ABSTRACT
Candida auris, a new multidrug-resistant Candida spp. which is associated with invasive infection and high rates of mortality, has recently emerged. Here, we determined the virulence factors (germination, adherence, biofilm formation, phospholipase and proteinase production) of 16 C. auris isolates and their susceptibilities to 11 drugs belonging to different antifungal classes, including a novel orally bioavailable 1,3-β-d-glucan synthesis inhibitor (SCY-078). We also examined the effect of SCY-078 on the growth, ultrastructure, and biofilm-forming abilities of C. auris. Our data showed that while the tested strains did not germinate, they did produce phospholipase and proteinase in a strain-dependent manner and had a significantly reduced ability to adhere and form biofilms compared to that of Candida albicans (P = 0.01). C. auris isolates demonstrated reduced susceptibility to fluconazole and amphotericin B, while, in general, they were susceptible to the remaining drugs tested. SCY-078 had an MIC90 of 1 mg/liter against C. auris and caused complete inhibition of the growth of C. auris and C. albicans. Scanning electron microscopy analysis showed that SCY-078 interrupted C. auris cell division, with the organism forming abnormal fused fungal cells. Additionally, SCY-078 possessed potent antibiofilm activity, wherein treated biofilms demonstrated significantly reduced metabolic activity and a significantly reduced thickness compared to the untreated control (P < 0.05 for both comparisons). Our study shows that C. auris expresses several virulence determinants (albeit to a lesser extent than C. albicans) and is resistant to fluconazole and amphotericin B. SCY-078, the new orally bioavailable antifungal, had potent antifungal/antibiofilm activity against C. auris, indicating that further evaluation of this antifungal is warranted.
KEYWORDS: Candida auris, SCY-078, virulence, biofilm
INTRODUCTION
The Centers for Disease Control and Prevention recently published an alert that emerging multidrug-resistant Candida auris is causing invasive infections (1). Originally isolated in 2008 from a Japanese patient's ear canal (2), C. auris has been reported to cause serious invasive infections (e.g., candidemia) with a high associated rate of mortality (approaching approximately 60%) (3). C. auris has caused serious infections globally, including in Japan, South Korea, India, Kuwait, South Africa, Pakistan, and the United Kingdom and, more recently, in Venezuela, Colombia, and the United States (2, 4–12). Of these C. auris infections, the majority have been secondary nosocomial infections (4, 9, 12, 13). A high percentage of clinical C. auris strains demonstrate resistance to fluconazole and show variable resistance to other antifungals belonging to the three major classes of clinically available antifungals (azoles, polyenes, echinocandins), thereby limiting treatment options (1, 3–6, 9–16).
To gain insight into this emerging Candida species, investigators conducted studies to characterize its virulence factors (e.g., phospholipase, proteinase, ability to form biofilms) (8, 17). Although these studies are informative, they examined only a limited number of C. auris strains. Here we studied 16 different C. auris isolates obtained from patients in Japan, India, South Korea, and Germany and (i) characterized their morphology and virulence factors (those that have been described for Candida species, particularly Candida albicans, e.g., germination, adherence, biofilm formation, and phospholipase and proteinase production), (ii) determined their susceptibilities to 11 antifungals belonging to different antifungal classes, including a novel orally bioavailable 1,3-β-d-glucan synthesis inhibitor (SCY-078) with demonstrated activity against multidrug-resistant Candida spp., and (iii) evaluated the effect of SCY-078 on the growth, ultrastructure, and biofilm-forming ability of C. auris (18).
RESULTS
C. auris does not form chlamydospores.
C. auris failed to form chlamydospores after growth on cornmeal agar for 3 days at 30°C. In contrast, C. albicans ATCC 14053 (the positive control) produced chlamydospores on cornmeal agar (data not shown).
Virulence factors.
Our data showed that the C. auris strains tested did not germinate when incubated with fetal bovine serum. In contrast, C. albicans ATCC 14053 germinated profusely (>90% within 2 h). Moreover, the evaluation of adherence using two representative C. auris isolates (MRL 31102 and MRL 31103) revealed that the C. auris strains exhibited a significantly reduced ability to adhere to catheter material compared to C. albicans (P < 0.05) (Fig. 1).
FIG 1.
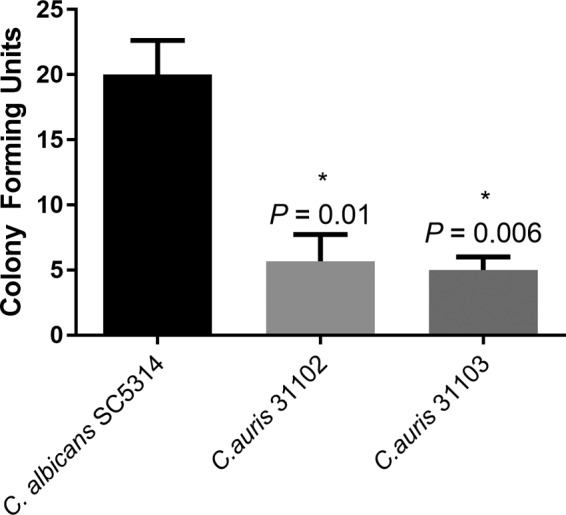
Comparison of adherence of C. auris strains. The ability of Candida species to adhere to a silicon elastomer catheter as a representative substrate was assessed. Cells were allowed to adhere to silicone elastomer discs, washed, and overlaid with Sabouraud dextrose agar, and the number of CFU adhering to the substrate was counted after incubation at 37°C for 18 to 24 h. The number of adherent C. auris cells was significantly less than that for C. albicans (positive control) (P ≤ 0.01). *, P value compared to the value for C. albicans.
The tested C. auris strains produced phospholipase and proteinase in a strain-dependent manner. As shown in Table 1, 37.5% of the tested C. auris strains (6/16 isolates) possessed phospholipase activity and 64% (9/14 isolates) tested positive for secreted proteinase activity (19). The level of proteinase production by the 9 strains ranged from 1.2 to 5.3 ng/ml (Table 1).
TABLE 1.
Phospholipase and proteinase activities of C. auris isolates
| Strain | Species | Phospholipase activity |
Proteinase activity (ng/ml) | |
|---|---|---|---|---|
| Class | Pz valuea | |||
| SC5314 | C. albicans | Control (++) | 0.66 | |
| MRL 31102 | C. auris | − | 1.00 | |
| MRL 31103 | C. auris | − | 1.00 | |
| CBS 10913 | C. auris | − | 1.00 | 0.0 |
| CBS 12372 | C. auris | + | 0.90 | 0.0 |
| CBS 12373 | C. auris | − | 1.00 | 1.4 |
| CBS 12766 | C. auris | + | 0.90 | 0.0 |
| CBS 12767 | C. auris | − | 1.00 | 0.0 |
| CBS 12768 | C. auris | + | 0.90 | 2.3 |
| CBS 12770 | C. auris | ++ | 0.78 | 1.8 |
| CBS 12771 | C. auris | − | 1.00 | 4.7 |
| CBS 12772 | C. auris | − | 1.00 | 0.0 |
| CBS 12773 | C. auris | + | 0.91 | 1.2 |
| CBS 12774 | C. auris | − | 1.00 | 2.8 |
| CBS 12775 | C. auris | + | 0.91 | 1.6 |
| CBS 12776 | C. auris | − | 1.00 | 5.3 |
| CBS 12777 | C. auris | − | 1.00 | 3.2 |
++, Pz = <0.89 (strong phospholipase activity); +, Pz = 0.90 to 0.99 (weak phospholipase activity); −, Pz = 1 (no phospholipase activity).
