ABSTRACT
Whole-genome sequencing of trimethoprim-resistant Escherichia coli clinical isolates identified a member of the trimethoprim-resistant type II dihydrofolate reductase gene family (dfrB). The dfrB4 gene was located within a class I integron flanked by multiple resistance genes. This arrangement was previously reported in a 130.6-kb multiresistance plasmid. The DfrB4 protein conferred a >2,000-fold increased trimethoprim resistance on overexpression in E. coli. Our results are consistent with the finding that dfrB4 contributes to clinical trimethoprim resistance.
KEYWORDS: type II dihydrofolate reductase, trimethoprim resistance, E. coli clinical isolates, dfrB4, antibiotic-resistant genes, class I integron, urinary tract infection
TEXT
Public health agencies worldwide rank trimethoprim (TMP) a broad-spectrum antibiotic of importance in human medicine (1). Widely used as a result of its low cost and effectiveness, TMP inhibits the activity of many microbial chromosomal dihydrofolate reductases (DHFRs); thus, DHFRs have long served as prioritized targets of antiproliferative drugs (2). Although the majority of living cells harbor a chromosomal member of the type I DHFR family, encoded by a dfrA homolog, the dfrB genes encode a family of plasmid-borne type II DHFRs that are evolutionarily unrelated to type I DHFRs. The dfrB genes have been found in pathogenic bacteria recovered from many food sources, including fish (3), pigs (4, 5), and cows (6), where they confer TMP resistance. Bacteria carrying dfrB genes have also been identified in wastewater samples (7). Over the past decade, dfrB genes have been tracked indirectly in antibiotic resistance studies through identification of integron-related elements (8–10). Therefore, the importance of dfrB genes in TMP resistance in human pathogens may be underappreciated (11, 12).
To date, only seven members of the dfrB gene family are known, and they are highly homologous (77% to 94% genetic identity, 77% to 99% amino acid identity) (Table 1). Among these, the DfrB1 protein (also known as R67 DHFR) is the best-studied type II DHFR (13–17). It is proposed to be recently evolved, and it confers a significant survival advantage under TMP exposure to microbes that harbor it (18). To date, the family of dfrB genes has consistently been reported to be contained within the following mobile genetic elements: a highly variable 57- to 141-bp recombination binding site (attC) and the 7-base canonical sequence (7-be), which is identical in all dfrB genes (5′-GTTRRRY) except dfrB2 (GTTAGGC) and includes a 7-be at the 3′ end, which is the reverse complement of the 7-be found upstream of the coding sequence (r7-be).
TABLE 1.
DNA sequence query coverage and identity of the main type II DHFRs, with the expected value
| DHFR | Query coverage (% identity) and expected valuea for: |
||||||
|---|---|---|---|---|---|---|---|
| dfrB1 | dfrB2 | dfrB3 | dfrB4 | dfrB5 | dfrB6 | dfrB7 | |
| Truncated dfrB1 | 100 | 95 (77) 2e−47 | 90 (88) 3e−77 | 89 (83) 5e−61 | 100 (89) 9e−90 | 100 (92) 7e−98 | 100 (92) 7e−98 |
| dfrB2 | 100 | 85 (86) 2e−66 | 72 (85) 2e−54 | 83 (79) 3e−45 | 97 (78) 4e−50 | 95 (77) 2e−47 | |
| dfrB3 | 100 | 94 (85) 2e−73 | 96 (86) 4e−75 | 97 (87) 7e−79 | 96 (86) 8e−78 | ||
| dfrB4 | 100 | 92 (81) 8e−59 | 94 (80) 1e−57 | 92 (80) 4e−56 | |||
| dfrB5 | 100 | 100 (91) 3e−96 | 100 (92) 1e−100 | ||||
| dfrB6 | 100 | 93 (94) 7e−111 | |||||
Expected values indicated by boldface (coding sequence only). The expected value (e) represents the probability of randomly matching two different sequences. The lower the e value, the more significant the match.
Clinical sample library.
We examined whole-genome shotgun sequencing data for 593 Escherichia coli isolates, including 380 E. coli isolates recovered from 324 individuals with hospital-associated human extraintestinal infections (19) and 189 E. coli isolates recovered from 189 women diagnosed with a community-acquired urinary tract infection (UTI) (A. R. Manges, unpublished data). Genomic E. coli DNA from women with UTIs (Manges collection) was extracted using the PureLink genomic DNA minikit (Thermo Fisher Scientific). Purified DNA was sheared in water using the Biorupter Pico (Diagenode), and sequencing libraries were prepared using the TruSeq DNA PCR-free library preparation kit (Illumina), according to the manufacturer's instructions. All E. coli isolates were sequenced on the Illumina HiSeq 2500 at the University of British Columbia's Pharmaceutical Sciences Sequencing Centre and British Columbia Genome Sciences Centre (Vancouver, BC).
