ABSTRACT
A novel transposon belonging to the Tn3-like family was identified on the chromosome of a commensal strain of Pseudomonas aeruginosa sequence type 2343 (ET02). Tn6350 is 7,367 bp long and harbors eight open reading frames (ORFs), an ATPase (IS481 family), a transposase (DDE catalytic type), a Tn3 resolvase, three hypothetical proteins, and genes encoding the new pyocin S8 with its immunity protein. We show that pyocin S8 displays activity against carbapenemase-producing P. aeruginosa, including IMP-1, SPM-1, VIM-1, GES-5, and KPC-2 producers.
KEYWORDS: transposon, S-type pyocins, bacteriocin, Pseudomonas aeruginosa, metallo-β-lactamases
TEXT
Pseudomonas aeruginosa is an opportunistic pathogen and a leading cause of nosocomial infections, which are very difficult to treat because hospital-associated lineages are usually multidrug resistant (MDR). Carbapenems have been considered the most effective drugs against MDR isolates. However, the emergence and dissemination of carbapenemase-producing P. aeruginosa strains have become a major health care problem (1, 2). Therefore, since therapeutic options are limited, the research of new therapeutic compounds is essential. In this regard, one alternative strategy has been the investigation of natural antibiotics (i.e., bacteriocins) produced by many bacteria for intraspecies competition (3–9). Specifically, bacteriocins produced by P. aeruginosa are called “pyocins.” Based on their structure, pyocins can be divided into three types, named R, F, and S. While R- and F-type pyocins are high-molecular-weight protein complexes that resemble phage tails, S-type pyocins are binary protein complexes consisting of a large protein that harbors the killing function and a smaller immunity protein that confers protection against the cognate bacteriocin (9, 10). Most S-type pyocins bind to specific receptors in the bacterial outer membrane and are translocated through it before killing their target. In P. aeruginosa, several S-type pyocins exhibiting different killing activities, such as DNase (S1, S2, S3, and AP41), tRNase (S4, S11, and S12), pore-forming (S5), rRNase (S6), and lipid II degradation activity (M1 and M4), have been described and functionally characterized. Currently, genes encoding S-type pyocins S7 through S10 have been identified in silico by analysis and comparison of draft and complete genome sequences of P. aeruginosa strains (9).
In this study, we performed a screening for strains producing pyocins with activity against MDR P. aeruginosa strains. In this regard, pyocins extracted from a commensal strain (ET02), belonging to sequence type ST2343, which was isolated from a healthy patient in Brazil, exhibited the highest antibacterial activity against MDR P. aeruginosa isolates, including carbapenemase-producing nosocomial lineages (Table 1).
TABLE 1.
Bacterial activity of pyocin S8 against MDR Pseudomonas aeruginosa strains isolated from clinical and environmental samples
| P. aeruginosa strain | Source | Resistance profile fora: |
β-Lactamaseb | Bacterial activity of pyocinsc: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPM | MEM | CAZ | FEP | AK | GEN | LVX | CIP | S8 | S2, S4, S5d | |||
| 48-1997A | Human | R | R | R | R | R | R | R | R | SPM-1 | + | – |
| 247-B | Human | R | R | R | I | S | R | R | R | VIM-1 | + | – |
| GIM | Human | R | R | R | R | I | R | R | R | GIM-1 | + | – |
| BH6 | Human | R | R | R | R | R | I | R | R | KPC-2 | + | – |
| PHB64 | Human | R | R | R | R | R | R | R | R | GES-5 | + | – |
| IMP | Human | R | R | R | R | R | R | R | I | IMP-1 | + | – |
| 298 | Human | I | R | R | R | S | I | S | S | OXA-18 | + | – |
| 395 | Human | R | R | R | R | R | R | R | R | IMP-18 | – | – |
| 179 | Human | R | R | R | R | R | R | R | R | VIM-1 | + | – |
| 1088 | Human | R | R | R | R | S | I | R | R | SPM-1 | + | – |
| 392 | Human | R | R | I | R | S | R | R | R | OXA-10 | – | – |
| 225 | Human | R | R | R | R | R | R | R | R | GES-1 | + | – |
| 44 | Human | R | R | R | R | R | R | R | R | SPM-1 | + | – |
| 19 | Environmental | R | R | R | R | R | R | R | R | SPM-1 | + | – |
| 141 | Environmental | R | R | R | R | R | R | R | R | SPM-1 | – | – |
| 151 | Environmental | R | R | R | R | R | R | R | R | SPM-1 | + | – |
| ET02 | Human | S | S | S | R | S | I | I | S | OXA-50 | – | + |
| PAO1 | Control | S | S | S | S | S | S | S | S | OXA-50 | + | – |
IPM, imipenem; MEM, meropenem; CAZ, ceftazidime; FEP, cefepime; AK, amikacin; GEN, gentamicin; LVX, levofloxacin; CIP, ciprofloxacin (21); R, resistant isolates; I, intermediate isolates; S, susceptible isolates.
