ABSTRACT
Enterococcus faecium is one of the primary causes of nosocomial infections. Disinfectants are commonly used to prevent infections with multidrug-resistant E. faecium in hospitals. Worryingly, E. faecium strains that exhibit tolerance to disinfectants have already been described. We aimed to identify and characterize E. faecium genes that contribute to tolerance to the disinfectant chlorhexidine (CHX). We used a transposon mutant library, constructed in a multidrug-resistant E. faecium bloodstream isolate, to perform a genome-wide screen to identify genetic determinants involved in tolerance to CHX. We identified a putative two-component system (2CS), composed of a putative sensor histidine kinase (ChtS) and a cognate DNA-binding response regulator (ChtR), which contributed to CHX tolerance in E. faecium. Targeted chtR and chtS deletion mutants exhibited compromised growth in the presence of CHX. Growth of the chtR and chtS mutants was also affected in the presence of the antibiotic bacitracin. The CHX- and bacitracin-tolerant phenotype of E. faecium E1162 was linked to a unique, nonsynonymous single nucleotide polymorphism in chtR. Transmission electron microscopy showed that upon challenge with CHX, the ΔchtR and ΔchtS mutants failed to divide properly and formed long chains. Normal growth and cell morphology were restored when the mutations were complemented in trans. Morphological abnormalities were also observed upon exposure of the ΔchtR and ΔchtS mutants to bacitracin. The tolerance to both chlorhexidine and bacitracin provided by ChtRS in E. faecium highlights the overlap between responses to disinfectants and antibiotics and the potential for the development of cross-tolerance for these classes of antimicrobials.
KEYWORDS: Enterococcus, bacitracin, biocides, chlorhexidine, disinfectants, tolerance
INTRODUCTION
Enterococcus faecium is a facultative anaerobic Gram-positive bacterium that naturally colonizes the gastrointestinal tract of humans and animals. Since the 1990s, E. faecium has also emerged as one of the leading causes of nosocomial infections (1, 2). The population of E. faecium is currently divided into clade A-1, containing most clinical isolates, clade A-2, with most animal-derived strains, and clade B, in which most isolates of healthy humans are clustered (3). Whether clade A-1 and clade A-2 are monophyletic and can be reliably distinguished from each other has recently been questioned (4). Nosocomial E. faecium strains are frequently resistant to glycopeptides and β-lactam antibiotics (5, 6), complicating the treatment of clinical infections. Since the late 1990s, and despite the worldwide spread of vancomycin-resistant enterococci (VRE), only two antibiotics (daptomycin and linezolid) have been approved by the FDA for use against VRE. Other antibiotics (quinupristin-dalfopristin, tigecycline, oritavancin, tedizolid, telavancin, and dalbavancin) have been suggested as alternatives for treatment of infections caused by VRE in clinical practice. They have, however, not been approved by the FDA for the treatment of VRE infections (7–9). The use of the polypeptide antibiotic bacitracin, in combination with other antibiotics, in the treatment of VRE infections has also been proposed (10).
While antibiotics are gradually losing their effectiveness against E. faecium, antiseptics and disinfectants are becoming increasingly important to prevent the spread of multidrug-resistant E. faecium in health care settings (11). Chlorhexidine (CHX) is a bisbiguanide agent and has diverse applications as a disinfectant for surfaces and as an antiseptic for topical applications (12). The mode of action of CHX is poorly understood. CHX, which is positively charged at neutral pH, is thought to be attracted to the bacterial cell surface, where it may electrostatically interact with negatively charged phospholipids. Depending on the concentration of CHX, it can reduce bacterial membrane fluidity or disrupt the structural integrity of the membrane, causing increased permeability and leakage of cell contents and, ultimately, cell death (13–15). In health care, CHX is often used in surgical scrubs for preoperative skin preparation, impregnated wash cloths for postoperative wound care, daily patient bathing, and oral care of intubated patients (16, 17). Regular bathing of patients with CHX significantly reduces the colonization by VRE and other multiresistant organisms in intensive care units and general medicine wards (18–23). Recently, increased tolerance to these compounds has been reported for Gram-positive cocci, and this could contribute to future co- or cross-selection for antibiotic resistance (24–30). In addition, subinhibitory concentrations of CHX induce the expression of genes involved in vancomycin and daptomycin resistance in enterococci (31).
In this study, we used microarray-based transposon mapping (M-TraM [32]) to perform a genome-wide screening of a transposon mutant library to identify genes involved in the tolerance to CHX in E. faecium. Two genes that were identified in the M-TraM screening were predicted to encode a two-component regulatory system (2CS). 2CSs are signal transduction systems in bacteria consisting of a sensor histidine kinase and its response regulator. They play important roles in the adaptation of bacteria to changes in the environment and have been implicated in orchestrating cellular responses that lead to increase of tolerance to antimicrobials in different Gram-positive bacteria, including enterococci (33–35). The two genes encoding the 2CS were further characterized to define their role in tolerance to CHX.
RESULTS
E. faecium strains from different phylogenetic backgrounds differ in their tolerance to CHX.
First, we assayed the tolerance to CHX of the E. faecium strain E1162, a multidrug-resistant bloodstream isolate, previously assigned to clade A-1 (3, 32), by measuring growth in Mueller-Hinton broth (MHB) supplemented with different concentrations of CHX (see Fig. S1 in the supplemental material). We found that at CHX concentrations of 1.7 μg ml−1 or higher, growth was essentially inhibited completely. In follow-up experiments, CHX was used at 1.2 μg ml−1, as this concentration led to an extended lag phase and lower growth rate of E. faecium E1162.