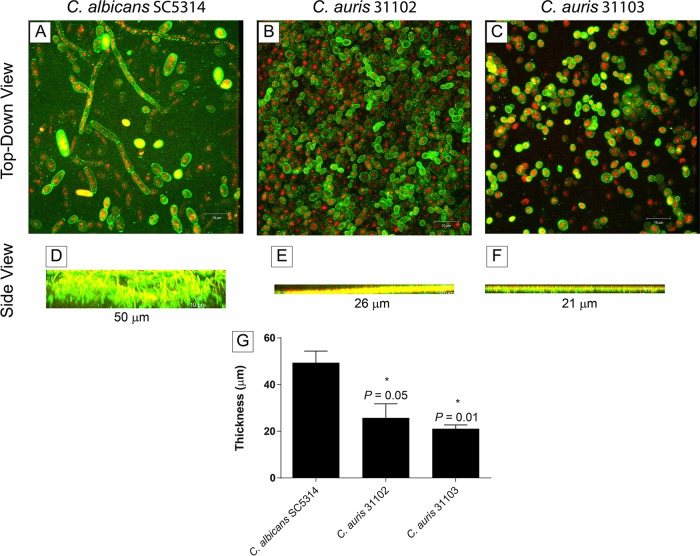
Analysis of biofilm formation by C. auris MRL 31102 and MRL 31103 demonstrated that the biofilms were mainly composed of yeast cells adhering to catheter material (Fig. 2B and C). In contrast, C. albicans SC5314 showed a highly heterogeneous architecture of biofilms with yeast cells and hyphae embedded within the extracellular matrix (Fig. 2A). Moreover, C. auris biofilms, unlike C. albicans biofilms, had a limited amount of extracellular matrix. Furthermore, the thickness of C. auris biofilms was significantly less than that of C. albicans biofilms (range, 21 to 26 μm for C. auris biofilms versus 50 μm for C. albicans biofilms; P ≤ 0.05) (Fig. 2D to G). Biofilm quantitation on the basis of metabolic activity and biomass revealed that the C. auris isolates tested (n = 15) formed significantly less biofilm than C. albicans SC5314 (Fig. 3A and B) (P < 0.05 for both comparisons).
FIG 2.
Formation of biofilms by C. albicans and C. auris strains. Confocal scanning laser micrographs show top-down three-dimensional views (A to C) and side views (D to F) of biofilms formed by C. albicans (A, D), C. auris MRL 31102 (B, E), and C. auris MRL 31103 (C, F). Magnifications, ×100. (G) Thickness of biofilms formed by the tested isolates. *, P value compared to the thickness of C. albicans biofilms. A P value of <0.05 was considered significant. All experiments were done in triplicate, and data represent means ± SDs. C. albicans SC5314 showed a highly heterogeneous architecture of biofilms with yeast cells and hyphae embedded within the extracellular matrix, while C. auris biofilms had minimal extracellular matrix and were significantly thinner than C. albicans biofilms.
FIG 3.
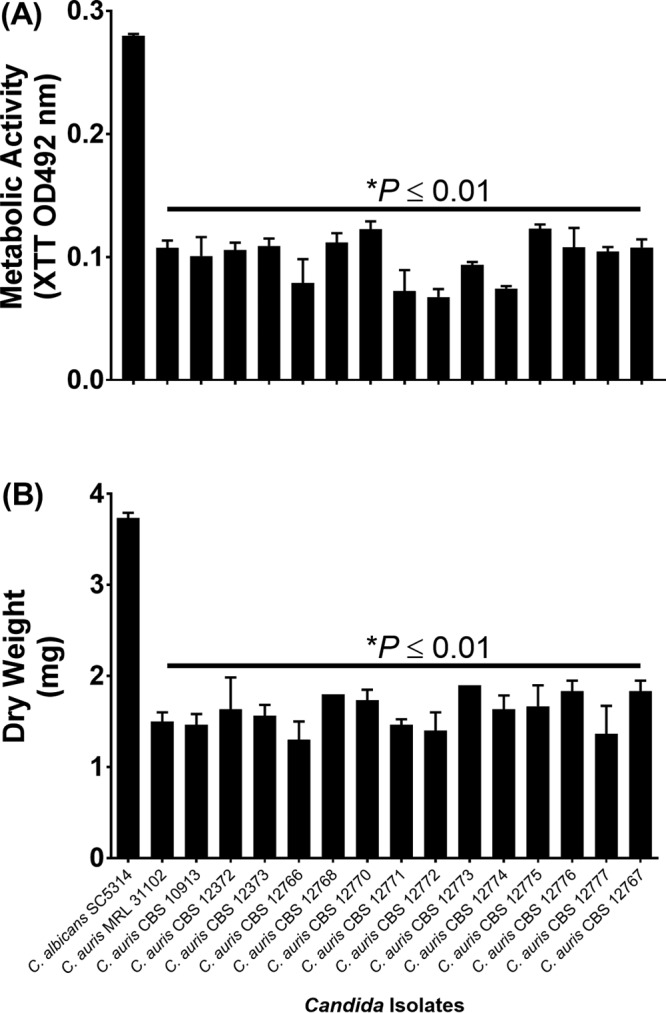
Quantification of biofilms formed by C. albicans and C. auris strains. The metabolic activity (A) and dry weight (B) of the biofilms formed by C. albicans, C. auris MRL 31102 (control), and 14 CBS C. auris strains are shown. *, P value compared to the results for C. albicans. A P value of <0.05 was considered significant. All experiments were done in triplicate, and the data in both plots represent means ± SDs. C. auris biofilms had significantly reduced metabolic activity and biomass compared to those of C. albicans biofilms.
C. auris growth is similar to C. albicans growth.
Growth curve analysis of the control C. auris and C. albicans isolates showed that the two Candida species had similar growth patterns, reaching stationary phase within approximately 20 h (Fig. 4). The doubling times for C. albicans SC5314, C. auris MRL 31102, and C. auris MRL 31103 were 101.22 ± 5.26, 105.62 ± 12.83, and 107.00 ± 7.14 min, respectively (mean ± standard deviation [SD]; P > 0.05 for all comparisons).
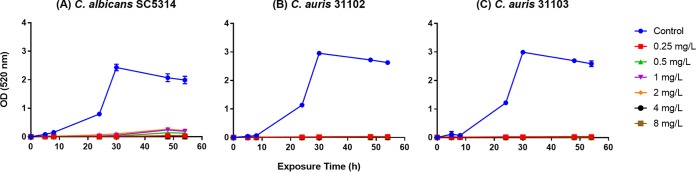
FIG 4.
Effect of different concentrations of the antifungal SCY-078 on the growth of C. albicans and C. auris isolates. C. albicans SC5314 (A), C. auris MRL 31102 (B), and C. auris MRL 31103 (C) cells were grown in the presence of the indicated concentrations of SCY-078. At different time points, aliquots were withdrawn and their ODs were determined spectrophotometrically. All experiments were done in triplicate, and the data in all three panels represent means ± SDs. SCY-078 inhibited the growth of the C. albicans and C. auris strains.
Antifungal susceptibility profile of C. auris.
