ABSTRACT
Twenty-seven colistin-resistant, carbapenemase-producing Klebsiella pneumoniae isolates were identified from hospitals in Serbia. All isolates were blaCTX-M-15 positive; ST101, ST888, ST437, ST336, and ST307 were blaOXA-48 positive; and ST340 was blaNDM-1 positive. ST307 had an insertion, and ST336 had a premature stop codon in the mgrB gene. Amino acid substitutions were detected in PmrAB of isolates ST101, ST888, ST336, and ST307. The mcr-1 and mcr-2 were not detected. An increase in phoP, phoQ, and pmrK gene transcription was detected for all sequence types.
KEYWORDS: Klebsiella pneumoniae, carbapenem resistance, colistin resistance, molecular epidemiology
TEXT
Polymyxins are the treatment cornerstone for infections caused by carbapenem-resistant Gram-negative bacilli, including Klebsiella pneumoniae. Thus, the emergence of colistin-resistant strains among the multidrug-resistant K. pneumoniae is an inevitable result of the increased use of this antimicrobial agent. Outbreaks of colistin-resistant, carbapenemase-producing K. pneumoniae are especially worrisome and have been described in hospitals in many countries, such as Greece, South Korea, the United States, and France (1–5).
Twenty seven colistin- and carbapenem-resistant K. pneumoniae isolates recovered in three Serbian tertiary care hospitals and one private laboratory between 2013 and 2016, were analyzed in this study (Table 1). Isolates Kc3 and Kc4 originated from the same patient, as well as K3 and K9, but were isolated from different specimens or within a time span of 6 months, respectively. Twelve K. pneumoniae isolates came from a single hospital, The Clinical Center of Vojvodina, a university-affiliated medical center in Novi Sad, in the northern part of Serbia (October 2015 to February 2016). Four isolates were from The Clinical Center Niš, an academic medical center in Niš, in the southern part of the country, and 10 from a private laboratory in Belgrade, in central Serbia. The majority of the isolates were from adult patients, and there was no evidence of prior colistin administration for these patients (Table 1). The only pediatric isolate was isolate 11070, which was obtained from the large university-affiliated tertiary care pediatric hospital in Belgrade, the Mother and Child Health Care Institute of Serbia Dr. Vukan Čupić, from a 3-year-old patient from Ukraine. The child had previously received multiple courses of antibiotics to treat recurrent episodes of acute pyelonephritis and had also been subjected to antibiotic prophylaxis, but there was no evidence of prior colistin administration.
TABLE 1.
Characteristics of colistin-resistant, carbapenemase-producing K. pneumoniae isolates from Serbiaa
| Case | Medical setting | Isolate | Isolation date (day/mo/yr) | Clinical sample | Colistin MIC (μg/ml) | Imipenem/meropenem MICs (μg/ml) | mgrB gene | PmrA/PmrB amino acid changes compared to colistin-susceptible strain | bla genes | PFGE genotype | MLST (ST) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (H) | CCN-N | Ni9 | 12/11/2013 | Urine | >16 | >8/>8 | WT | –/– | blaNDM-1, blaCTX-M-15 | III | ST340 |
| 2 (H) | CCN-N | Ni21 | 09/01/2014 | Urine | >16 | >8/>8 | WT | –/– | blaNDM-1, blaCTX-M-15 | III | ST340 |
| 3 (H) | CCN-N | Ni34 | 21/03/2014 | Blood | >16 | >8/>8 | WT | –/– | blaNDM-1, blaCTX-M-15 | III | ST340 |
| 4 (H) | CCN-N | DM5 | 05/02/2014 | Urine | >16 | >8/>8 | WT | –/– | blaNDM-1, blaCTX-M-15 | III | ST340 |
| 5 (H) | CCV-NS | Kc1 | 27/12/2015 | Skin | 16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | II | ST101 |
| 6 (H) | CCV-NS | Kc2 | 10/11/2015 | Wound | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | II | ST101 |
| 7 (H) | CCV-NS | Kc3 | 27/12/2015 | Wound | >16 | >8/>8 | WT | Ala217Val/Thr157Pro; Gly256Arg | blaOXA-48, blaCTX-M-15 | II | ST101 |
| 8 (H) | CCV-NS | Kc4 | 18/12/2015 | Skin | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | II | ST101 |
| 9 (H) | CCV-NS | Kc5 | 06/01/2016 | Skin | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | II | ST101 |