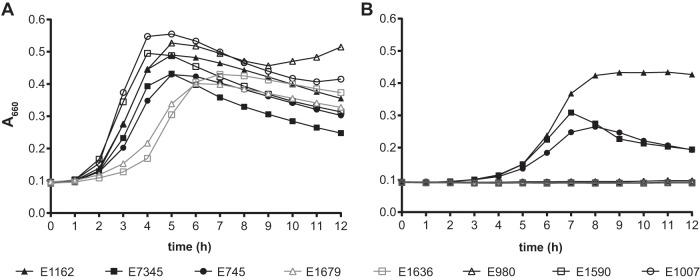
Next, we compared the abilities of seven other E. faecium strains (two strains from clade A-1, two strains from clade A-2, and three strains from clade B [3]) to grow in the presence of CHX (Fig. 1). We found that the strains from clade A-1, all of which were isolated from bloodstream infections in hospitalized patients, were able to grow in medium containing 1.2 μg ml−1 CHX, while the strains from clade A-2 or clade B could not (Fig. 1). E1162 had the highest growth rate in the presence of the disinfectant, and therefore it was chosen for the follow-up experiments into the mechanism of CHX tolerance in E. faecium.
FIG 1.
Growth of E. faecium strains challenged with CHX. Shown are the growth curves for different E. faecium strains in MHB alone (A) and MHB in the presence of 1.2 μg ml−1 CHX (B). Clade A-1 strains are indicated by solid black symbols, and clade A-2 and clade B are indicated by open gray and open black symbols, respectively. The growth curves represent the averages from three replicates.
Identification of a CHX tolerance locus in E. faecium E1162 by M-TraM.
We identified conditionally essential genes in the E. faecium E1162 transposon mutant library, during growth in the presence of 1.2 μg ml−1 CHX, through microarray-based transposon mapping (M-TraM) (32). While the M-TraM analysis hinted at a functional contribution of several genes in CHX tolerance (Table 1), we focused on the gene with locus tag EfmE1162_2203, of which the transposon mutant was significantly affected (8.7-fold lower abundance in the CHX-exposed library compared to the untreated library) during growth in the presence of the disinfectant. EfmE1162_2203 is annotated as encoding a DNA-binding response regulator. EfmE1162_2203 is located adjacent to a gene (Efm E1162_2202) encoding a histidine kinase of which the transposon mutant abundance was moderately (3.0-fold) reduced upon exposure to CHX. The proteins encoded by EfmE1162_2202 and EfmE1162_2203 likely form a 2CS in E. faecium E1162. We have renamed EfmE1162_2203 and EfmE1162_2202 chtR and chtS, for chlorhexidine tolerance response regulator and chlorhexidine tolerance sensor histidine kinase, respectively.
TABLE 1.
E. faecium genes implicated in tolerance to chlorhexidine by M-TraM analysisa
| Locus tagb | Gene name | Annotation | Avg fold changec |
|---|---|---|---|
| EfmE1162_2203 | chrR | DNA-binding response regulator | 8.7 |
| EfmE1162_0264 | Permease protein, putative | 4.6 | |
| EfmE1162_0996 | Hypothetical protein | 4.2 | |
| EfmE1162_2026 | Lactose phosphotransferase system repressor | 3.9 | |
| EfmE1162_0997 | Conserved hypothetical protein | 3.9 | |
| EfmE1162_2510 | Holliday junction DNA helicase RuvA | 3.8 | |
| EfmE1162_0300 | PrgW | 3.6 | |
| EfmE1162_2635 | Esterase | 3.6 | |
| EfmE1162_0431 | Conserved hypothetical protein | 3.2 | |
| EfmE1162_0394 | PrgO | 3.2 | |
| EfmE1162_2202 | chrS | Sensor histidine kinase | 3.0 |
Boldface type indicates the chtRS system targeted for further analysis.
The locus tag represents the gene containing the transposon insertion.
Fold change in expression of the gene as determined by a ratio of the library grown under the control condition to that under the CHX-challenged condition. Results were averaged from four replicates.
To determine the distribution of chtR and chtS among E. faecium strains, we assessed the presence of these two genes in all of the strains tested for CHX tolerance and in 85 previously published genome sequences, which were previously assigned to clade A-1 (n = 41), clade A-2 (n = 31), and clade B (n = 13) (3, 36). The analysis showed that the ChtRS 2CS is conserved in all analyzed E. faecium strains. By analyzing the nucleotide sequences of the chtR and chtS genes of the eight E. faecium strains tested for their tolerance to CHX (Fig. 1), a single nonsynonymous nucleotide change, leading to an amino acid substitution (P102H), was found in the ChtR protein of all CHX-tolerant clade A-1 strains, compared to the non-CHX-tolerant clade A-1 and B strains. This amino acid is located in a predicted dimerization interface located in the signal receiver domain of ChtR.
The ChtRS 2CS contributes to CHX and bacitracin tolerance.