As shown in Table 2, the MIC90 of SCY-078 was 1 mg/liter, which was similar to the MIC90 of caspofungin (CAS) and micafungin (MFG) (1 mg/liter for both) and within 2 dilutions of the MIC90 of anidulafungin (AFG; 0.25 mg/liter). The MIC90 of SCY-078 was also similar to that of flucytosine (5FC; MIC90 = 1 mg/liter) and lower than that of amphotericin B (AMB; MIC90 = 4 mg/liter). Antifungal susceptibility testing of azoles showed that isavuconazole (ISA) was the most active agent tested (MIC90 = 0.125 mg/liter), followed by posaconazole (POS; MIC90 = 0.5 mg/liter), itraconazole (ITC; MIC90 = 1 mg/liter), and voriconazole (VRC; MIC90 = 2 mg/liter), while fluconazole (FLC) was the least active azole (MIC90 > 64 mg/liter) against the tested isolates (Table 3).
TABLE 2.
In vitro susceptibilities of C. auris isolates to 1,3-β-d-glucan synthesis inhibitors
| Strain | MICa (mg/liter) |
|||
|---|---|---|---|---|
| SCY-078 | AFG | CAS | MFG | |
| MRL 31102 | 0.5 | 0.25 | 0.5 | 0.25 |
| MRL 31103 | 0.5 | 0.25 | 0.5 | 0.25 |
| CBS 10913 | 1 | 0.125 | 0.5 | 2 |
| CBS 12372 | 1 | 0.125 | 0.5 | 1 |
| CBS 12373 | 1 | 0.125 | 0.5 | 1 |
| CBS 12766 | 0.5 | 0.125 | 0.5 | 1 |
| CBS 12767 | 1 | 0.125 | 0.5 | 1 |
| CBS 12768 | 1 | 0.125 | 0.5 | 2 |
| CBS 12770 | 1 | 0.125 | 0.5 | 1 |
| CBS 12771 | 1 | 0.25 | 0.5 | 1 |
| CBS 12772 | 1 | 0.25 | 1 | 1 |
| CBS 12773 | 1 | 0.25 | 1 | 1 |
| CBS 12774 | 2 | 0.125 | 0.25 | 1 |
| CBS 12775 | 1 | 0.125 | 0.25 | 1 |
| CBS 12776 | 1 | 0.125 | 1 | 1 |
| CBS 12777 | 1 | 0.125 | 1 | 1 |
| Range | 0.5–2 | 0.125–0.25 | 0.25–1 | 0.25–2 |
| MIC50 | 1 | 0.125 | 0.5 | 1 |
| MIC90 | 1 | 0.25 | 1 | 1 |
MICs were determined at 24 h.
TABLE 3.
In vitro susceptibilities of C. auris isolates to azoles, 5FC, and AMB
| Strain | MICa (mg/liter) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5FC (48 h) | AMB |
FLC |
ISA (24 h) | ITC (48 h) | POS (48 h) | VRC |
||||
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |||||
| MRL 31102 | 0.5 | 4 | 4 | >64 | >64 | 0.031 | 0.5 | 0.5 | 0.125 | 0.5 |
| MRL 31103 | 0.5 | 4 | 4 | >64 | >64 | 0.031 | 0.5 | 0.25 | 0.125 | 0.5 |
| CBS 10913 | 1 | 1 | 2 | 1 | 2 | 0.004 | <0.063 | 0.25 | <0.063 | <0.063 |
| CBS 12372 | 0.5 | 1 | 2 | 2 | >64 | 0.25 | 1 | 1 | 0.5 | 2 |
| CBS 12373 | 0.5 | 1 | 2 | 16 | >64 | 0.125 | 1 | 0.25 | 0.25 | 2 |
| CBS 12766 | 0.5 | 4 | 4 | 32 | >64 | 0.125 | 1 | 0.5 | 0.5 | 0.5 |
| CBS 12767 | 1 | 2 | 4 | 2 | >64 | 0.016 | 0.5 | 0.25 | 0.5 | 0.5 |
| CBS 12768 | 0.5 | 4 | 4 | 32 | >64 | 0.125 | 1 | 0.25 | 0.5 | 0.5 |
| CBS 12770 | 0.5 | 4 | 4 | 32 | >64 | 0.25 | 0.5 | 0.5 | 0.5 | 2 |
| CBS 12771 | 0.5 | 4 | 4 | 8 | >64 | 0.063 | 0.5 | 0.5 | 1 | 1 |
| CBS 12772 | 1 | 8 | 8 | >64 | >64 | 0.125 | 0.5 | 0.5 | 0.5 | 1 |
| CBS 12773 | 0.5 | 2 | 8 | >64 | >64 | 0.063 | 1 | 1 | 0.5 | 0.5 |
| CBS 12774 | 0.5 | 2 | 4 | >64 | >64 | 0.063 | 0.5 | 0.5 | 1 | 2 |
| CBS 12775 | 0.5 | 2 | 4 | 1 | >64 | 0.016 | 0.5 | 0.25 | 0.5 | 2 |
| CBS 12776 | 0.5 | 2 | 4 | 1 | >64 | 0.063 | 0.5 | 0.25 | 0.5 | 0.5 |
| CBS 12777 | 0.5 | 0.5 | 4 | 1 | >64 | 0.063 | 0.5 | 0.25 | 1 | 1 |
| Range | 0.5 to 1 | 0.5 to 8 | 2 to 4 | 1 to >64 | 2 to >64 | 0.004 to 0.25 | <0.063 to 1 | 0.25 to 1 | <0.063 to 1 | <0.063 to 2 |
| MIC50 | 0.5 | 2 | 4 | 16 | >64 | 0.063 | 0.5 | 0.25 | 0.5 | 0.5 |
| MIC90 | 1 | 4 | 4 | >64 | >64 | 0.125 | 1 | 0.5 | 1 | 2 |
SCY-078 inhibits the growth of C. auris and C. albicans.
Having shown that C. auris had low MIC values of SCY-078, we examined the ability of this drug to inhibit the growth of this yeast. Exposure of C. auris and C. albicans to SCY-078 (at concentrations ranging from 0.25 to 8 mg/liter) led to the complete inhibition of growth of this pathogenic fungus (Fig. 4).
SCY-078 disrupted the ultrastructure of C. auris.
We used scanning electron microscopy to determine the effect of SCY-078 on the ultrastructure of C. auris. As can be seen in Fig. 5A, untreated control C. auris cells had a well-defined, oval-shaped yeast morphology as well as several budding cells. In contrast, cells exposed to SCY-078 (at a concentration of 1× MIC [0.5 mg/liter]) exhibited a severely distorted yeast cell topography with cells fused together, indicating that the cells were unable to divide (Fig. 5B).
FIG 5.
Scanning electron micrograph of C. auris treated with no drug (control) (A) and with SCY-078 at 1× MIC (0.5 mg/liter) (B). Cells were exposed to no drug (control) or SCY-078 at 1× MIC overnight at 35°C and then fixed in 2% glutaraldehyde and processed for scanning electron microscopy. Untreated control C. auris cells had a well-defined, oval-shaped yeast morphology as well as several budding yeasts (A). In contrast, cells exposed to SCY-078 (at a concentration of 1× MIC) exhibited a severely distorted yeast cell topography with cells fused together, indicating that the cells were unable to divide (B). Magnifications, ×6,000.
SCY-078 possesses activity against C. auris biofilms.