In silico screening.
Seven members of the dfrB gene family were used as templates for in silico screening, including two variants of the dfrB1 gene that differ by the absence or presence of 19 additional N-terminal amino acids not essential for the reductase activity (20; see http://www.esi.umontreal.ca/~pelletjo/ToulouseSupplemental-material.pdf). The in silico dfrB screening was performed by aligning paired-end reads from 569 whole-genome-sequencing data sets to the set of dfrB genes in FASTA format with the Burrows-Wheeler aligner (BWA), using standard alignment parameters. Sequencing reads from E. coli isolates recovered from blood samples of an individual taken 2 days apart aligned exactly over the entire dfrB4 gene sequence (19 [https://www.ncbi.nlm.nih.gov/sra/SRX560289 and https://www.ncbi.nlm.nih.gov/sra/SRX560290]). The DNA consensus sequences were identical to the previously deposited 237-bp dfrB4 sequence (GenBank AY968808 and KP314737.1). Three further samples held one read that overlapped with the dfrB4 integron with 0 to 2 mismatches. Because no contig within assemblies of these three data sets aligned with dfrB4, they were not considered further.
Reconstruction of dfrB4 mobile genetic element and contiguous DNA segments.
Read sets with significant alignment to any reference dfrB gene were assembled into contigs, and the dfrB genes were aligned with BWA to these assembled contigs. The contigs from both samples have an r7-be (GTTGGGC) 61 bp upstream of the dfrB4 coding sequence and an attC sequence downstream (Fig. 1). The 7-be found in the attC (GCCCAAC) is 53 bp downstream from the coding sequence. These flanking sequences are identical to those of a previously reported dfrB4 mobile genetic element (GenBank accession no. AY970968.1) from Klebsiella pneumoniae (21). The attC sequence differs by one nucleotide from another dfrB4 mobile genetic element from E. coli (GenBank accession no. KP314737.1) (22; http://www.esi.umontreal.ca/~pelletjo/ToulouseSupplemental-material.pdf).
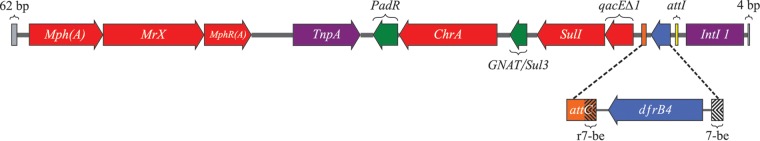
FIG 1.
Scheme of the 9,029-bp DNA segment derived from sample SRX560289. Purple, indispensable genes for integration of mobile genetic elements. Yellow, intI (integrase/recombinase), tnpA (transposase), and the integron-associated recombination site (attI). Red, resistance genes (qacEΔ1, biocide resistance; sulI, sulfonamide resistance; the mphR(A)/mphA/mrx gene cluster, erythromycin resistance; and chrA, chromate resistance), except dfrB4 (dihydrofolate reductase, TMP resistance), which is blue. Orange, recombination binding site (attC). Dashed lines, 7-be sequences. Green, DNA sequences sharing similarity with known genes: GNAT/sulI3 (acetyltransferase/sulfonamide resistance) and padR (transcriptional regulator). Gray, the 62- and 4-bp sequence differences between DNA contigs.
Contiguous to the mobile genetic element, we identified a 8,963-bp segment derived from sample SRX560290 and a 9,029-bp segment derived from sample SRX560289 (Fig. 1) (19). The 66-bp difference did not belong to an open reading frame (ORF) and lay outside the dfrB4 mobile genetic element. The dfrB4 gene was carried with intI1 (class 1 integron integrase/recombinase) and tnpA (transposase), which are indispensable for integration of mobile genetic elements (23). Moreover, multiple antibiotic, biocide, and metal resistance genes were identified: qacEΔ1 (biocide resistance [24, 25]), sulI (sulfonamide resistance [26]), the mphR(A)/mphA/mrx gene cluster (macrolide/erythromycin resistance [27]), and chrA (chromate resistance [28, 29]). The qacEΔ1 gene encodes a truncated and less-efficient cation efflux pump version of qacE (24).