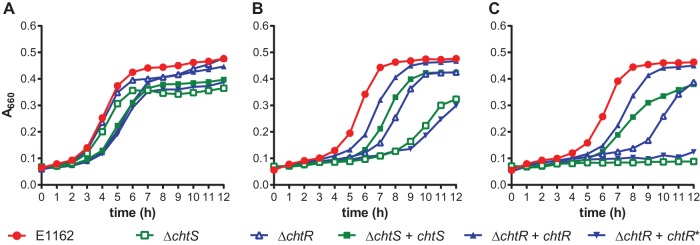
To validate the M-TraM results, we constructed chtR and chtS markerless deletion (ΔchtR and ΔchtS) mutants. We also constructed two strains in which these mutations were complemented in trans, which were named the ΔchtR + chtR and ΔchtS + chtS strains. No differences in growth were observed between E1162 and the two targeted mutants or the complemented strains when grown in MHB (Fig. 2A). In the presence of 1.2 μg ml−1 CHX, E1162 had a lag phase of approximately 4 h. Under the same conditions, the ΔchtR and ΔchtS mutants had a lag phase of almost 8 h and exhibited slower exponential growth than E1162 (Fig. 2B). The ΔchtR + chtR and ΔchtS + chtS complemented strains exhibited wild-type levels of growth in the presence of CHX. These results confirm that both chtR and chtS are involved in CHX tolerance of E. faecium E1162. When the in trans copy of chtR was mutated (resulting in chtR*) to engineer a proline at position 102 of ChtR, as is characteristic for ChtR in CHX-susceptible strains, it could no longer complement the growth defect caused by the chtR deletion.
FIG 2.
Effect of targeted mutations in the ΔchtR and ΔchtS mutants upon challenge with CHX and bacitracin. The growth curves shown in panel A correspond to strains growing in MH broth. Panels B and C correspond to the same strains growing in MH broth supplemented with 1.2 μg ml−1 CHX and 4 μg ml−1 bacitracin, respectively. Wild-type strain E1162 is shown in red, while the chtS and chtR targeted deletion mutants are shown in green and blue open symbols, respectively. Solid green and blue symbols represent the growth curves of the in trans complemented strains (the ΔchtS + chtS, ΔchtR + chtR, and ΔchtR + chtR* strains). The growth curves represent the averages from three experiments.
In addition, we decided to test the effect of the deletions in chrR and chrS on the tolerance of E. faecium E1162 to the antibiotic bacitracin, as a homologous 2CS (EF0926-EF0927) has previously been described to have a minor role in bacitracin tolerance in Enterococcus faecalis V583 (37). Upon exposure of the ΔchtR and ΔchtS mutants to 4 μg ml−1 bacitracin, the growth of both mutants was completely inhibited, while in the in trans complemented strains, growth was restored to near wild-type levels (Fig. 2C). The chtR deletion mutant complemented with the chtR* allele remained inhibited in its growth in the presence of bacitracin.
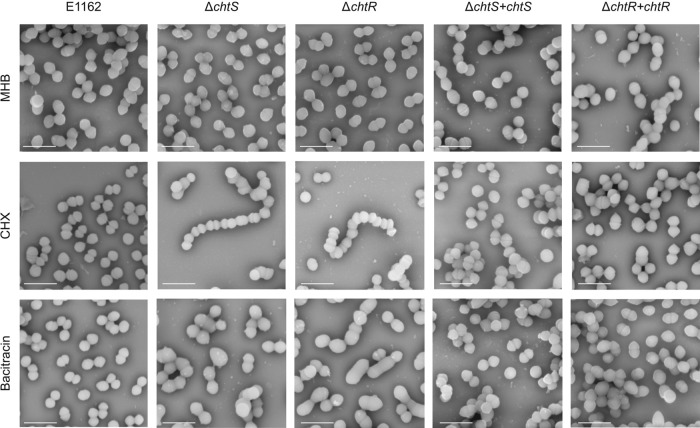
CHX and bacitracin challenge affects cellular morphology in the ΔchtR and ΔchtS mutants but not in E. faecium E1162.
In order to further characterize the effects of CHX and bacitracin on E. faecium E1162 and its chtR and chtS mutants, cells were analyzed by scanning electron microscopy (SEM) (Fig. 3). In these experiments, bacitracin was added to the medium at 1 μg ml−1, as this concentration is permissive for growth of the chtR and the chtS deletion mutants. No apparent changes in cellular morphology were found when E1162 was challenged with CHX or bacitracin, compared to growth in MHB. However, when the ΔchtS and ΔchtR mutants were challenged with CHX, the cells failed to divide properly and formed chains. Exposure of the ΔchtS and ΔchtR mutants to bacitracin resulted in swollen cells with various cellular abnormalities. The chaining phenotype and the cellular abnormalities found in CHX and bacitracin-challenged ΔchtS and ΔchtR mutants, respectively, were not observed in the ΔchtR + chtR and ΔchtS + chtS in trans complemented strains.
FIG 3.
Cell morphology of E. faecium E1162, the ΔchtS and ΔchtR mutants, and the ΔchtS + chtS and ΔchtR + chtR complemented strains upon exposure to CHX and bacitracin. E1162, the ΔchtS and ΔchtR mutants, and the ΔchtS + chtS and ΔchtR + chtR complemented strains were grown to an OD660 of 0.2. The cells were grown in Mueller-Hinton broth (MHB) alone, in MHB with 1.2 μg ml−1 chlorhexidine (CHX), or in MHB with 1 μg ml−1 bacitracin. Specimens were coated with 1-nm-diameter gold particles. Images were taken at a magnification of 35,000×. The scale bars correspond to 2 μm.