| 10 (H) | CCV-NS | Kc6 | 28/02/2016 | Wound | 16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | II | ST101 |
| 11 (H) | CCV-NS | Kc7 | 17/02/2016 | Wound | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | II | ST101 |
| 12 (H) | CCV-NS | Kc8 | 20/11/2015 | Wound | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | II | ST101 |
| 13 (H) | CCV-NS | Kc9 | 19/10/2015 | BA | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | II | ST101 |
| 14 (H) | CCV-NS | Kc10 | 27/11/2015 | Urine | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | II | ST101 |
| 15 (H) | CCV-NS | Kc11 | 10/08/2015 | Skin | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | II | ST101 |
| 16 (H) | CCV-NS | Kc12 | 17/07/2015 | Urine | 16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | II | ST101 |
| 17 (O) | K-B | K1 | 28/10/2015 | Urine | >16 | 8/8 | WT | –/– | blaOXA-48, blaCTX-M-15 | IV | ST437 |
| 18 (O) | K-B | K2 | 28/10/2015 | Urine | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | VI | ST888 |
| 19 (O) | K-B | K3 | 05/12/2015 | Blood/CVC | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | VI | ST888 |
| 20 (H) | K-B | K4 | 26/12/2015 | Blood | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | VI | ST888 |
| 21 (H) | K-B | K5 | 30/12/2015 | Urine | >16 | >8/>8 | 29 aa, truncated | Glu57Gly/Gly256Arg | blaOXA-48, blaCTX-M-15 | V | ST336 |
| 22 (O) | K-B | K6 | 03/01/2016 | Urine | 16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | VI | ST888 |
| 23 (O) | K-B | K7 | 2015 | Urine | >16 | 8/8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | VI | ST888 |
| 24 (O) | K-B | K8 | 2015 | Urine | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | VI | ST888 |
| 25 (O) | K-B | K9 | 01/02/2016 | BA | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | VI | ST888 |
| 26 (O) | K-B | K10 | 28/02/2016 | Urine | >16 | >8/>8 | WT | Ala217Val/Gly256Arg | blaOXA-48, blaCTX-M-15 | VI | ST888 |
| 27 (H) | MCHCIS-B | 11070 | 26/10/2015 | TA | >16 | >8/>8 | ISKpn26 | Ala41Thr/Leu213Met; Gly256Arg | blaOXA-48, blaCTX-M-15 | I | ST307 |
Abbreviations: H, hospitalized; O, outpatient; CCN-N, Clinical Center of Niš, Niš; CCV-NS, Clinical Center of Vojvodina, Novi Sad; K-B, Institute for Laboratory Diagnostics Konzilijum, Belgrade; MCHCIS-B, Mother and Child Health Care Institute of Serbia Dr. Vukan Čupić, Belgrade; BA, bronchial aspirate; CVC, central venous catheter; TA, tracheal aspirate.
Genetic relatedness among isolates was analyzed by pulsed-field gel electrophoresis (PFGE) of XbaI-restricted total genomic DNA according to a previously described protocol (6). PFGE testing revealed presence of six different genotypes. All isolates from northern Serbia (The Clinical Center of Vojvodina; Kc1-12) belonged to genotype II, and isolates from southern Serbia (The Clinical Center Niš; Ni9, Ni21, Ni34, and DM5) belonged to genotype III. Isolates from the private laboratory (Konzilijum, Belgrade) clustered in three different genotypes: genotype IV (K1), genotype V (K5), and genotype VI (K2, K3, K4, K6, K7, K8, K9, and K10). Pediatric isolate 11070 singled out and was designated genotype I. Multilocus sequence typing for representatives of each genotype was performed using primers and conditions described by Diancourt et al. (7). The determination of specific sequence types (STs) according to the obtained allelic profiles was accomplished using the database (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html) of the Institut Pasteur, Paris, France, and six different STs were identified (Table 1). Based on this analysis, the dominant ST was ST101 (genotype II), which encompassed 44.44% of the colistin-resistant isolates. These isolates were outbreak related and recovered from The Clinical Center of Vojvodina, Novi Sad, in the northern Serbia. This ST was followed by ST888 (genotype VI), which encompassed 29.63% of isolates. Isolates belonging to genotype III were designated ST340, those belonging to genotype IV were designated ST437, those belonging to genotype V were designated ST336, and those belonging to genotype I were designated ST307.