Genes encoding β-lactamases were confirmed by PCR and sequencing.
Bacterial activity of pyocins was evaluated by the presence (+) or absence (–) of inhibition zones measuring ≥10 mm, using a pyocin assay (5).
Pyocins produced by P. aeruginosa strain PAO1 (9).
The antimicrobial activity of pyocins from the ET02 strain was evaluated using a previously described pyocin assay method (5). In brief, the pyocin production in ET02 was induced by adding a 3 μg/ml final concentration of mitomycin C in culture medium. Pyocin molecules were then precipitated by ammonium sulfate. The high-molecular-weight R- and F-type pyocins were sedimented by ultracentrifugation, whereas S-type pyocins were recovered from the supernatant (11). Both high- and low-molecular-weight pyocins were screened against carbapenemase-producing P. aeruginosa isolates, where the fraction containing the S-type pyocins from the ET02 strain displayed the highest antibacterial activity against clinically significant β-lactamase (SPM-1, GIM-1, VIM-1, IMP-1, KPC-2, OXA-18, GES-1, and GES-5)-producing P. aeruginosa strains (12–17) (Table 1). In contrast, S-type pyocins produced by P. aeruginosa PAO1 (i.e., S2, S4, S5) (9), which were used as a control, showed no activity against the strain panel (Table 1). S-type pyocins from the ET02 strain were submitted to proteomic analysis by mass spectrometry, and data were searched against the Swiss-Prot database, resulting in 19% coverage of the pyocin AP41 sequence (GenBank accession number Q51502).
Total genomic DNA of P. aeruginosa ET02 was extracted to construct a mate-paired library, which was sequenced using the MiSeq platform (Illumina, Inc.). The sequence reads were de novo assembled using an A5-miseq pipeline (18), and after automatic annotation using Prokka (www.github.com/tseemann/prokka), the sequence was manually curated using the GenBank database and InterPro (www.ebi.ac.uk/interpro). Curiously, during whole-genome sequencing (WGS) analysis, we noted that a DNA region of 7,367 bp containing the S-type pyocin nucleotide sequence from strain ET02 displayed 100% identity with a region analyzed from the complete genome sequence of P. aeruginosa strain BAMCPA07-48, recently submitted to the NCBI GenBank database (accession number CP015377.1). This region contained a transposon DNA-invertase, hypothetical proteins, and an uncharacterized S-type pyocin. Additionally, a translated search of GenBank (blastx) revealed that the S-type pyocin from ET02 displayed 90% identity to AP41 pyocin (GenBank accession number Q51502).
Primers were designed to confirm the nucleotide sequence of ET02 pyocin by Sanger sequencing, and then the translated sequence was deduced and analyzed; 772 amino acids were obtained. Although attempts to perform a comparative protein analysis using different databases were unsuccessful, we found a recent review of antibacterial proteins and peptides of Pseudomonas (9), in which new S-type pyocins from P. aeruginosa were identified based on an in silico analysis, where a pyocin named S8 was identical to the pyocin produced by P. aeruginosa ET02.