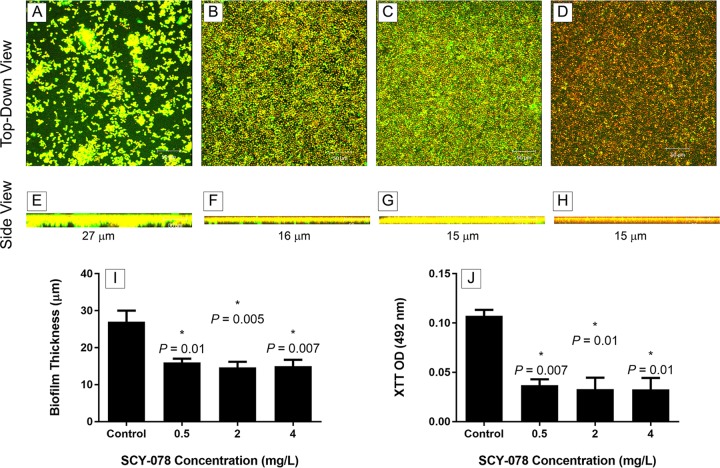
Resistance to antifungals is a hallmark phenotype of biofilms. To determine whether SCY-078 possesses antibiofilm activity, we exposed mature-phase C. auris biofilms to a range of drug concentrations (0.5, 2, and 4 mg/liter). Metabolic activity and confocal scanning laser microscopy data showed that SCY-078 reduced C. auris biofilms significantly at all concentrations tested (P < 0.05) (Fig. 6) (19). Unlike untreated cells, which showed an intense green fluorescence (resulting from concanavalin A [ConA] binding to polysaccharides), C. auris biofilms treated with SCY-078 showed yeast cells with reduced fluorescence, particularly at an SCY-078 concentration of 4 mg/liter (Fig. 6D and H). Additionally, at all tested drug concentrations, the metabolic activity and thickness of the biofilms were reduced significantly compared to those for untreated control biofilms (P < 0.05) (Fig. 6I and J).
FIG 6.
Confocal scanning laser microscopy analyses of the effect of SCY-078 on biofilms formed by C. auris. Biofilms formed by C. auris MRL 31102 were exposed to no drug (control) (A, E) or SCY-078 at different concentrations: 0.5 mg/liter (B, F), 2 mg/liter (C, G), or 4 mg/liter (D, H). Top-down views (A to D) and side views (E to H) of untreated and treated biofilms are shown. Magnifications, ×25. (I and J) The thickness (I) and metabolic activity (J) of untreated (control) and SCY-078-treated biofilms. *, P value compared to the results for the untreated control (no drug). A P value of <0.05 was considered significant. All experiments were done in triplicate, and the bars represent means ± SDs. SCY-078 exhibited potent activity against biofilms formed by C. auris strains.
DISCUSSION
In the current study, we examined 16 different C. auris isolates obtained from different parts of the world; characterized the major Candida virulence factors (including germination, adherence, biofilm formation, and phospholipase and proteinase production); determined their susceptibilities to 11 drugs belonging to different antifungal classes, including a novel orally bioavailable 1,3-β-d-glucan synthesis inhibitor (SCY-078); and evaluated the effect of SCY-078 on the growth, morphology, and ultrastructure of C. auris and its ability to form biofilms.
Germination, adherence, phospholipase and proteinase production, and biofilm formation are major virulence factors known to contribute to Candida pathogenesis and have been well characterized in C. albicans by a number of investigators, including our group (20–24). Few studies have investigated these virulence factors in C. auris, and those that have been conducted have been limited to strains from a single geographical areas or have focused on one strain (7, 17). Our data showed that C. auris, unlike C. albicans, is devoid of the ability to germinate, form hyphae, or produce chlamydospores. Our data confirm and extend the observations by others since we analyzed isolates from different geographic regions (3, 4, 17). Additionally, we showed for the first time that C. auris exhibits a minimal ability to adhere to silicone elastomer relative to C. albicans. Since the ability of Candida to adhere to catheter surfaces is important in causing catheter-associated candidiasis, the weak adherence ability of C. auris suggests that it likely plays some role in catheter-associated candidiasis but not a large one, in contrast to C. albicans and Candida parapsilosis, which are known to cause such infections (25).
Our findings demonstrated that C. auris phospholipase production was strain dependent and that phospholipase production was detected in 37.5% of the isolates tested. In general, the tested C. auris strains that produced this enzyme tended to have weak phospholipase activity. Only one C. auris strain (CBS 12770) had phospholipase activity comparable to that of C. albicans, a known high phospholipase producer (26). Kumar et al. (17) recently reported the isolation of an azole-resistant C. auris strain from a vulvovaginitis patient and showed that this strain exhibited strong phospholipase activity (ratio of the colony diameter to the colony diameter plus the precipitation zone [Pz] = 0.72), proteinase activity, and hemolysin activity. Ours is the first study to demonstrate that the production of phospholipase by C. auris is strain dependent, with the majority of isolates being non-phospholipase producers. In general, the number of C. auris strains tested that produced secreted proteinase was higher than the number that produced phospholipase (64% versus 37.5%, respectively). However, similar to the ability to produce phospholipase, the ability of C. auris to produce secreted proteinase was strain dependent. Interestingly, we observed that strains that were high proteinase producers were not necessarily high phospholipase producers and vice versa (Table 1).
Our results showed that the tested C. auris strains had the ability to form biofilms. However, these biofilms were significantly thinner than the biofilms formed by C. albicans (the C. auris biofilms were approximately 50% the thickness of the C. albicans biofilms). A variation in the ability of different C. auris strains to form biofilms was noted, though these differences were not statistically significant. Our results are in disagreement with those of Oh et al. (7), who tested 15 C. auris isolates and reported that they did not produce biofilms. The differences between our data and those of Oh et al. (7) could be attributed to the use of different media and the fact that they did not use silicon elastomer as a substrate, nor did the investigators treat the plastic surfaces with fetal bovine serum (27). Alternatively, all the isolates used in the study of Oh et al. (7) were obtained from patients' ears, while the strains used in this study were mostly from patients with disseminated candidiasis. In addition, Oh et al. (7) used a semiquantitative subjective scale (range, negative result to a score of +5) to report biofilm formation, while we used three separate quantitative measures (metabolic activity, dry weight, and biofilm thickness).
In general, the ability of C. auris to express the various virulence factors is much weaker than that of C. albicans, suggesting that this emerging species is not as virulent as the latter species. Borman et al. (8) attempted to address the virulence of C. auris by using a Galleria mellonella larva model. Their data showed that C. auris had the same pathogenicity as C. albicans and C. tropicalis and was more virulent (P < 0.05) than other Candida spp. (e.g., C. glabrata, C. parapsilosis, C. krusei). However, unpublished data from our center confirm the poor ability of C. auris to infect and disseminate in mice compared to C. albicans (data not shown). We showed that it is critical to immunocompromise the mice and use a large inoculum (3 × 107 yeast cells/animal) to successfully infect the mice (unpublished data). Therefore, our data suggest that the multidrug-resistant phenotype of C. auris comes with a major fitness cost.
The clinical significance of C. auris appears to reside in its ability to develop resistance to multiple commonly used antifungal agents, leading to infections with high rates of mortality (2–6, 8, 9, 13–16). Therefore, the identification of agents that are effective against this species is critical. Our data demonstrated that C. auris responded differently to various antifungals. Among the azoles, all but one isolate showed reduced susceptibility to fluconazole (MIC, >64 mg/liter), with the other azoles showing variable antifungal activity and isavuconazole being the most active. The only isolate tested in this study to have a low fluconazole MIC was CBS 10913, which, interestingly, was the only strain isolated from a nonblood source (2). Additionally, caspofungin, micafungin, and anidulafungin showed similar activities against the tested isolates. These findings are in agreement with those of recent studies from other investigators, who also reported that C. auris isolates were generally resistant or less susceptible to azoles but susceptible to echinocandins (2–6, 8, 9, 13, 14, 16). Our finding that amphotericin B demonstrated less activity against C. auris strains also agrees with those of others reporting high MICs for this agent against C. auris (9, 16, 28). Our data show that the C. auris isolates tested in our study exhibited multidrug resistance against fluconazole and amphotericin B. Moreover, some isolates also exhibited high MIC values for voriconazole and itraconazole.