Antimicrobial susceptibility was determined by microdilution method according to the European Committee on Antimicrobial Susceptibility Testing recommendations (http://www.eucast.org) (8). The colistin MIC for all isolates was ≥16 μg/ml (Table 1). Antimicrobial susceptibility testing revealed that analyzed isolates were also resistant to carbapenems and that the MICs for imipenem and meropenem were ≥8 μg/ml. A PCR method was used to detect the carbapenemase-encoding genes blaKPC, blaVIM, blaIMP, blaNDM, and blaOXA-48 (9–11), as well as blaCTX-M-15 (12). blaCTX-M-15 was detected in all isolates. Among the carbapenemase genes, the blaOXA-48 determinant was the most prevalent, being detected in 23 of 27 isolates (ST101, ST888, ST437, ST336, and ST307) (Table 1). blaOXA-48 has been commonly associated with ST101 worldwide and, according to the results of an 11-year (2001 to 2011) molecular epidemiologic study of blaOXA-48 in Europe and North Africa, ST101 is the most frequently observed sequence type (13). Among the carbapenem-resistant K. pneumoniae STs identified in this study, the emergence of colistin resistance had been already reported in KPC-2-producing ST101 (14) and blaKPC-2- and blaCTX-M-producing ST307 (15, 16). Carbapenem- and colistin-resistant isolates of ST437 have been reported previously (17). The blaNDM gene was detected in ST340 (Table 1). Although ST340 strains carrying the blaNDM-1 gene had been described (18), colistin-resistant, NDM-1-producing isolates of this ST, to the best of our knowledge, have not yet been reported. The acquisition of colistin resistance by a NDM-1-producing K. pneumoniae strain highlights the risk of the emergence of panresistant strains. Colistin-resistant strains of OXA-48-producing ST888 and ST336 have not yet been found.
In order to reveal the molecular mechanism(s) of colistin resistance, the presence of mcr-1 and mcr-2 genes was analyzed in all colistin-resistant isolates from the collection by a previously described method (19, 20). Since the mcr-1 and mcr-2 genes were not found, we focused on other mechanisms of colistin resistance, specifically mgrB gene inactivation; the presence of the mutations in the pmrA, pmrB, phoP, phoQ, crrA, and crrB genes; and the upgraded expression of phoP, phoQ, and pmrK genes. The amplification of the mgrB gene was performed in all isolates by a previously described method (21). Sequence analysis of the mgrB gene showed that one isolate (ST307) generated amplicon that was larger than the one from K. pneumoniae IT977 (a control, colistin-susceptible isolate). Amplicon sequencing revealed that insertional inactivation had occurred in the coding region of the K. pneumoniae ST307 mgrB gene. Insertional inactivation occurred at nucleotide 75 and was raised by insertional sequence that shared 99% of identity at the nucleotide level with ISKpn26 insertion sequence (IS5 family of insertion sequences). The insertional sequence was identified using the ISfinder database (http://www-is.biotoul.fr) (22). Insertional inactivation was not detected in other STs from the study. However, ST336 had premature amber stop codon (TAG) due to a C-to-T change at position 88, which generates a truncated MgrB protein of 29 amino acids. Other STs had the wild-type mgrB gene, without any changes in nucleotide sequence that could result in change of protein synthesis or activity. Nucleotide sequences of genes and corresponding amino acid sequences of PmrA and PmrB proteins from all isolates were compared to those of the colistin-susceptible strain K. pneumoniae IT977, and the changes detected are shown in Table 1. The observed amino acid substitutions could have role in development of colistin resistance, but only Thr157Pro in the PmrB protein has been previously described (23). No amino acid substitutions were detected in PhoP or PhoQ protein. The crrA and crrB genes were found only in ST340, ST336 and ST307, but amino acid substitutions were not detected compared to K. pneumoniae available in GenBank.