Additionally, both segments included similarity to the GNAT (acetyltransferase) and sulI3 genes (30). The DNA sequence was 96% identical to GNAT from Aeromonas hydrophila with 72% query coverage (GenBank accession no. JX141473.1). The sulI3 gene was 100% identical with 55% query coverage (GenBank accession no. EF382672.1) yet included a 140-nucleotide 3′ deletion. The segments included the little-known padR transcriptional regulator. Nine additional ORFs were identified as hypothetical proteins.
A BLAST alignment of the contig derived from SRX560289 against all organisms resulted in 957 hits with an E value of ≤1 × e−24 (31, 32). Among these, a single hit (GenBank accession no. CP014320.1) included the dfrB4 mobile genetic element. It was a 130.6-kb plasmid from a clinical isolate of a patient with recurrent E. coli sequence type 131 (ST131) cystitis, the most prevalent E. coli lineage found in recurrent UTIs (33). Other hits included mismatches (≤15) and gaps (≤38) and excluded the dfrB4 gene sequence. This suggests that the dfrB4 mobile genetic element has recently been incorporated into the CP014320.1 multiresistance plasmid.
The DfrB4 protein confers TMP resistance.
The high sequence identity of the DfrB4 protein to the well-characterized TMP-resistant DfrB1 protein (Table 1) and the 6-mm TMP zone of inhibition for these clinical samples (S. Salipante, personal communication) corresponding to an MIC of >16 μg/ml strongly argue that DfrB4 should confer TMP resistance to E. coli (34). To corroborate this hypothesis, the consensus dfrB4 coding sequence was obtained (GeneArt gene synthesis; Thermo Fisher Scientific) with an N-terminal hexahistidine affinity tag for eventual purification. It was cloned into pET24-cTEM19m between NdeI and HindIII, under IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible overexpression in E. coli BL21(DE3). The MIC value was determined at least in duplicate through broth microdilution after incubation at 37°C for 14 to 16 h. We observed high resistance to TMP when expressing DfrB4 (>600 μg/ml, which is the maximal concentration of TMP that is soluble in 5% methanol) (35). The MICs for all negative controls were at least 2,000-fold lower, specifically E. coli BL21(DE3) (0.30 μg/ml) and E. coli BL21(DE3)/pET24-cTEM19m expressing an IPTG-inducible β-lactamase (0.075 μg/ml) (36). Thus, the MIC value of DrfB4 greatly surpasses the E. coli TMP resistance threshold (2 μg/ml) and is associated with 2.5% of all E. coli strains considered to exhibit dangerously high TMP resistance (≥512 μg/ml) (35, 37). In these experiments, DfrB4 was overexpressed; the expression level of DfrB4 in its native form is unknown.
To our knowledge, this work constitutes the first report of the type II TMP-resistant dfrB4 gene identified in a clinical sample by whole-genome sequencing. The 2,000-fold increase in MIC for TMP that we observed in a transformed E. coli isolate points to TMP resistance in the clinical isolate identified carrying the dfrB4 gene. The observation of this highly TMP-resistant DfrB4 within a known class I integron indicates that it is being disseminated, although the extent and rate of its propagation are currently unknown as it has not been systematically tracked. Our results highlight the importance of tracking the presence of this gene family in TMP-resistant clinical samples.
ACKNOWLEDGMENTS
We thank Steve Salipante for providing the zone of inhibition data for the original clinical samples and Simon Toulouse for assistance with drafting Fig. 1.
This work was supported by NSERC Discovery Grant 227853 (J.N.P). J.L.T. received a scholarship from Fonds Québécois pour la Recherche sur la Nature et le Technologies (FRQ-NT). This work was supported by funds from CIHR (A.R.M) (MOP 114879).
L.A. received a scholarship from PROTEO, the Québec Network for Research on Protein Structure, Function and Engineering, which is funded by FRQ-NT. J.L.T. and L.A. received scholarships from Faculté des études supérieures et post-doctorales de l'Université de Montréal (FESP).