To identify new antifungals that may be affective against C. auris, in this study we tested SCY-078, a new 1,3-β-d-glucan synthesis inhibitor, which has been shown to possess potent activity against various Candida spp. An added advantage of SCY-078 is that it is the first orally bioavailable 1,3-β-d-glucan synthesis inhibitor (29). Our data showed that SCY-078 has potent antifungal activity against the C. auris isolates tested. Further, SCY-078 showed growth inhibition and antibiofilm activity against this emerging species. Moreover, C. auris cells exposed to SCY-078 exhibited a severely distorted yeast cell topography and failed to divide. Other investigators showed that increasing concentrations of caspofungin treatment altered the morphology of various Candida species (30, 31). These authors showed that caspofungin treatment of cells affected the morphology of Candida, resulting in cells with an increased size, a lack of distinct rings around the budding site, and the absence of filamentation in C. albicans. Unlike caspofungin, treatment with SCY-078 led to the inhibition of cell division, suggesting that, in addition to the inhibition of 1,3-β-d-glucan synthesis, this drug may have a separate target or may affect this enzyme through yet undefined unique interactions.
In summary, our study showed that C. auris expresses several virulence factors, albeit to a lesser extent than C. albicans and in a strain-dependent manner. We demonstrated that SCY-078 is a potent drug and could be an important addition to the antifungal armamentarium to treat patients with infections caused by this multiresistant species.
MATERIALS AND METHODS
Isolates tested.
The following C. auris strains were used in this study: 2 C. auris isolates (MRL 31102 and MRL 31103) obtained from a 68-year-old male enrolled in a recent candidemia trial at a German site and 14 C. auris isolates from the CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands (CBS 10913, CBS 12372, CBS 12373, CBS 12766 to CBS 12768, and CBS 12770 to CBS 12777) that were isolated from patients living in Japan, India, and South Korea (Table 4). To identify the MRL 31102 and MRL 31103 isolates, we originally utilized the yeast identification tool API 20C AUX. However, we were unable to identify the strains with a high percentage (>85%) of certainty. Moreover, since C. auris is also often misidentified as Candida haemulonii, Candida famata, Candida sake, or Rhodotorula glutinis using typical identification methods, including the API 20C AUX tool and the Vitek 2 system, we used DNA typing, employing the internal transcribed spacer 1 (ITS1) and ITS4 regions of the fungal ribosome (2–6, 12–15). Identification was performed using a BLAST algorithm search against the sequences in the UNITE database (32). This analysis allowed us to identify MRL 31102 and MRL 31103, which had 89 and 99% homology with the C. auris strains in the UNITE database, respectively, as C. auris. In the current study, two strains of C. albicans (SC5314 and ATCC 14053), obtained from the American Type Culture Collection (Manassas, VA), were used as comparators. C. albicans was selected as a comparator because its virulence factors have been well characterized and it is responsible for nearly 50% of all Candida infections.
TABLE 4.
C. auris isolates used in this study
| Strain | Species | Isolation source | Country of isolation |
|---|---|---|---|
| MRL 31102 | C. auris | Blood | Germany |
| MRL 31103 | C. auris | Blood | Germany |
| CBS 10913 | C. auris | Ear | Japan |
| CBS 12372 | C. auris | Blood | South Korea |
| CBS 12373 | C. auris | Blood | South Korea |
| CBS 12766 | C. auris | Blood | India |
| CBS 12767 | C. auris | Blood | India |
| CBS 12768 | C. auris | Blood | India |
| CBS 12770 | C. auris | Blood | India |
| CBS 12771 | C. auris | Blood | India |
| CBS 12772 | C. auris | Blood | India |
| CBS 12773 | C. auris | Blood | India |
| CBS 12774 | C. auris | Blood | India |
| CBS 12775 | C. auris | Blood | India |
| CBS 12776 | C. auris | Blood | India |
| CBS 12777 | C. auris | Blood | India |
Ability of C. auris to form chlamydospores.
Since chlamydospores are utilized to help distinguish fungal types (33), we evaluated the ability of the C. auris isolates to exhibit this morphotype. Briefly, an isolated colony of C. auris was inoculated onto a cornmeal agar plate (Becton Dickinson, Sparks, MD, USA) to make a lawn. A coverslip was applied to the inoculated area in order to create microaerophilic conditions, and the plate was incubated at 30°C for 3 days with protection from light. The plate was subsequently examined for chlamydospore formation under white light at a ×20 magnification. C. albicans ATCC 14053 was used as a positive control.
Characterization of C. auris virulence factors.
The ability of Candida species to cause infection is attributed to the possession of virulence factors, including the ability to germinate, adhere to host cells, secrete extracellular enzymes such as phospholipase and proteinase, and form biofilms (18, 34, 35). To date, the characterization of C. auris virulence factors is limited.
(i) Ability to germinate.
The ability of C. auris to form germ tubes was assessed using the germ tube test (36–38). Briefly, an isolated colony of C. auris, taken from a 24-h culture grown on potato-dextrose agar (Becton Dickinson, Sparks, MD, USA), was inoculated into 0.5 ml of fetal bovine serum (Fisher Thermo Scientific, Cleveland, OH, USA) and incubated for 2 h at 37°C. Next, 10 μl of fetal bovine serum was then transferred onto a glass slide, a coverslip was added, and the slide was examined using phase-contrast microscopy at a ×20 magnification.
(ii) Adherence assay.
The ability of Candida species to adhere to indwelling medical devices and host tissues is a known virulence factor. Since silicon elastomer is used extensively in central venous catheters, in this study, we evaluated the ability of C. auris isolates to adhere to silicon elastomer, which was used as a representative catheter material.
To determine the ability of C. auris to adhere to silicon elastomer surfaces, yeast cells were inoculated into 10 ml yeast nitrogen base broth (Becton Dickinson, Sparks, MD, USA) and incubated overnight at 37°C. The cells were then washed twice with Hanks' buffered salt solution, adjusted to 5 × 103 cells/ml, and further diluted to obtain a cell suspension of 250 cells/ml using the same buffer. Silicon elastomer discs (diameter, 1.2 cm) were transferred to 12-well tissue culture plates, to which 2 ml of cell suspension (prepared as described above) was added. Next, the plates were incubated at 37°C for 30 min to allow the cells to adhere to the discs. After incubation, discs containing adherent cells were transferred to a fresh plate containing 2 ml Hanks' buffered salt solution. Subsequently, the discs were washed twice with Hanks' buffered salt solution and, following aspiration of the buffer, were overlaid with 3 ml of Sabouraud dextrose agar (Becton Dickinson, Sparks, MD, USA). The plates were incubated at 37°C for 18 to 24 h, and the number of C. auris cells adhering to the catheter material was assessed by counting the number of CFU.
(iii) Phospholipase assay.