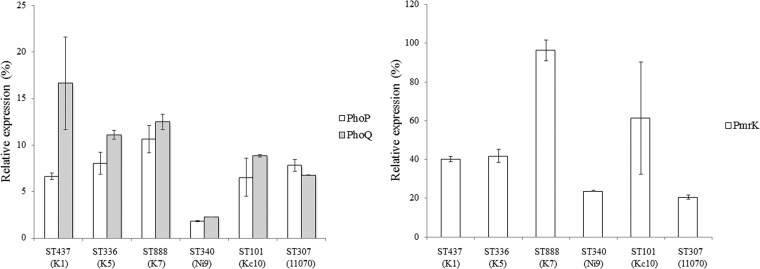
Reverse transcription-quantitative PCR (RT-qPCR) was used to determine the expression levels of the phoP, phoQ, and pmrK genes. Expression of the rpsL gene represented an internal control. The primers and conditions used for the RT-qPCR analyses are listed in Table 2 (21). Normalization was done against the rpsL gene using the ΔΔCT method (relative) (24), and the values obtained were then normalized against those detected in the colistin-susceptible isolate IT977. Analysis of phoP and phoQ transcription in ST307 with the inactivated mgrB gene revealed a 7.8-fold increase for the phoP gene and a 6.8-fold increase for the phoQ gene (Fig. 1). ST336, with a truncated MgrB, underwent 8- and 11-fold increases, respectively, in phoP and phoQ gene transcription. Although other analyzed strains did not undergo insertional inactivation of the mgrB gene, the expression of the phoP and phoQ genes was elevated and ranged from an 1.8-fold increase in phoP gene expression in ST340 up to a 16.3-fold increase in phoQ gene expression in ST437 (Fig. 1). Moreover, analysis of transcription of the pmrK gene, which belongs to the pmrHFIJKLM operon, revealed a significant increase in transcription levels that varied from a 20.6-fold increase in ST307 to a 96.2-fold increase in ST888 (Fig. 1).
TABLE 2.
Primers and conditions used in RT-qPCRa
| Primer | Sequence (5′-3′) | Cycling conditions |
|---|---|---|
| phoP_F | ATTGAAGAGGTTGCCGCCCGC | 95°C for 1 s, 52°C for 5 s, 72°C for 7 s |
| phoP_R | GCTTGATCGGCTGGTCATTCACC | 95°C for 1 s, 52°C for 5 s, 72°C for 7 s |
| phoQ_F | ATATGCTGGCGAGATGGGAAAACGG | 95°C for 1 s, 52°C for 5 s, 72°C for 7 s |
| phoQ_R | CCAGCCAGGGAACATCACGCT | 95°C for 1 s, 52°C for 5 s, 72°C for 7 s |
| pmrK_FT | GCGGGCCATCAGGATCGACAGCG | 95°C for 1 s, 65°C for 5 s, 72°C for 7 s |
| pmrK_RT | CGTTCTGGTACTACATCCCGTTCCTGA | 95°C for 1 s, 65°C for 5 s, 72°C for 7 s |
| rpsL13_F | GCCGTACTTGGAGCGAGCCTG | 95°C for 1 s, 52°C for 5 s, 72°C for 7 s |
| rpsL14_F | CCGTGGCGGTCGTGTTAAAGA | 95°C for 1 s, 52°C for 5 s, 72°C for 7 s |
Reprinted from reference 21 with permission.
FIG 1.
Relative expression of the phoP, phoQ, and pmrK genes in different colistin-resistant, carbapenemase-producing K. pneumonia STs isolated in Serbia. The values and standard deviations represent means from three independent experiments. The percent value represents the increase in gene expression relative to values observed for colistin-susceptible K. pneumoniae IT977.