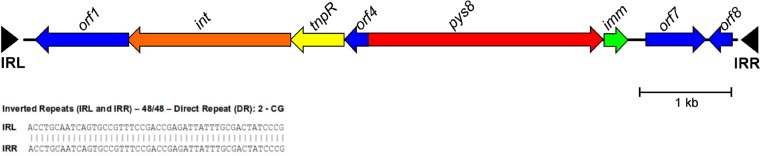
The genetic environment of S8 pyocin from ET02 was submitted to the Tn Number Registry database (http://www.ucl.ac.uk/eastman/research/departments/microbial-diseases/tn), confirming a new transposon, which was designed Tn6350. In this regard, Tn6350 is a 7,367-bp transposon (GenBank accession number KY347015) belonging to the Tn3-like family, which was located on the chromosome of ET02. This transposon carries genes encoding eight ORFs, an ATPase (IS481 family), a transposase (DDE catalytic type), a resolvase (Tn3 family), an S-type pyocin, an immunity protein, and three hypothetical proteins (Fig. 1).
FIG 1.
Genetic organization of Tn6350 (GenBank accession number KY347015), which harbors genes encoding pyocin S8. Predicted coding regions are represented by thick arrows indicating the direction of transcription. This element contains eight ORFs, including an ATPase (orf1), a transposase (int), a resolvase (tnpR), a cytotoxic subunit of pyocin S8 (pys8), an immunity subunit of pyocin S8 (imm), and three more hypothetical proteins (ofr4, orf7, and orf8). The inverted repeat (IR) sequences flanking the element are marked by black triangles (IRL, inverted repeat left; IRR, inverted repeat right). They have 48 bp and 100% identity. The element is flanked by short direct repeats (DRs) rich in CG dinucleotide.
S-Type pyocins carried by transposon elements have been previously described (19). Transposons are genetic elements able to move within and among genomes. These elements are important modulators of bacterial genomes, in many cases improving the environmental adaptation (20). In recent years, WGS approaches have allowed the in silico comparative analysis of bacterial genomes. In this context, novel transposable elements have been identified. So, although Tn6350 has not been previously described and characterized, it might be present in other P. aeruginosa strains.
Interestingly, pyocin S8 (harbored by Tn6350) exhibited a wide bactericidal activity against clinically significant carbapenemase-producing P. aeruginosa strains. For the host strain, this bacteriocin production can confer a fitness advantage, whereas the potency and targeted action of this pyocin could be developed into a clinically useful antibiotic against MDR pathogens for which there are few therapeutic options (4).
In summary, we hereby describe a new transposon carrying S8 pyocin genes encoding a bacteriocin presenting activity against carbapenemase-producing P. aeruginosa strains. The clinical therapeutic potential of this new pyocin is worthy of further investigation.
Accession number(s).
This whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number LYLY00000000.
ACKNOWLEDGMENTS
This work was supported by research grants from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (grant 2016/08593-9), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (grant 462042/2014-6). N.L. is a research grant fellow of CNPq.
We thank A. L. Darini and M. C. Nogueira for kindly providing P. aeruginosa strains BH6 and PHB64, respectively.
We have no conflicts of interest to declare.