The ability of the C. auris strains to secrete phospholipase was determined using a phospholipase activity assay that employed Sabouraud dextrose agar as a medium supplemented with 58.4 g/liter NaCl, 5.5 g/liter CaCl2, and 10% egg yolk emulsion (30% stock; Hi-Media, India) as previously described (26, 39). Medium without egg yolk was sterilized at 121°C for 15 min and cooled to 50°C. Egg yolk emulsion was then added, and the agar was dispensed into petri dishes (20 ml/plate). A 10-μl aliquot containing 1 × 107 yeast cells/ml was added to the center of the plates, and the plates were incubated at 37°C for 5 days. Following incubation, the production of phospholipase (indicated by the appearance of a whitish zone of precipitation around the yeast colony) was assessed by measuring the ratio of the colony diameter to the colony diameter plus the precipitation zone (the Pz value). Pz values of ≤0.89, 0.90 to 0.99, and 1 indicate strong, weak, and no phospholipase activity, respectively (26).
(iv) Proteinase assay.
Proteinase activity assays were performed using a protease fluorescent detection kit (catalog number PF0100; Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer's specifications (40). The C. auris strains were cultured in 20 ml yeast nitrogen base for 48 h. Following incubation, the supernatant containing secreted proteinases was concentrated, and the volumes were adjusted to the proportion of the total number of CFU. Subsequently, 10 μl of the normalized protein-rich supernatants was tested for proteinase activity using the kit described above.
(v) Biofilm formation.
The ability of Candida to form biofilms has been linked to catheter-associated infections. Therefore, we evaluated whether C. auris can produce biofilms using our in vitro biofilm model as described previously (19). In the current study, we used silicone elastomer as a substrate for biofilm formation since catheters are commonly constructed using this material and silicone elastomer has been used as a substrate in numerous studies investigating catheter-associated biofilms performed by our group and others (41–51). Briefly, silicone elastomer discs (diameter, 1.2 cm) were placed in 12-well tissue culture plates and incubated in fetal bovine serum at 37°C on a rocker for 24 h. The discs were then removed from the fetal bovine serum, immersed in a 4-ml cell suspension with a concentration of 1 × 107 cells/ml, and incubated for 90 min at 37°C (adhesion phase). Subsequently, the discs were transferred to 4 ml yeast nitrogen base medium and incubated for 24 h at 37°C to form mature biofilms (mature phase) (19).
The quantification of the biofilms formed by different strains was performed using a colorimetric metabolic assay (to measure mitochondrial dehydrogenase activity) and dry weight analysis (to measure total biofilm mass, which includes fungal cells and matrix). For evaluation of metabolic activity, discs with biofilms were transferred to 12-well tissue culture plates containing 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT; 12.5 μg/ml) and menadione (1 μM) in phosphate-buffered saline and incubated at 37°C for 3 h as described previously (19). Next, the biofilms were scraped, transferred into a tube, and centrifuged at 3,000 rpm for 15 min. The absorbance of the resulting supernatant was measured spectrophotometrically at 492 nm. To determine dry weight (biomass), the pellet obtained from the centrifugation step described above was resuspended in phosphate-buffered saline and filtered through a preweighed filter (pore size, 0.2 μm; Millipore, Billerica, MA, USA), dried at 37°C for 48 h, and weighed (19). The biomass of the biofilms was calculated from the difference in the weight of the preweighed filters.
To examine the morphology and thickness of the formed biofilms, confocal scanning laser microscopy was used. Briefly, following biofilm formation, the silicon elastomer discs on which the biofilms were developed were transferred to a 35-mm-diameter glass-bottom petri dish (MatTek Corp., Ashland, MA, USA) and incubated for 45 min at 37°C in 2 ml of phosphate-buffered saline containing the fluorescent stains FUN-1 (10 μM) and concanavalin A (ConA; 25 mg/liter)-Alexa Fluor 488 conjugate (both dyes from Molecular Probes, Inc., Eugene, OR). FUN-1 (excitation wavelength, 543 nm; emission wavelength, 560 nm) is converted to orange-red cylindrical intravacuolar structures by metabolically active cells, and ConA (excitation wavelength, 488 nm; emission wavelength, 505 nm) fluoresces green when bound to glucose and mannose residues of fungal polysaccharides (present in the cell wall and biofilm matrix). After incubation with the dyes, the silicon elastomer discs were flipped and the biofilms were examined using a Zeiss LSM510 confocal scanning laser microscope (Carl Zeiss, Inc.). To determine the structure of the biofilms, a series of horizontal (x-y) optical sections with a thickness of 0.9 μm were taken at 0.44-μm intervals throughout the full length of the biofilm. Confocal images of green (ConA) and red (FUN-1) fluorescence were simultaneously collected using a multitrack mode (19).
Determination of susceptibility profiles of C. auris strains.
To establish the susceptibility profiles of the C. auris strains, broth microdilution susceptibility testing was performed in accordance with the guidelines in the Clinical and Laboratory Standards Institute (CLSI) M27-A3 document (52). RPMI 1640 medium with 3-(N-morpholino)propanesulfonic acid (RPMI), an inoculum of 0.5 × 103 to 2.5 × 103 cells/ml, and incubation at 35°C were used. The activities of 11 antifungals against the C. auris isolates were tested, including SCY-078 (Scynexis, Jersey City, NJ, USA), a novel orally bioavailable 1,3-β-d-glucan synthesis inhibitor, and currently available antifungals amphotericin B and anidulafungin (Pfizer Pharmaceuticals, New York, NY, USA), caspofungin (Merck Co., Kenilworth, NJ, USA), fluconazole, flucytosine, and isavuconazole (Astellas Pharma US, Inc., Northbrook, IL, USA), itraconazole and micafungin (Astellas Pharma, Inc., Tokyo, Japan), posaconazole, and voriconazole (AMB, FLC, 5FC, ITC, POS, and VRC were obtained from Sigma-Aldrich, St. Louis, MO, USA). MIC panels were incubated for 24 h (SCY-078, AMB, AFG, CAS, FLC, ISA, MFG, and VRC) or 48 h (5FC, AMB, FLC, ITC, POS, and VRC). MIC endpoints were determined visually as the lowest concentration of drug that resulted in the complete inhibition of growth (AMB) or a decrease of growth by ≥50% relative to that of the growth control (SCY-078, AFG, CAS, FLC, 5FC, ISA, ITC, MFG, POS, VRC) (52). In all instances, MIC plates were prepared using reagent-grade powders, as directed by CLSI (52).
Effect of SCY-078 on the growth of C. auris and C. albicans.
Evaluation of the ability of a drug to inhibit microbial growth is conventionally determined either by counting the number of CFU, which is an indicator of cell viability, or by turbidity measurement (using a spectrophotometer), which indicates cell density but does not differentiate between live and dead cells (52, 53). In this study, we used the spectrophotometric approach to evaluate the ability of SCY-078 to inhibit Candida growth as described previously (54). Briefly, cells were harvested from 18- to 24-h-old cultures, washed twice with phosphate-buffered saline, and adjusted to 5 × 105 cells/ml in 50-ml conical tubes containing RPMI alone (with no drug as a growth control) or 0.5×, 1×, 2×, 4×, 8×, or 16× MIC of SCY-078. Medium alone with no drug or yeast cells was used as a blank. All tubes were incubated at 37°C. At different time points (0, 5, 8, 24, 30, 48, and 54 h), aliquots were taken and their optical densities (ODs) were determined spectrophotometrically at 520 nm. A growth curve showing cell inhibition temporally was constructed.
Scanning electron microscopy.
