Pappas PG, Patterson TF, Reinhardt J, Chin-Hong P, Tobin E, Kett DH, Biswas P, Swanson R. 2014. Evaluation of an early step-down strategy from intravenous anidulafungin to oral azole therapy for the treatment of candidemia and other forms of invasive candidiasis: results from an open-label trial. BMC Infect Dis 14:97. doi: 10.1186/1471-2334-14-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perlin DS. 2015. Echinocandin resistance in Candida. Clin Infect Dis 61(Suppl 6):S612–S617. doi: 10.1093/cid/civ791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfaller MA, Boyken L, Hollis RJ, Kroeger J, Messer SA, Tendolkar S, Diekema DJ. 2008. In vitro susceptibility of invasive isolates of Candida spp. to anidulafungin, caspofungin, and micafungin: six years of global surveillance. J Clin Microbiol 46:150–156. doi: 10.1128/JCM.01901-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaller MA, Boyken L, Hollis RJ, Kroeger J, Messer SA, Tendolkar S, Jones RN, Turnidge J, Diekema DJ. 2010. Wild-type MIC distributions and epidemiological cutoff values for the echinocandins and Candida spp. J Clin Microbiol 48:52–56. doi: 10.1128/JCM.01590-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Effron G, Katiyar SK, Park S, Edlind TD, Perlin DS. 2008. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob Agents Chemother 52:2305–2312. doi: 10.1128/AAC.00262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sambrook J, Fritsch EF, Maniatis T. 1998. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard—3rd ed CLSI document M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2009. Method for antifungal disk diffusion susceptibility testing of yeast; approved guideline—2nd ed CLSI document M44-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2012. Reference method for broth dilution antifungal susceptibility testing of yeast; 4th informational supplement. CLSI document M27-S4. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Johnson ME, Katiyar SK, Edlind TD. 2011. New Fks hot spot for acquired echinocandin resistance in Saccharomyces cerevisiae and its contribution to intrinsic resistance of Scedosporium species. Antimicrob Agents Chemother 55:3774–3781. doi: 10.1128/AAC.01811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katiyar SK, Edlind TD. 2009. Role for Fks1 in the intrinsic echinocandin resistance of Fusarium solani as evidenced by hybrid expression in Saccharomyces cerevisiae. Antimicrob Agents Chemother 53:1772–1778. doi: 10.1128/AAC.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douglas CM, Foor F, Marrinan JA, Morin N, Nielsen JB, Dahl AM, Mazur P, Baginsky W, Li W, el-Sherbeini M. 1994. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-D-glucan synthase. Proc Natl Acad Sci U S A 91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edlind TD, Henry KW, Vermitsky JP, Edlind MP, Raj S, Katiyar SK. 2005. Promoter-dependent disruption of genes: simple, rapid, and specific PCR-based method with application to three different yeast. Curr Genet 48:117–125. doi: 10.1007/s00294-005-0008-3. [DOI] [PubMed] [Google Scholar]
- 17.Gietz RD, Schiestl RH, Willems AR, Woods RA. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 18.Cordoba S, Vivot W, Bosco-Borgeat ME, Taverna C, Szusz W, Murisengo O, Isla G, Davel G. 2011. Species distribution and susceptibility profile of yeasts isolated from blood cultures: results of a multicenter active laboratory-based surveillance study in Argentina. Rev Argent Microbiol 43:176–185. [DOI] [PubMed] [Google Scholar]
- 19.Pfaller M, Neofytos D, Diekema D, Azie N, Meier-Kriesche HU, Quan SP, Horn D. 2012. Epidemiology and outcomes of candidemia in 3648 patients: data from the Prospective Antifungal Therapy (PATH Alliance®) registry, 2004–2008. Diagn Microbiol Infect Dis 74:323–331. doi: 10.1016/j.diagmicrobio.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Pfaller MA, Diekema DJ, Mendez M, Kibbler C, Erzsebet P, Chang SC, Gibbs DL, Newell VA. 2006. Candida guilliermondii, an opportunistic fungal pathogen with decreased susceptibility to fluconazole: geographic and temporal trends from the ARTEMIS DISK antifungal surveillance program. J Clin Microbiol 44:3551–3556. doi: 10.1128/JCM.00865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pappas PG, Kauffman CA, Andes D, Benjamin DK Jr, Calandra TF, Edwards JE Jr, Filler SG, Fisher JF, Kullberg BJ, Ostrosky-Zeichner L, Reboli AC, Rex JH, Walsh TJ, Sobel JD. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis 48:503–535. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabbara N, Lacroix C, Peffault de Latour R, Socie G, Ghannoum M, Ribaud P. 2008. Breakthrough C. parapsilosis and C. guilliermondii blood stream infections in allogeneic hematopoietic stem cell transplant recipients receiving long-term caspofungin therapy. Haematologica 93:639–640. doi: 10.3324/haematol.11149. [DOI] [PubMed] [Google Scholar]