DISCUSSION
Enterococci have recently become important nosocomial pathogens (4). The ability of E. faecium to rapidly acquire drug resistance determinants threatens the treatment of infections caused by this organism. Antiseptics and disinfectants, including CHX, have been used for decades to prevent the spread of multidrug-resistant pathogens, including enterococci, in health care settings (19). In the present study, we found that clinical multidrug-resistant E. faecium isolates belonging to a distinct subpopulation of hospital-associated strains that are contained in clade A-1 were able to tolerate CHX, while E. faecium strains belonging to the other clades (A-2 and B) were more susceptible to CHX. The increased tolerance to CHX in clade A-1 strains, compared to strains from other E. faecium clades, may have been selected by the exposure to antiseptics and disinfectants, which are commonly used in health care settings. As clade A-1 strains appear to have specifically evolved to thrive in hospitalized patients, the increased tolerance to disinfectants may form an additional adaptation to this specific niche and could contribute to the success of these isolates as hospital-acquired opportunistic pathogens. Using M-TraM, we identified the 2CS ChtRS as being essential for CHX tolerance in the drug-resistant clinical isolate E. faecium E1162.
The chtS and chtR genes putatively encode a histidine kinase and a response regulator, together forming a 2CS. 2CSs regulate the expression of genes as a response to environmental cues (38–40). The signal is received at the extracellular sensor domain, and its transduction occurs via ATP-dependent phosphorylation. The phosphoryl group is transferred from the histidine phosphotransfer domain to the conserved signal receiver domain of the response regulator (41). The single amino acid substitution (P102H) that distinguishes ChtR in CHX-tolerant clade A-1 strains from ChtR in CHX-susceptible clade A-2 and clade B strains is located in the predicted dimerization interface of the REC domain in ChtR. The activation and regulation of response regulators by dimerization through receiver domains have previously been studied in other bacteria (42–44), including Gram-positive organisms (45), and changes in the dimerization interface of the signal receiver domain of ChtR could affect the function of the response regulator, thereby altering the control of gene expression upon CHX exposure by ChtRS. The inability of the construct with the mutated allele of chtR (encoding the ChtR protein with a proline residue at position 102) to complement a chtR deletion suggests a crucial role for this SNP in the CHX-tolerant phenotype of clade A-1 strains. Mechanistic studies of the proteins encoded by the different chtR alleles, including their dimerization and phosphotransfer characteristics, may be the topic of future studies.
In the present study, E1162 mutants deficient in chtRS exhibited decreased tolerance to CHX and, in addition, were more susceptible to bacitracin. In other Gram-positive bacteria, including E. faecalis, 2CSs also contribute to the protective response against low concentrations of bacitracin (46–50). Bacitracin is an antibiotic that targets peptidoglycan biosynthesis by binding to undecaprenol pyrophosphate (UPP), blocking its recycling during peptidoglycan synthesis. This, in turn, interferes with the transport of new peptidoglycan building blocks, leading to disruption of cell wall synthesis (51). Since 2CSs exert their effect through the regulation of expression of effector genes, the observed loss of CHX and bacitracin tolerance in the chtR and chtS deletion mutants is most likely due to altered expression of genes regulated by ChtRS. The effector genes regulated by ChtRS remain to be elucidated.
Bacitracin can decrease colonization by vancomycin-resistant E. faecium in the gastrointestinal tract (52, 53) and may be used orally to control the dissemination of vancomycin-resistant enterococci (10). However, therapeutic failure of bacitracin to treat VRE colonization has also been reported (54, 55) and may be caused by intrinsic bacitractin resistance in enterococci, to which ChtRS contributes in E. faecium. E. faecium has a multitude of intrinsic and acquired resistance mechanisms that allow it to survive the selective pressures imposed by the nosocomial environment, including antibiotic therapy and the use of disinfectants. Further studies should be performed to characterize the mechanisms by which different antiseptics and disinfectants may cross-select or coselect for clinically relevant antibiotics. Information resulting from this line of inquiry may be used to develop efficient disinfection protocols, while minimizing the risk of further resistance development in E. faecium.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table S1 in the supplemental material. Escherichia coli was grown in lysogeny broth (LB), and E. faecium strains were grown in Muller-Hinton broth (MHB) at 37°C with shaking at 200 rpm, unless mentioned otherwise. When appropriate, antibiotics were added at the following concentrations: spectinomycin at 300 μg ml−1 for E. faecium and 100 μg ml−1 for E. coli and gentamicin at 300 μg ml−1 for E. faecium and 25 μg ml−1 for E. coli. CHX was used at 1.2 μg ml−1, unless mentioned otherwise. Media were obtained from Oxoid (Basingstoke, United Kingdom). Antibiotics and disinfectants were obtained from Sigma-Aldrich (Saint Louis, MO).
M-TraM to identify genes involved in CHX tolerance.
The mariner transposon mutant library of E. faecium E1162 and the M-TraM method have been described previously (32). In brief, four overnight cultures of the E1162 mutant library were cultured at 37°C in MHB, diluted to an optical density at 660 nm (OD660) of 0.025 in 20 ml of prewarmed MHB supplemented with CHX, and then grown at 37°C until the mid-exponential phase (OD660 of 0.3). M-TraM was performed with four biological replicates following previously described procedures (32). Statistical analysis of hybridization signals between the conditions was performed using Cyber-T (56). Genes were considered differentially expressed when all four probes showed a Bayesian P value of <0.001 and the abundance of a gene was ≤0.2 or ≥5.0 compared to the untreated control (32).