The effect of SCY-078 on C. auris morphology and ultrastructure was determined using scanning electron microscopy as described previously (19). Briefly, the C. auris strains were exposed overnight to 1× MIC (0.5 mg/liter) of SCY-078 at 35°C. Next, 200 μl of a cell suspension was fixed in 2% glutaraldehyde, and the fixed cell suspension was incubated at 4°C for 48 h. After fixation, the samples were processed and dried. The processed samples were coated with palladium for 60 s and viewed with a Nova NanoLab 200 FEG-SEM/FIB scanning electron microscope in high-vacuum mode at 2.00 kV.
Effect of SCY-078 on C. auris biofilms.
To evaluate the activity of SCY-078 against C. auris biofilms, discs with mature biofilms were transferred to wells containing different concentrations of SCY-078 (range, 0.5 to 4 mg/liter). Following 48 h of incubation, the metabolic activities of the biofilms were measured using the XTT reduction assay as described above. Images and the thicknesses of biofilms growing in the presence or absence of drug were captured using confocal scanning laser microscopy, also as described above (19).
Statistical analysis.
Statistical analyses of all the data were performed using GraphPad Prism (version 6) software. The treated groups were compared to the untreated control groups using unpaired t tests, and a P value of <0.05 was considered statistically significant. All experiments were done in triplicate. Doubling times were calculated using R, the statistical programming language (https://cran.r-project.org/).
Accession number(s).
The sequences of C. auris strains MRL 31102 and MRL 31103 have been deposited in GenBank under accession numbers KY514262 and KY514058, respectively.
ACKNOWLEDGMENTS
Partial support for this study was provided by Scynexis Inc. and a grant from NIH (grant number R01DE024228) to M.G. and P.K.M. We also acknowledge support from the NIH-funded Skin Diseases Research Center (NIAMS P30 AR039750).
We thank Nanthawan Avishai for technical support with the scanning electron microscopy experiments. We also acknowledge support from the Swagelok Center for Surface Analysis of Materials, Case Western Reserve University, for scanning electron microscopy analyses. We also acknowledge the Confocal Scanning Laser Microscopy Core at Genetics Department, Case Western Reserve University.
REFERENCES
- 1.Centers for Disease Control and Prevention. 24 June 2016, posting date. Global emergence of invasive infections caused by the multidrug-resistant yeast Candida auris. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/fungal/diseases/candidiasis/candida-auris-alert.html. Accessed 1 July 2016. [Google Scholar]
- 2.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 3.Lee WG, Shin JH, Uh Y, Kang MG, Kim SH. 2011. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 49:3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, Singh PK, Jain S, Kathuria S, Randhawa HS, Hagen F, Meis JF. 2013. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 19:1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magobo RE, Corcoran C, Seetharam S, Govender NP. 2014. Candida auris-associated candidemia, South Africa. Emerg Infect Dis 20:1250–1251. doi: 10.3201/eid2007.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emara M, Ahmad S, Khan Z, Joseph L, Al-Obaid I, Purohit P, Bafna R. 2015. Candida auris candidemia in Kuwait, 2014. Emerg Infect Dis 21:1091–1092. doi: 10.3201/eid2106.150270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh BJ, Shin JH, Kim M-N, Sung H, Lee K, Joo MY, Shin MG, Suh SP, Ryang DW. 2011. Biofilm formation and genotyping of Candida haemulonii, Candida pseudohaemulonii, and a proposed new species (Candida auris) isolates from Korea. Med Mycol 49:98–102. doi: 10.3109/13693786.2010.493563. [DOI] [PubMed] [Google Scholar]
- 8.Borman AM, Szekely A, Johnson EM. 2016. Comparative pathogenicity of United Kingdom isolates of the emerging pathogen Candida auris and other key pathogenic Candida species. mSphere 1:e00189-16. doi: 10.1128/mSphere.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calvo B, Melo ASA, Perozo-Mena A, Hernandez M, Francisco EC, Hagen F, Meis JF, Colombo AL. 2016. First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J Infect 73:369–374. doi: 10.1016/j.jinf.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Morales-Lopez SE, Parra-Giraldo CM, Ceballos-Garzon A, Martinez HP, Rodriguez GJ, Alvarez-Moreno CA, Rodriguez JY. 2017. Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg Infect Dis 23:162–164. doi: 10.3201/eid2301.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, Hall A, Ryan L, Shackleton J, Trimlett R, Meis JF, Armstrong-James D, Fisher MC. 2016. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, Kemble SK, Pacilli M, Black SR, Landon E, Ridgway J, Palmore TN, Zelzany A, Adams EH, Quinn M, Chaturvedi S, Greenko J, Fernandez R, Southwick K, Furuya EY, Calfee DP, Hamula C, Patel G, Barrett P, Lafaro P, Berkow EL, Moulton-Meissner H, Noble-Wang J, Fagan RP, Jackson BR, Lockhart SR, Litvintseva AP, Chiller TM. 2017. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus—United States, May 2013-August 2016. Am J Transplant 17:296–299. doi: 10.1111/ajt.14121. [DOI] [PubMed] [Google Scholar]
- 13.Chowdhary A, Kumar VA, Sharma C, Prakash A, Agarwal K, Babu R, Dinesh KR, Karim S, Singh SK, Hagen F, Meis JF. 2014. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis 33:919–926. doi: 10.1007/s10096-013-2027-1. [DOI] [PubMed] [Google Scholar]
- 14.Khillan V, Rathore N, Kathuria S, Chowdhary A. 2014. A rare case of breakthrough fungal pericarditis due to fluconazole-resistant Candida auris in a patient with chronic liver disease, p 1–5. JMM Case Rep doi: 10.1099/jmmcr.0.T00018. [DOI] [Google Scholar]
- 15.Kim M-N, Shin JH, Sung H, Lee K, Kim E-C, Ryoo N, Lee J-S, Jung S-I, Park KH, Kee SJ, Kim SH, Shin MG, Suh SP, Ryang DW. 2009. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis 48:e57–e61. doi: 10.1086/597108. [DOI] [PubMed] [Google Scholar]
- 16.Chakrabarti A, Sood P, Rudramurthy SM, Chen S, Kaur H, Capoor M, Chhina D, Rao R, Eshwara VK, Xess I, Kindo AJ, Umabala P, Savio J, Patel A, Ray U, Mohan S, Iyer R, Chander J, Arora A, Sardana R, Roy I, Appalaraju B, Sharma A, Shetty A, Khanna N, Marak R, Biswas S, Das S, Harish BN, Joshi S, Mendiratta D. 2015. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med 41:285–295. doi: 10.1007/s00134-014-3603-2. [DOI] [PubMed] [Google Scholar]
- 17.Kumar D, Banerjee T, Pratap CB, Tilak R. 2015. Itraconazole-resistant Candida auris with phospholipase, proteinase and hemolysin activity from a case of vulvovaginitis. J Infect Dev Ctries 9:435–437. doi: 10.3855/jidc.4582. [DOI] [PubMed] [Google Scholar]
- 18.Ghannoum MA, Radwan SS. 1990. Candida adherence to epithelial cells, vol 1 CRC Press, Inc, Boca Raton, FL. [Google Scholar]
- 19.Chandra J, Mukherjee PK, Ghannoum MA. 2008. In vitro growth and analysis of Candida biofilms. Nat Protoc 3:1909–1924. doi: 10.1038/nprot.2008.192. [DOI] [PubMed] [Google Scholar]
- 20.Berman J, Sudbery PE. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet 3:918–930. [DOI] [PubMed] [Google Scholar]