Although alterations in the mgrB gene nucleotide sequence are the most common cause of colistin resistance in K. pneumoniae (21, 25–28), we detected here the presence of such changes in only two isolates (ST336 and ST307), while the others had a wild-type nucleotide mgrB gene sequence. In addition, mutations leading to amino acid substitutions in PmrA and/or PmrB could have role in colistin resistance development in 22 of 27 isolates. However, the absence of changes in genes associated with colistin resistance for ST340 and ST437 could indicate that there are other regulators of PhoPQ regulatory system in K. pneumoniae, considering that such proteins have already been identified in Escherichia coli, Shigella sp., and Salmonella enterica serovar Typhimurium. Since these regulators are not conserved among Enterobacteriaceae, PhoPQ regulator(s) specific for K. pneumoniae may exist (29).
Accession numbers.
The nucleotide sequence of the mgrB gene obtained from K. pneumoniae ST307 and ST336 are available in the European Nucleotide Archive under accession numbers LT635644 and LT635643, respectively. Other nucleotide sequences analyzed in this study are available from GenBank (KY586987 to KY587110).
ACKNOWLEDGMENTS
K.N., A.T., S.B., Z.V., D.M., and I.Ć. were involved in the acquisition of laboratory and medical data, analysis of data, and final approval of the manuscript. M.K. was involved in data analysis, drafting of the article, critical revision of the article, and the final approval of the manuscript. B.J. designed the study, was involved in analysis of the data, drafting of article, and final approval of the manuscript.
This study was supported by grant 173019 from the Ministry of Education, Science, and Technological Development of the Republic of Serbia.
REFERENCES
- 1.Kontopoulou K, Protonotariou E, Vasilakos K, Kriti M, Koteli A, Antoniadou E, Sofianou D. 2010. Hospital outbreak caused by Klebsiella pneumoniae producing KPC-2 β-lactamase resistant to colistin. J Hosp Infect 76:70–73. doi: 10.1016/j.jhin.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 2.Suh JY, Son JS, Chung DR, Peck KR, Ko KS, Song JH. 2010. Nonclonal emergence of colistin-resistant Klebsiella pneumoniae isolates from blood samples in South Korea. Antimicrob Agents Chemother 54:560–562. doi: 10.1128/AAC.00762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elemam A, Rahimian J, Mandell W. 2009. Infection with panresistant Klebsiella pneumoniae: a report of two cases and a brief review of the literature. Clin Infect Dis 49:271–274. doi: 10.1086/600042. [DOI] [PubMed] [Google Scholar]
- 4.Bogdanovich T, Adams-Haduch JM, Tian GB, Nquyen MH, Kwak EJ, Muto CA, Doi Y. 2011. Colistin-resistant, Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin Infect Dis 53:373–376. doi: 10.1093/cid/cir401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jayol A, Poirel L, Dortet L, Nordmann P. 2016. National survey of colistin resistance among carbapenemase-producing Enterobacteriaceae and outbreak caused by colistin-resistant OXA-48-producing Klebsiella pneumoniae, France, 2014. Euro Surveill 21:30339. doi: 10.2807/1560-7917.ES.2016.21.37.30339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kojic M, Strahinic I, Topisirovic L. 2005. Proteinase PI and lactococcin A genes are located on the largest plasmid in Lactococcus lactis subsp. lactis bv diacetylactis S50. Can J Microbiol 51:305–314. doi: 10.1139/w05-009. [DOI] [PubMed] [Google Scholar]
- 7.Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The European Committee on Antimicrobial Susceptibility Testing. 2017. Breakpoint tables for interpretation of MICs and zone diameters, version 7.0. EUCAST, Copenhagen, Denmark. [Google Scholar]
- 9.Bratu S, Tolaney P, Karumudi U, Quale J, Mooty M, Nichani S, Landman D. 2005. Carbapenemase-producing Klebsiella pneumoniae in Brooklyn, NY: molecular epidemiology and in vitro activity of polymyxin B and other agents. J Antimicrob Chemother 56:128–132. doi: 10.1093/jac/dki175. [DOI] [PubMed] [Google Scholar]
- 10.Ellington MJ, Kistler J, Livermore DM, Woodford N. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J Antimicrob Chemother 59:321–322. [DOI] [PubMed] [Google Scholar]