REFERENCES
- 1.Government of Canada. 2015. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2013 annual report, chapter 1. Design and Methods Public Health Agency of Canada, Guelph, ON, Canada. [Google Scholar]
- 2.Ho JM, Juurlink DN. 2011. Considerations when prescribing trimethoprim-sulfamethoxazole. Can Med Assoc J 183:1851–1858. doi: 10.1503/cmaj.111152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.L'Abée-Lund TM, Sørum H. 2001. Class 1 integrons mediate antibiotic resistance in the fish pathogen Aeromonas salmonicida worldwide. Microb Drug Resist 7:263–272. doi: 10.1089/10766290152652819. [DOI] [PubMed] [Google Scholar]
- 4.Kadlec K, Kehrenberg C, Schwarz S. 2005. Molecular basis of resistance to trimethoprim, chloramphenicol and sulphonamides in Bordetella bronchiseptica. J Antimicrob Chemother 56:485–490. doi: 10.1093/jac/dki262. [DOI] [PubMed] [Google Scholar]
- 5.Sunde M. 2005. Prevalence and characterization of class 1 and class 2 integrons in Escherichia coli isolated from meat and meat products of Norwegian origin. J Antimicrob Chemother 56:1019–1024. doi: 10.1093/jac/dki377. [DOI] [PubMed] [Google Scholar]
- 6.Barlow RS, Pemberton JM, Desmarchelier PM, Gobius KS. 2004. Isolation and characterization of integron-containing bacteria without antibiotic selection. Antimicrob Agents Chemother 48:838–842. doi: 10.1128/AAC.48.3.838-842.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tennstedt T, Szczepanowski R, Braun S, Pühler A, Schlüter A. 2003. Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant. FEMS Microbiol Ecol 45:239–252. doi: 10.1016/S0168-6496(03)00164-8. [DOI] [PubMed] [Google Scholar]
- 8.Kehrenberg C, Schwarz S. 2011. Trimethoprim resistance in a porcine Pasteurella aerogenes isolate is based on a dfrA1 gene cassette located in a partially truncated class 2 integron. J Antimicrob Chemother 66:450–452. doi: 10.1093/jac/dkq461. [DOI] [PubMed] [Google Scholar]
- 9.Partridge SR, Tsafnat G, Coiera E, Iredell JR. 2009. Gene cassettes and cassette arrays in mobile resistance integrons: review article. FEMS Microbiol Rev 33:757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 10.Levings RS, Lightfoot D, Elbourne LDH, Djordjevic SP, Hall RM. 2006. New integron-associated gene cassette encoding a trimethoprim-resistant DfrB-type dihydrofolate reductase. Antimicrob Agents Chemother 50:2863–2865. doi: 10.1128/AAC.00449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller LG, Tang AW. 2004. Treatment of uncomplicated urinary tract infections in an era of increasing antimicrobial resistance. Mayo Clinic Proc 79:1048–1053. doi: 10.4065/79.8.1048. [DOI] [PubMed] [Google Scholar]
- 12.Sundqvist M, Geli P, Andersson DI, Sjolund-Karlsson M, Runehagen A, Cars H, Abelson-Storby K, Cars O, Kahlmeter G. 2010. Little evidence for reversibility of trimethoprim resistance after a drastic reduction in trimethoprim use. J Antimicrob Chemother 65:350–360. doi: 10.1093/jac/dkp387. [DOI] [PubMed] [Google Scholar]
- 13.Bastien D, Ebert MCCJC, Forge D, Toulouse J, Kadnikova N, Perron F, Mayence A, Huang TL, Vanden Eynde JJ, Pelletier JN. 2012. Fragment-based design of symmetrical bis-benzimidazoles as selective inhibitors of the trimethoprim-resistant, type II R67 dihydrofolate reductase. J Med Chem 55:3182–3192. doi: 10.1021/jm201645r. [DOI] [PubMed] [Google Scholar]
- 14.Narayana N. 2006. High-resolution structure of a plasmid-encoded dihydrofolate reductase: pentagonal network of water molecules in the D2-symmetric active site. Acta Crystallogr Sect D Biol Crystallogr 62:695–706. doi: 10.1107/S0907444906014764. [DOI] [PubMed] [Google Scholar]
- 15.Duff MR, Chopra S, Strader MB, Agarwal PK, Howell EE. 2016. Tales of dihydrofolate binding to R67 dihydrofolate reductase. Biochemistry 55:133–145. doi: 10.1021/acs.biochem.5b00981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yachnin BJ, Colin DY, Volpato JP, Ebert M, Pelletier JN, Berghuis AM. 2011. Novel crystallization conditions for tandem variant R67 DHFR yield a wild-type crystal structure. Acta Crystallogr Sect F Struct Biol Cryst Commun 67:1316–1322. doi: 10.1107/S1744309111030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fling ME, Elwell LP. 1980. Protein expression in Escherichia coli minicells containing recombinant plasmids specifying trimethoprim-resistant dihydrofolate reductases. J Bacteriol 141:779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alonso H, Gready JE. 2006. Integron-sequestered dihydrofolate reductase: a recently redeployed enzyme. Trends Microbiol 14:236–242. doi: 10.1016/j.tim.