Construction of markerless deletion mutant in chtS and chtR and in trans complementation.
chtS and chtR markerless deletion mutants were generated by previously described methods (32). Brain heart infusion broth or agar was used as the growth medium for E. faecium during all genetic manipulations. In brief, the 5′ and 3′ flanking regions (approximately 500 bp) of each gene were PCR amplified with two sets of primers as follows: Up-chtS_2202-F-XhoI and Up-chtS_2202-R-EcoRI for the upstream fragment of chtS and Down-chtS_2202-F-EcoRI and Down-chtS_2202-R-XmaI for the downstream fragment of chtS and Up-chtR_2203-F-XhoI and Up-chtR_2203-R-EcoRI for the upstream fragment of chtR and Down-chtR_2203-F-EcoRI and Down-chtR_2203-R-XmaI for the downstream fragment of chtR. The primer sequences are listed in Table S2 in the supplemental material. The two flanking regions of each gene were then fused together by PCR and cloned into pWS3 (57). A gentamicin resistance cassette flanked by lox66 and lox71 sites was amplified by PCR using the primers pAT392_EcoRI_lox66_genta_F and pAT392_EcoRI_lox71_genta_R and cloned into the EcoRI site that was generated between the 5′ and 3′ flanking regions of each gene in the pWS3 construct, as described previously (32). Finally, the two constructs, named pWJ1 and pWJ2, were electrotransformed individually into E. faecium E1162, and the chtS (ΔchtS) and chtR (ΔchtR) markerless deletion mutants were generated as described before (32).
For in trans complementation of the ΔchtS and ΔchtR mutants, the full-length genes and upstream regions of 356 and 390 nucleotides, respectively, were amplified by PCR using Accuprime high-fidelity Taq polymerase (Life Technologies, Bleiswijk, The Netherlands). The primers for these PCRs were named Comp2202_Fw_SacI and Comp2202_Rv_SmaI for the amplification of chtS and Comp2203_Fw_SacI and Comp2203_Rv_SmaI for the amplification of chtR. These primers introduce SacI and SmaI restriction sites, and after digestion with these enzymes, the resulting products were cloned into pEF25. The constructs were sequenced to confirm the absence of mutations and electrotransformed into the ΔchtS and ΔchtR mutants as described previously (32), generating the ΔchtS + chtS and ΔchtR + chtR complemented strains.
To determine the role of a nonsynonymous SNP in chtR, leading to a P102H amino acid substitution in ChtR in clade A-1 strains, we ordered the chtR gene of E1162 and its promoter as a genomic block (gBlock [Integrated DNA Technologies, Leuven, Belgium]) but made a specific base change leading to a proline residue at position 102 in the translated protein. The construct was otherwise identical (confirmed by sequencing) to the PCR product used to complement the ΔchtR mutant in E1162. SacI and SmaI sites at the end of the gBlock were used to clone the fragment into pEF25. The construct was then electrotransformed into the ΔchtR mutant, resulting in the ΔchtR + chtR* strain.
Growth inhibition assays.
The MIC of CHX was determined by broth microdilution in MHB, according to standard methodologies (58). MICs of CHX were not more than one dilution step different from each other for all strains in this study (data not shown). For this reason, we focused on kinetic growth assays, which provide more quantitative information than the endpoint measurements used in MIC determinations. Growth curves were determined using a BioScreen C instrument (Oy Growth Curves AB, Helsinki, Finland). Cultures of E. faecium E1162, the ΔchtR and ΔchtS mutants, and the in trans complemented strains were inoculated in MHB, with appropriate antibiotics, and incubated overnight at 37°C. Overnight cultures were then diluted to an OD660 of 0.0025 in 300 μl MHB and challenged with CHX or bacitracin. The cultures were incubated in the BioScreen C system at 37°C with continuous shaking. The absorbance at 660 nm (A660) was recorded every 15 min for 12 h. Each experiment was performed in triplicate.
SEM.
E1162 and the ΔchtS and ΔchtR mutants were grown overnight in MHB. Subsequently, they were diluted to an OD660 of 0.0025 in MHB and MHB supplemented with 1.2 μg ml−1 CHX or 1 μg ml−1 bacitracin and further grown until the OD660 reached 0.2. Bacteria were immediately fixed with 1% glutaraldehyde (Sigma) onto poly-l-lysine-coated glass slides and prepared for SEM, as previously described (59). In brief, the cells were serially dehydrated by consecutive incubations of 5 min in 25% ethanol in phosphate-buffered saline (PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 [pH 7.4]) and 50% ethanol in PBS, 75% and 90% ethanol, and twice in 100% ethanol, followed by 15- to 20-min incubations in 50% ethanol–hexamethyldisilazane (HMDS) and 100% HMDS. After overnight evaporation of HMDS at room temperature, samples were mounted onto specimen mounts and coated with 1-nm gold particles, using a Quorum Q150R sputter coater at 20 mA. Microscopy was performed using the Phenom PRO tabletop scanning electron microscope (PhenomWorld, Eindhoven, The Netherlands).
Accession number(s).
The microarray data generated in the M-TraM experiment have been deposited in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession no. E-MTAB-4173.