- 11.Teo J, Ngan G, Balm M, Jureen R, Krishnan P, Lin R. 2012. Molecular characterization of NDM-1 producing Enterobacteriaceae isolates in Singapore hospitals. Western Pac Surveill Response J 3:19–24. doi: 10.5365/wpsar.2011.2.4.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conceicao T, Brizio A, Duarte A. 2005. First description of CTX-M-15-producing Klebsiella pneumoniae in Portugal. Antimicrob Agents Chemother 49:477–478. doi: 10.1128/AAC.49.1.477-478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potron A, Poirel L, Rondinaud E, Nordmann P. 2013. Intercontinental spread of OXA-48 β-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill 18:20549 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20549. [DOI] [PubMed] [Google Scholar]
- 14.Del Franco M, Paone L, Novati R, Giacomazzi CG, Bagattini M, Galotto C, Montanera PG, Triassi M, Zarrili R. 2015. Molecular epidemiology of carbapenem-resistant Enterobacteriaceae in Valle d'Aosta region, Italy, shows the emergence of KPC-2 producing Klebsiella pneumoniae clonal complex 101 (ST101 and ST1789). BMC Microbiol 15:260. doi: 10.1186/s12866-015-0597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonura C, Giuffrè M, Aleo A, Fasciana T, Di Bernardo F, Stampone T, Giammanco A, Palma MD, Mammina C, MDR-GN Working Group . 2015. An update of the evolving epidemic of blaKPC carrying Klebsiella pneumoniae in Sicily, Italy, 2014: emergence of multiple non-ST258 clones. PLoS One 10:e0132936. doi: 10.1371/journal.pone.0132936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Habeeb MA, Haque A, Nematzadeh S, Iversen A, Giske CG. 2013. High prevalence of 16S rRNA methylase RmtB among CTX-M extended-spectrum β-lactamase-producing Klebsiella pneumoniae from Islamabad, Pakistan. Int J Antimicrob Agents 41:524–526. doi: 10.1016/j.ijantimicag.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 17.Bartolleti F, Seco BMS, dos Santos CC, Felipe CB, Lemo MEB, da Silva Alves T, Passadore LF, Mimica MJ, Sampaio SCF, Zavascki AP, Sampaio JLM. 2016. Polymyxin B resistance in carbapenem-resistant Klebsiella pneumoniae, Sao Paulo, Brazil. Emerg Infect Dis 22:1849–1851. doi: 10.3201/eid2210.160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother 55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 20.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21: 30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 21.Cannatelli A, D'Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, Gaibani P, Rossolini GM. 2013. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother 57:5521–5526. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayol A, Poirel L, Brink A, Villegas MV, Yilmaz M, Nordmann P. 2014. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Chemother 58:4762–4766. doi: 10.1128/AAC.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Wright MS, Suzuki Y, Jones MB, Marshal SH, Rudin SD, van Duin D, Kaye K, Jacobs MR, Bonomo RA, Adams MD. 2015. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother 59:536–543. doi: 10.1128/AAC.04037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel L, Jayol A, Bontron S, Villegas MV, Ozdamar M, Türkoglu S, Nordmann P. 2015. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother 70:75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 27.Cannatelli A, Giani T, D'Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM, COLGRIT Study Group . 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC carbapenemase-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5603. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Camacho E, Gómez-Gil R, Tobes R, Manrique M, Lorenzo M, Galvan B, Salvarelli E, Moatassim Y, Salanueva IJ, Pareja E, Codoner FM, Alvarez-Tejado M, Garcillan-Barcia MP, De La Cruz F, Mingorance J. 2014. Genomic analysis of the emergence and evolution of multidrug resistance during a Klebsiella pneumonia outbreak including carbapenem and colistin resistance. J Antimicrob Chemother 69:632–636. doi: 10.1093/jac/dkt419. [DOI] [PubMed] [Google Scholar]
- 29.Eguchi Y, Itou J, Yamane M, Demizu R, Yamato F, Okada A, Mori H, Kato A, Utsumi R. 2007. B1500, a small membrane protein, connects the two-component systems EvgS/EvgA and PhoQ/PhoP in Escherichia coli. Proc Natl Acad Sci U S A 104:18712–18717. doi: 10.1073/pnas.0705768104. [DOI] [PMC free article] [PubMed] [Google Scholar]