- 21.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell 2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moyes DL, Richardson JP, Naglik JR. 2015. Candida albicans-epithelial interactions and pathogenicity mechanisms: scratching the surface. Virulence 6:338–346. doi: 10.1080/21505594.2015.1012981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghannoum MA. 2000. Potential role of phospholipases in virulence and fungal pathogenesis. Clin Microbiol Rev 13:122–143. doi: 10.1128/CMR.13.1.122-143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fanning S, Mitchell AP. 2012. Fungal biofilms. PLoS Pathog 8:e1002585. doi: 10.1371/journal.ppat.1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn DM, Ghannoum MA. 2004. Candida biofilms: antifungal resistance and emerging therapeutic options. Curr Opin Investig Drugs 5:186–197. [PubMed] [Google Scholar]
- 26.Price MF, Wilkerson ID, Gentry LO. 1982. Plate method for detection of phospholipase activity in Candida albicans. Sabouraudia 20:7–14. doi: 10.1080/00362178285380031. [DOI] [PubMed] [Google Scholar]
- 27.Shin JH, Kee SJ, Shin MG, Kim SH, Shin DH, Lee SK, Suh SP, Ryang DW. 2002. Biofilm production by isolates of Candida species recovered from nonneutropenic patients: comparison of bloodstream isolates with isolates from other sources. J Clin Microbiol 40:1244–1248. doi: 10.1128/JCM.40.4.1244-1248.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, Meis JF, Chowdharya A. 2015. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization–time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol 53:1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiménez-Ortigosa C, Paderu P, Motyl MR, Perlina DS. 2014. Enfumafungin derivative MK-3118 shows increased in vitro potency against clinical echinocandin-resistant Candida species and Aspergillus species isolates. Antimicrob Agents Chemother 58:1248–1251. doi: 10.1128/AAC.02145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rueda C, Cuenca-Estrella M, Zaragoza O. 2014. Paradoxical growth of Candida albicans in the presence of caspofungin is associated with multiple cell wall rearrangements and decreased virulence. Antimicrob Agents Chemother 58:1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bizerra FC, Melo AS, Katchburian E, Freymuller E, Straus AH, Takahashi HK, Colombo AL. 2011. Changes in cell wall synthesis and ultrastructure during paradoxical growth effect of caspofungin on four different Candida species. Antimicrob Agents Chemother 55:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abarenkov K, Henrik Nilsson R, Larsson KH, Alexander IJ, Eberhardt U, Erland S, Hoiland K, Kjoller R, Larsson E, Pennanen T, Sen R, Taylor AF, Tedersoo L, Ursing BM, Vralstad T, Liimatainen K, Peintner U, Koljalg U. 2010. The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol 186:281–285. doi: 10.1111/j.1469-8137.2009.03160.x. [DOI] [PubMed] [Google Scholar]
- 33.Odds FC. 1988. Candida and candidosis: a review and bibliography, 2nd ed, vol 1 Bailliere Tindall, London, England. [Google Scholar]
- 34.Hofs S, Mogavero S, Hube B. 2016. Interaction of Candida albicans with host cells: virulence factors, host defense, escape strategies, and the microbiota. J Microbiol 54:149–169. doi: 10.1007/s12275-016-5514-0. [DOI] [PubMed] [Google Scholar]
- 35.Tsui C, Kong EF, Jabra-Rizk MA. 2016. Pathogenesis of Candida albicans biofilm. Pathog Dis 74:ftw018. doi: 10.1093/femspd/ftw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon-Chung KJ, Bennett JE. 1992. Medical mycology. Lea & Febiger, Philadelphia, PA. [Google Scholar]
- 37.McGinnis MR. 1980. Laboratory handbook of medical mycology. Academic Press, New York, NY. [Google Scholar]
- 38.Larone DH. 1995. Medically important fungi: a guide to identification, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 39.Ghannoum MA. 1998. Extracellular phospholipases as universal virulence factor in pathogenic fungi. Nihon Ishinkin Gakkai Zasshi 39:55–59. doi: 10.3314/jjmm.39.55. [DOI] [PubMed] [Google Scholar]
- 40.Cupp-Enyard C. 2009. Use of the protease fluorescent detection kit to determine protease activity. J Vis Exp 2009:1514. doi: 10.3791/1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daniels KJ, Srikantha T, Pujol C, Park YN, Soll DR. 2015. Role of Tec1 in the development, architecture, and integrity of sexual biofilms of Candida albicans. Eukaryot Cell 14:228–240. doi: 10.1128/EC.00224-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emerson RJ IV, Camesano TA. 2004. Nanoscale investigation of pathogenic microbial adhesion to a biomaterial. Appl Environ Microbiol 70:6012–6022. doi: 10.1128/AEM.70.10.6012-6022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fazly A, Jain C, Dehner AC, Issi L, Lilly EA, Ali A, Cao H, Fidel PL Jr, Rao RP, Kaufman PD. 2013. Chemical screening identifies filastatin, a small molecule inhibitor of Candida albicans adhesion, morphogenesis, and pathogenesis. Proc Natl Acad Sci U S A 110:13594–13599. doi: 10.1073/pnas.1305982110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawser SP, Douglas LJ. 1994. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun 62:915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holmes AR, van der Wielen P, Cannon RD, Ruske D, Dawes P. 2006. Candida albicans binds to saliva proteins selectively adsorbed to silicone. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 102:488–494. doi: 10.1016/j.tripleo.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 46.Katragkou A, Chatzimoschou A, Simitsopoulou M, Georgiadou E, Roilides E. 2011. Additive antifungal activity of anidulafungin and human neutrophils against Candida parapsilosis biofilms. J Antimicrob Chemother 66:588–591. doi: 10.1093/jac/dkq466. [DOI] [PubMed] [Google Scholar]
- 47.Kuhn DM, Chandra J, Mukherjee PK, Ghannoum MA. 2002. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect Immun 70:878–888. doi: 10.1128/IAI.70.2.878-888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuhn DM, George T, Chandra J, Mukherjee PK, Ghannoum MA. 2002. Antifungal susceptibility of Candida biofilms: unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob Agents Chemother 46:1773–1780. doi: 10.1128/AAC.46.6.1773-1780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodger G, Taylor RL, Pearson GJ, Verran J. 2010. In vitro colonization of an experimental silicone by Candida albicans. J Biomed Mater Res B Appl Biomater 92:226–235. doi: 10.1002/jbm.b.31509. [DOI] [PubMed] [Google Scholar]
- 50.Zhao X, Daniels KJ, Oh SH, Green CB, Yeater KM, Soll DR, Hoyer LL. 2006. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 152:2287–2299. doi: 10.1099/mic.0.28959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao X, Oh SH, Yeater KM, Hoyer LL. 2005. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiology 151:1619–1630. doi: 10.1099/mic.0.27763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts, 3rd ed Approved standard M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 53.Canton E, Peman J, Viudes A, Quindos G, Gobernado M, Espinel-Ingroff A. 2003. Minimum fungicidal concentrations of amphotericin B for bloodstream Candida species. Diagn Microbiol Infect Dis 45:203–206. doi: 10.1016/S0732-8893(02)00525-4. [DOI] [PubMed] [Google Scholar]
- 54.Mukherjee PK, Chandra J, Retuerto M, Sikaroodi M, Brown RE, Jurevic R, Salata RA, Lederman MM, Gillevet PM, Ghannoum MA. 2014. Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog 10:e1003996. doi: 10.1371/journal.ppat.1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]