Supplementary Material
ACKNOWLEDGMENTS
We thank Antoni P. A. Hendrickx for the training he provided for scanning electron microscopy.
This work was supported by the European Union Seventh Framework Programme (FP7-HEALTH-2011-single-stage) “Evolution and Transfer of Antibiotic Resistance” (EvoTAR) under grant agreement no. 282004 and by an NWO-VIDI grant (917.13.357) to W.v.S. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The authors declare they have no competing financial interests.
A.M.G.P, R.J.L.W. and W.v.S. designed the study. A.M.G.P., J.W., J.C.B., M.R., E.C.B., E.M., and X.Z. performed experiments. J.R.B. contributed bioinformatic analyses. All authors contributed to data interpretation. The manuscript was written by A.M.G.P., M.J.M.B., R.J.L.W., and W.v.S.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02122-16.
REFERENCES
- 1.Gilmore MS, Lebreton F, van Schaik W. 2013. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol 16:10–16. doi: 10.1016/j.mib.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guzman Prieto AM, van Schaik W, Rogers MRC, Coque TM, Baquero F, Corander J, Willems RJL. 2016. Global emergence and dissemination of enterococci as nosocomial pathogens: attack of the clones? Front Microbiol 7:788. doi: 10.3389/fmicb.2016.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lebreton F, van Schaik W, Manson McGuire A, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJL, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raven KE, Reuter S, Reynolds R, Brodrick HJ, Russell JE, Török ME, Parkhill J, Peacock SJ. 2016. A decade of genomic history for healthcare-associated Enterococcus faecium, in the United Kingdom and Ireland. Genome Res 26:1388–1396. doi: 10.1101/gr.204024.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattoir V, Leclercq R. 2013. Twenty-five years of shared life with vancomycin-resistant enterococci: is it time to divorce? J Antimicrob Chemother 68:731–742. doi: 10.1093/jac/dks469. [DOI] [PubMed] [Google Scholar]
- 6.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linden P. 2007. Optimizing therapy for vancomycin-resistant enterococci (VRE). Semin Respir Crit Care Med 28:632–645. doi: 10.1055/s-2007-996410. [DOI] [PubMed] [Google Scholar]
- 8.Arias CA, Contreras GA, Murray BE. 2010. Management of multi-drug resistant enterococcal infections. Clin Microbiol Infect 16:555–562. doi: 10.1111/j.1469-0691.2010.03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Driscoll T, Crank C. 2015. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist 8:217–230. doi: 10.2147/IDR.S54125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tran TT, Palmer HR, Weimar MR, Arias CA, Cook GM, Murray BE. 2015. Oral bacitracin: a consideration for suppression of intestinal vancomycin-resistant enterococci (VRE) and for VRE bacteremia from an apparent gastrointestinal tract source. Clin Infect Dis 60:1726–1728. doi: 10.1093/cid/civ130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grare M, Dibama HM, Lafosse S, Ribon A, Mourer M, Regnouf-de-Vains J-B, Finance C, Duval RE. 2010. Cationic compounds with activity against multidrug-resistant bacteria: interest of a new compound compared with two older antiseptics, hexamidine and chlorhexidine. Clin Microbiol Infect 16:432–438. doi: 10.1111/j.1469-0691.2009.02837.x. [DOI] [PubMed] [Google Scholar]
- 12.Lim K-S, Kam PCA. 2008. Chlorhexidine—pharmacology and clinical applications. Anaesth Intensive Care 36:502–512. [DOI] [PubMed] [Google Scholar]
- 13.Hugo WB, Longworth AR. 1964. Some aspects of the mode of action of chlorhexidine. J Pharm Pharmacol 16:655–662. doi: 10.1111/j.2042-7158.1964.tb07384.x. [DOI] [PubMed] [Google Scholar]
- 14.Weinstein RA, Milstone AM, Passaretti CL, Perl TM. 2008. Chlorhexidine: expanding the armamentarium for infection control and prevention. Clin Infect Dis 46:274–281. doi: 10.1086/524736. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert P, Moore LE. 2005. Cationic antiseptics: diversity of action under a common epithet. J Appl Microbiol 99:703–715. doi: 10.1111/j.1365-2672.2005.02664.x. [DOI] [PubMed] [Google Scholar]
- 16.Lin YJ, Xu L, Huang XZ, Jiang F, Li SL, Lin F, Ye QY, Chen ML, Lin JL. 2015. Reduced occurrence of ventilator-associated pneumonia after cardiac surgery using preoperative 0.2% chlorhexidine oral rinse: results from a single-centre single-blinded randomized trial. J Hosp Infect 91:362–366. doi: 10.1016/j.jhin.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 17.Climo MW, Yokoe DS, Warren DK, Perl TM, Bolon M, Herwaldt LA, Weinstein RA, Sepkowitz KA, Jernigan JA, Sanogo K, Wong ES. 2013. Effect of daily chlorhexidine bathing on hospital-acquired infection. N Engl J Med 368:533–542. doi: 10.1056/NEJMoa1113849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saatchi M, Shokraneh A, Navaei H, Maracy MR, Shojaei H. 2014. Antibacterial effect of calcium hydroxide combined with chlorhexidine on Enterococcus faecalis: a systematic review and meta-analysis. J Appl Oral Sci 22:356–365. doi: 10.1590/1678-775720140032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derde LPG, Dautzenberg MJD, Bonten MJM. 2012. Chlorhexidine body washing to control antimicrobial-resistant bacteria in intensive care units: a systematic review. Intensive Care Med 38:931–939. doi: 10.1007/s00134-012-2542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karki S, Cheng AC. 2012. Impact of non-rinse skin cleansing with chlorhexidine gluconate on prevention of healthcare-associated infections and colonization with multi-resistant organisms: a systematic review. J Hosp Infect 82:71–84. doi: 10.1016/j.jhin.