ABSTRACT
The recently discovered colistin resistance element, mcr-1, adds to the list of antimicrobial resistance genes that rapidly erode the antimicrobial efficacy of not only the commonly used antibiotics but also the last-line agents of carbapenems and colistin. This study investigated the prevalence of the mobile colistin resistance determinant mcr-1 in Salmonella strains recovered from clinical settings in China and the transmission potential of mcr-1-bearing mobile elements harbored by such isolates. The mcr-1 gene was recoverable in 1.4% of clinical isolates tested, with the majority of them belonging to Salmonella enterica serotype Typhimurium. These isolates exhibited diverse pulsed-field gel electrophoresis (PFGE) profiles and high resistance to antibiotics other than colistin and particularly to cephalosporins. Plasmid analysis showed that mcr-1 was carried on a variety of plasmids with sizes ranging from ∼30 to ∼250 kb, among which there were conjugative plasmids of ∼30 kb, ∼60 kb, and ∼250 kb and nonconjugative plasmids of ∼140 kb, ∼180 kb, and ∼240 kb. Sequencing of representative mcr-1-carrying plasmids revealed that all conjugative plasmids belonged to the IncX4, IncI2, and IncHI2 types and were highly similar to the corresponding types of plasmids reported previously. Nonconjugative plasmids all belonged to the IncHI2 type, and the nontransferability of these plasmids was attributed to the loss of a region carrying partial or complete tra genes. Our data revealed that, similar to the situation in Escherichia coli, mcr-1 transmission in Salmonella was accelerated by various plasmids, suggesting that transmission of mcr-1-carrying plasmids between different species of Enterobacteriaceae may be a common event.
KEYWORDS: Salmonella, mcr-1, transmission, IncX4, IncI2, IncHI2, plasmid
INTRODUCTION
Polymyxins are considered the last-resort antibiotics used to treat carbapenem-resistant Enterobacteriaceae (CRE)-related infections due to their high efficacy and low resistance rate among clinical CRE strains. Polymyxins were discovered more than 50 years ago; however, due to their nephrotoxic and neurotoxic side effects, they are rarely used clinically (1). In recent years, due to the increasing prevalence of carbapenem-resistant Enterobacteriaceae and emergence of extensively drug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa strains, polymyxins have been reevaluated and are considered among the few remaining agents which exhibit bactericidal effects on multidrug-resistant (MDR) Gram-negative bacterial pathogens (2, 3).
The rate of bacterial resistance to polymyxins had previously been thought to be extremely low and mainly attributed to chromosomal mutations in specific two-component regulatory systems (e.g., pmrAB and phoPQ and their negative regulator mgrB in the case of Klebsiella pneumoniae), leading to structural modification of lipid A (4, 5). Recently, a new plasmid-mediated colistin resistance mechanism, mediated by the MCR-1 protein, a phosphoethanolamine transferase which can add the phosphoethanolamine moiety to lipid A, was discovered (6). The mcr-1 gene was detectable in not only Escherichia coli but also other bacterial species such as Klebsiella pneumonia and Salmonella in isolates recovered from various countries (6, 7). The mcr-1-bearing plasmids were found to possess the ability to self-transmit between animal and human bacterial isolates and were also highly stable in bacteria even in the absence of polymyxin selection pressure (6). However, the mcr-1 gene was found in only ∼1% of clinical E. coli isolates, whereas the carriage rate could be as high as ∼20% in bacteria isolated from farm animals and meat products (6). This discrepancy prompted us to make the hypothesis that only a small proportion of mcr-1-bearing strains can cause human infections.