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Salipante SJ, Roach DJ, Kitzman JO, Snyder MW, Stackhouse B, Butler-Wu SM, Lee C, Cookson BT, Shendure J. 2015. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res 25:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reece LJ, Nichols R, Ogden RC, Howell EE. 1991. Construction of a synthetic gene for an R-plasmid-encoded dihydrofolate reductase and studies on the role of the N-terminus in the protein. Biochemistry 30:10895–10904. doi: 10.1021/bi00109a013. [DOI] [PubMed] [Google Scholar]
- 21.Tortola MT, Lavilla S, Miro E, Gonzalez JJ, Larrosa N, Sabate M, Navarro F, Prats G. 2005. First detection of a carbapenem-hydrolyzing metalloenzyme in two Enterobacteriaceae isolates in Spain. Antimicrob Agents Chemother 49:3492–3494. doi: 10.1128/AAC.49.8.3492-3494.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delport TC, Harcourt RG, Beaumont LJ, Webster KN, Power ML. 2015. Molecular detection of antibiotic-resistance determinants in Escherichia coli isolated from the endangered Australian sea lion (Neophoca cinerea). J Wildlife Dis 51:555–563. doi: 10.7589/2014-08-200. [DOI] [PubMed] [Google Scholar]
- 23.Woodford N, Carattoli A, Karisik E, Underwood A, Ellington MJ, Livermore DM. 2009. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob Agents Chemother 53:4472–4482. doi: 10.1128/AAC.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helal ZH, Khan MI. 2015. QacE and QacEΔ1 genes and their correlation to antibiotics and biocides resistance Pseudomonas aeruginosa. Am J Biomed Sci 7:52–62. [Google Scholar]
- 25.Jaglic Z, Cervinkova D. 2012. Genetic basis of resistance to quaternary ammonium compounds–the qac genes and their role: a review. Vet Med 57:275–281. [Google Scholar]
- 26.Gillings MR. 2014. Integrons: past, present, and future. Microbiol Mol Biol Rev 78:257–277. doi: 10.1128/MMBR.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J, Sagar V, Smolinsky A, Bourke C, LaRonde-LeBlanc N, Cropp TA. 2009. Structure and function of the macrolide biosensor protein, MphR(A), with and without erythromycin. J Mol Biol 387:1250–1260. doi: 10.1016/j.jmb.2009.02.058. [DOI] [PubMed] [Google Scholar]
- 28.Caballero-Flores GG, Acosta-Navarrete YM, Ramirez-Diaz MI, Silva-Sanchez J, Cervantes C. 2012. Chromate-resistance genes in plasmids from antibiotic-resistant nosocomial enterobacterial isolates. FEMS Microbiol Lett 327:148–154. doi: 10.1111/j.1574-6968.2011.02473.x. [DOI] [PubMed] [Google Scholar]
- 29.Ramirez-Diaz MI, Diaz-Perez C, Vargas E, Riveros-Rosas H, Campos-Garcia J, Cervantes C. 2008. Mechanisms of bacterial resistance to chromium compounds. BioMetals 21:321–332. doi: 10.1007/s10534-007-9121-8. [DOI] [PubMed] [Google Scholar]
- 30.Wachino JI, Shibayama K, Kurokawa H, Kimura K, Yamane K, Suzuki S, Shibata N, Ike Y, Arakawa Y. 2007. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother 51:4401–4409. doi: 10.1128/AAC.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. 2012. GenBank. Nucleic Acids Res 40:D48–53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Johnson TJ, Aziz M, Liu CM, Sokurenko E, Kisiela DI, Paul S, Andersen P, Johnson JR, Price B. 2016. Complete genome sequence of a CTX-M-15-producing Escherichia coli strain from the H30Rx subclone of sequence type 131 from a patient with recurrent urinary tract infections, closely related to a lethal urosepsis isolate from the patient's sister. Genome Announc 4:15–16. doi: 10.1128/genomeA.00334-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clinical and Laboratory Standards Institute. 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement, vol. 31. CLSI document M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 35.Wiegand I, Hilpert K, Hancock REW. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 36.Gobeil S, Gagné D, Doucet N, Pelletier J. 2016. 15N, 13C and 1H backbone resonance assignments of an artificially engineered TEM-1/PSE-4 class A β-lactamase chimera and its deconvoluted mutant. Biomol NMR Assign 10:93–99. doi: 10.1007/s12104-015-9645-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.European Committee on Antimicrobial Susceptibility Testing. 2016. International MIC distribution. https://mic.eucast.org/Eucast2/regShow.jsp?Id=4125.