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Kassakian SZ, Mermel LA, Jefferson JA, Parenteau SL, Machan JT. 2011. Impact of chlorhexidine bathing on hospital-acquired infections among general medical patients. Infect Control Hosp Epidemiol 32:238–243. doi: 10.1086/658334. [DOI] [PubMed] [Google Scholar]
- 22.Popovich KJ, Hota B, Hayes R, Weinstein RA, Hayden MK. 2009. Effectiveness of routine patient cleansing with chlorhexidine gluconate for infection prevention in the medical intensive care unit. Infect Control Hosp Epidemiol 30:959–963. doi: 10.1086/605925. [DOI] [PubMed] [Google Scholar]
- 23.Climo MW, Sepkowitz KA, Zuccotti G, Fraser VJ, Warren DK, Perl TM, Speck K, Jernigan JA, Robles JR, Wong ES. 2009. The effect of daily bathing with chlorhexidine on the acquisition of methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus, and healthcare-associated bloodstream infections: results of a quasi-experimental multicenter trial. Crit Care Med 37:1858–1865. doi: 10.1097/CCM.0b013e31819ffe6d. [DOI] [PubMed] [Google Scholar]
- 24.Suller MT, Russell AD. 1999. Antibiotic and biocide resistance in methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococcus. J Hosp Infect 43:281–291. doi: 10.1016/S0195-6701(99)90424-3. [DOI] [PubMed] [Google Scholar]
- 25.Horner C, Mawer D, Wilcox M. 2012. Reduced susceptibility to chlorhexidine in staphylococci: is it increasing and does it matter? J Antimicrob Chemother 67:2547–2559. doi: 10.1093/jac/dks284. [DOI] [PubMed] [Google Scholar]
- 26.Russell A. 2004. Bacterial adaptation and resistance to antiseptics, disinfectants and preservatives is not a new phenomenon. J Hosp Infect 57:97–104. doi: 10.1016/j.jhin.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry L, Merlin T, Rupp J, Griffith J. 1996. Reduced susceptibility of methicillin-resistant Staphylococcus aureus to cetylpyridinium chloride and chlorhexidine. Chemotherapy 42:248–252. doi: 10.1159/000239451. [DOI] [PubMed] [Google Scholar]
- 28.Block C, Robenshtok E, Simhon A, Shapiro M. 2000. Evaluation of chlorhexidine and povidone iodine activity against methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus faecalis using a surface test. J Hosp Infect 46:147–152. doi: 10.1053/jhin.2000.0805. [DOI] [PubMed] [Google Scholar]
- 29.Fraise AP. 2002. Susceptibility of antibiotic-resistant cocci to biocides. J Appl Microbiol 92(Suppl):158S–162S. doi: 10.1046/j.1365-2672.92.5s1.2.x. [DOI] [PubMed] [Google Scholar]
- 30.Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ. 2015. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics 16:964. doi: 10.1186/s12864-015-2153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhardwaj P, Ziegler E, Palmer KL. 2016. Chlorhexidine Induces VanA-type vancomycin resistance genes in enterococci. Antimicrob Agents Chemother 60:2209–2221. doi: 10.1128/AAC.02595-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Paganelli FL, Bierschenk D, Kuipers A, Bonten MJM, Willems RJL, van Schaik W. 2012. Genome-wide identification of ampicillin resistance determinants in Enterococcus faecium. PLoS Genet 8:e1002804. doi: 10.1371/journal.pgen.1002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawada-Matsuo M, Yoshida Y, Nakamura N, Komatsuzawa H. 2011. Role of two-component systems in the resistance of Staphylococcus aureus to antibacterial agents. Virulence 2:427–430. doi: 10.4161/viru.2.5.17711. [DOI] [PubMed] [Google Scholar]
- 34.Comenge Y, Quintiliani R, Li L, Dubost L, Brouard J-P, Hugonnet J-E, Arthur M. 2003. The CroRS two-component regulatory system is required for intrinsic beta-lactam resistance in Enterococcus faecalis. J Bacteriol 185:7184–7192. doi: 10.1128/JB.185.24.7184-7192.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong H-J, Hutchings MI, Buttner MJ, Biotechnology and Biological Sciences Research Council, UK. 2008. Vancomycin resistance VanS/VanR two-component systems. Adv Exp Med Biol 631:200–213. doi: 10.1007/978-0-387-78885-2_14. [DOI] [PubMed] [Google Scholar]