REFERENCES
- 1.Zavascki AP, Carvalhaes CG, Picão RC, Gales AC. 2010. Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: resistance mechanisms and implications for therapy. Expert Rev Anti Infect Ther 8:71–93. doi: 10.1586/eri.09.108. [DOI] [PubMed] [Google Scholar]
- 2.Oliver A, Mulet X, López-Causapé C, Juan C. 2015. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21-22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 4.McCaughey LC, Ritchie ND, Douce GR, Evans TJ, Walker D. 2016. Efficacy of species-specific protein antibiotics in a murine model of acute Pseudomonas aeruginosa lung infection. Sci Rep 6:30201. doi: 10.1038/srep30201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith K, Martin L, Rinaldi A, Rajendran R, Ramage G, Walker D. 2012. Activity of pyocin S2 against Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 56:1599–1601. doi: 10.1128/AAC.05714-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ritchie JM, Greenwich JL, Davis BM, Bronson RT, Gebhart D, Williams SR, Martin D, Scholl D, Waldor MK. 2011. An Escherichia coli O157-specific engineered pyocin prevents and ameliorates infection by E. coli O157:H7 in an animal model of diarrheal disease. Antimicrob Agents Chemother 55:5469–5474. doi: 10.1128/AAC.05031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scholl D, Martin DW Jr. 2008. Antibacterial efficacy of R-type pyocins towards Pseudomonas aeruginosa in a murine peritonitis model. Antimicrob Agents Chemother 52:1647–1652. doi: 10.1128/AAC.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scholl D, Cooley M, Williams SR, Gebhart D, Martin D, Bates A, Mandrell R. 2009. An engineered R-type pyocin is a highly specific and sensitive bactericidal agent for the food-borne pathogen Escherichia coli O157:H7. Antimicrob Agents Chemother 53:3074–3080. doi: 10.1128/AAC.01660-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghequire MG, De Mot R. 2014. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev 38:523–568. doi: 10.1111/1574-6976.12079. [DOI] [PubMed] [Google Scholar]
- 10.Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499–510. doi: 10.1016/S0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 11.Williams SR, Gebhart D, Martin DW, Scholl D. 2008. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microbiol 74:3868–3876. doi: 10.1128/AEM.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castanheira M, Toleman M, Jones R, Schmidt F, Walsh T. 2004. Molecular characterization of a β-lactamase gene, blaGIM-1, encoding a new subclass of metallo-β-lactamase. Antimicrob Agents Chemother 48:4654–4661. doi: 10.1128/AAC.48.12.4654-4661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontes LC, Neves PR, Oliveira S, Silva KC, Hachich EM, Sato MI, Lincopan N. 2011. Isolation of Pseudomonas aeruginosa coproducing metallo-β-lactamase SPM-1 and 16S rRNA methylase RmtD1 in an urban river. Antimicrob Agents Chemother 55:3063–3064. doi: 10.1128/AAC.00138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turano H, Gomes F, Medeiros M, Oliveira S, Fontes LC, Sato MI, Lincopan N. 2016. Presence of high-risk clones of OXA-23-producing Acinetobacter baumannii (ST79) and SPM-1-producing Pseudomonas aeruginosa (ST277) in environmental water samples in Brazil. Diagn Microbiol Infect Dis 86:80–82. doi: 10.1016/j.diagmicrobio.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Toleman MA, Simm AM, Murphy TA, Gales AC, Biedenbach DJ, Jones RN, Walsh TR. 2002. Molecular characterization of SPM-1, a novel metallo-beta-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J Antimicrob Chemother 50:673–679. doi: 10.1093/jac/dkf210. [DOI] [PubMed] [Google Scholar]
- 16.Polotto M, Casella T, de Lucca Oliveira MG, Rúbio FG, Nogueira ML, de Almeida MT, Nogueira MC. 2012. Detection of P. aeruginosa harboring blaCTX-M-2, blaGES-1 and blaGES-5, blaIMP-1 and blaSPM-1 causing infections in Brazilian tertiary-care hospital. BMC Infect Dis 12:176. doi: 10.1186/1471-2334-12-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galetti R, Andrade LN, Chandler M, de Mello Varani A, Darini AL. 2016. New small plasmid harboring blaKPC-2 in Pseudomonas aeruginosa. Antimicrob Agents Chemother 60:3211–3214. doi: 10.1128/AAC.00247-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 19.Sano Y, Kageyama M. 1993. A novel transposon-like structure carries the genes for pyocin AP41, a Pseudomonas aeruginosa bacteriocin with a DNase domain homology to E2 group colicins. Mol Gen Genet 237:161–170. 10.1007/BF00282797. [DOI] [PubMed] [Google Scholar]
- 20.Casacuberta E, González J. 2013. The impact of transposable elements in environmental adaptation. Mol Ecol 22:1503–1517. doi: 10.1111/mec.12170. [DOI] [PubMed] [Google Scholar]
- 21.CLSI. 2016. Performance standards for antimicrobial susceptibility testing; 26th informational supplement. CLSI document M100-S26. CLSI, Wayne, PA. [Google Scholar]