- 36.Paganelli FL, de Been M, Braat JC, Hoogenboezem T, Vink C, Bayjanov J, Rogers MRC, Huebner J, Bonten MJM, Willems RJL, Leavis HL. 2015. Distinct SagA from hospital-associated clade A1 Enterococcus faecium strains contributes to biofilm formation. Appl Environ Microbiol 81:6873–6882. doi: 10.1128/AEM.01716-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gebhard S, Fang C, Shaaly A, Leslie DJ, Weimar MR, Kalamorz F, Carne A, Cook GM. 2014. Identification and characterization of a bacitracin resistance network in Enterococcus faecalis. Antimicrob Agents Chemother 58:1425–1433. doi: 10.1128/AAC.02111-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bourret RB, Silversmith RE. 2010. Two-component signal transduction. Curr Opin Microbiol 13:113–115. doi: 10.1016/j.mib.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capra EJ, Laub MT. 2012. Evolution of two-component signal transduction systems. Annu Rev Microbiol 66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitrophanov AY, Groisman EA. 2008. Signal integration in bacterial two-component regulatory systems. Genes Dev 22:2601–2611. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casino P, Rubio V, Marina A. 2010. The mechanism of signal transduction by two-component systems. Curr Opin Struct Biol 20:763–771. doi: 10.1016/j.sbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 42.Toro-Roman A, Wu T, Stock AM. 2005. A common dimerization interface in bacterial response regulators KdpE and TorR. Protein Sci 14:3077–3088. doi: 10.1110/ps.051722805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachhawat P, Swapna GVT, Montelione GT, Stock AM. 2005. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure 13:1353–1363. doi: 10.1016/j.str.2005.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menon S, Wang S. 2011. Structure of the response regulator PhoP from Mycobacterium tuberculosis reveals a dimer through the receiver domain. Biochemistry 50:5948–5957. doi: 10.1021/bi2005575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boudes M, Sanchez D, Graille M, van Tilbeurgh H, Durand D, Quevillon-Cheruel S. 2014. Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae. Nucleic Acids Res 42:5302–5313. doi: 10.1093/nar/gku110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kesel S, Mader A, Höfler C, Mascher T, Leisner M. 2013. Immediate and heterogeneous response of the LiaFSR two-component system of Bacillus subtilis to the peptide antibiotic bacitracin. PLoS One 8:e53457. doi: 10.1371/journal.pone.0053457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida Y, Matsuo M, Oogai Y, Kato F, Nakamura N, Sugai M, Komatsuzawa H. 2011. Bacitracin sensing and resistance in Staphylococcus aureus: bacitracin sensing and resistance in S. aureus. FEMS Microbiol Lett 320:33–39. doi: 10.1111/j.1574-6968.2011.02291.x. [DOI] [PubMed] [Google Scholar]
- 48.Dintner S, Heermann R, Fang C, Jung K, Gebhard S. 2014. A sensory complex consisting of an ATP-binding cassette transporter and a two-component regulatory system controls bacitracin resistance in Bacillus subtilis. J Biol Chem 289:27899–27910. doi: 10.1074/jbc.M114.596221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuda H, Yamashita Y, Shibata Y, Nakano Y, Koga T. 2002. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob Agents Chemother 46:3756–3764. doi: 10.1128/AAC.46.12.3756-3764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manson JM, Keis S, Smith JMB, Cook GM. 2004. Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob Agents Chemother 48:3743–3748. doi: 10.1128/AAC.48.10.3743-3748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stone KJ, Strominger JL. 1971. Mechanism of action of bacitracin: complexation with metal ion and C55-isoprenyl pyrophosphate. Proc Natl Acad Sci U S A 68:3223–3227. doi: 10.1073/pnas.68.12.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chia JK, Nakata MM, Park SS, Lewis RP, McKee B. 1995. Use of bacitracin therapy for infection due to vancomycin-resistant Enterococcus faecium. Clin Infect Dis 21:1520. doi: 10.1093/clinids/21.6.1520. [DOI] [PubMed] [Google Scholar]
- 53.O'Donovan CA, Fan-Havard P, Tecson-Tumang FT, Smith SM, Eng RH. 1994. Enteric eradication of vancomycin-resistant Enterococcus faecium with oral bacitracin. Diagn Microbiol Infect Dis 18:105–109. doi: 10.1016/0732-8893(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 54.Weinstein MR, Dedier H, Brunton J, Campbell I, Conly JM. 1999. Lack of efficacy of oral bacitracin plus doxycycline for the eradication of stool colonization with vancomycin-resistant Enterococcus faecium. Clin Infect Dis 29:361–366. doi: 10.1086/520216. [DOI] [PubMed] [Google Scholar]
- 55.Hachem R, Raad I. 2002. Failure of oral antimicrobial agents in eradicating gastrointestinal colonization with vancomycin-resistant enterococci. Infect Control Hosp Epidemiol 23:43–44. doi: 10.1086/501968. [DOI] [PubMed] [Google Scholar]
- 56.Baldi P, Long AD. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 57.Zhang X, Vrijenhoek JEP, Bonten MJM, Willems RJL, van Schaik W. 2011. A genetic element present on megaplasmids allows Enterococcus faecium to use raffinose as carbon source. Environ Microbiol 13:518–528. doi: 10.1111/j.1462-2920.2010.02355.x. [DOI] [PubMed] [Google Scholar]
- 58.Andrews JM. 2001. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48(Suppl 1):S5–S16. [DOI] [PubMed] [Google Scholar]
- 59.Hendrickx APA, Top J, Bayjanov JR, Kemperman H, Rogers MRC, Paganelli FL, Bonten MJM, Willems RJL. 2015. Antibiotic-driven dysbiosis mediates intraluminal agglutination and alternative segregation of Enterococcus faecium from the intestinal epithelium. mBio 6:e01346-15. doi: 10.1128/mBio.01346-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.