In Salmonella, the mcr-1 gene was first described through analysis of whole-genome sequences of Salmonella available in GenBank, in which mcr-1-bearing plasmids were identified in 10 clinical Salmonella enterica isolates submitted between 2012 and 2015, including 8 S. enterica serotype Typhimurium, 1 S. enterica serotype Paratyphi B var. Java, and 1 S. enterica serotype Virchow strain (8). The mcr-1 gene was subsequently reported to be recoverable from Salmonella strains isolated from food, animals, and clinical specimens in Europe, the United States, and China (9–14) and was found to be harbored mainly by IncX4 and IncI1 plasmids of various sizes (10, 11). However, these data remain scattered and do not provide a comprehensive view on either the prevalence of the mcr-1 gene in Salmonella or the transmission kinetics of the different types of mobile elements that harbor such resistance determinants. To address these important issues, we conducted a nationwide surveillance on the prevalence of the mcr-1 gene in Salmonella strains recovered from clinical specimens and investigated the transmission potential of the mcr-1 gene recovered from these Salmonella isolates. Our data provide a comprehensive view of the extent of mcr-1 contamination in Salmonella in China and essential insights into the features and routes of transmission of mcr-1 among strains of this important foodborne pathogen.
RESULTS
Prevalence and characteristics of mcr-1-positive Salmonella.
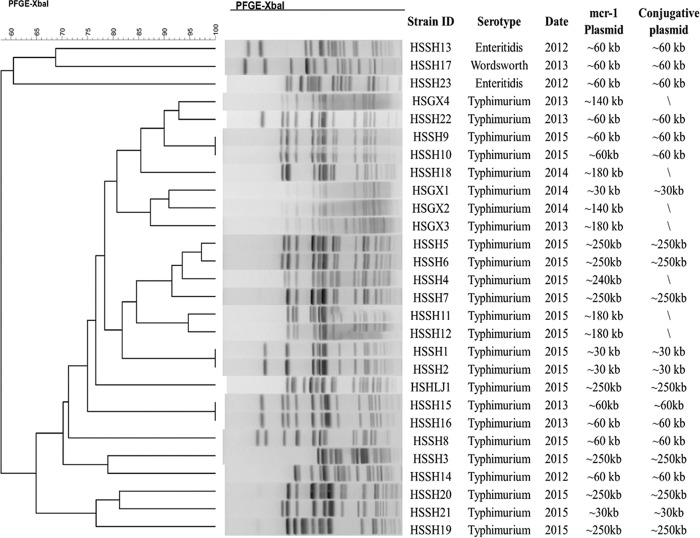
A total of 2,034 nonrepeated human clinical Salmonella isolates collected nationwide during the period of 2012 to 2015 by the Chinese Center for Disease Control and Prevention (CDC) were subjected to screening of the mcr-1 gene, and 28 out of 2,034 (1.4%) isolates were found to contain this resistance gene. Among these 28 mcr-1-positive Salmonella isolates, 25 were S. Typhimurium and 3 were S. enterica serotype Enteritidis. All three S. Enteritidis isolates were collected in 2012, while S. Typhimurium was the only serotype found to be carrying the mcr-1 gene afterward. All mcr-1-positive Salmonella isolates showed very diverse pulsed-field gel electrophoresis (PFGE) patterns, suggesting that transmission of mcr-1-carrying plasmids may be more common than clonal spread in mcr-1-positive Salmonella (Fig. 1). Antimicrobial susceptibility analysis revealed that these 28 mcr-1-positive Salmonella isolates generally displayed a high rate of resistance to other antibiotics in addition to colistin, in particular, to ceftriaxone (78%) (Table 1).
FIG 1.
Summary of genetic characteristics of mcr-1-bearing Salmonella strains isolated from different sources.
TABLE 1.
Antimicrobial susceptibility of mcr-1-positive Salmonella strains isolated from different sources
| Antibiotic | Breakpoint (μg/ml) | % of resistancea |
|
|---|---|---|---|
| WT (n = 28) | TC (n = 21) | ||
| Ampicillin | ≥32 | 89 | 52 |
| Amoxicillin-clavulanic acid | ≥32/16 | 50 | 14 |
| Ceftriaxone | ≥4 | 64 | 62 |
| Chloramphenicol | ≥32 | 64 | 19 |
| Gentamicin | ≥16 | 54 | 24 |
| Nalidixic acid | 32 | 68 | 24 |
| Ciprofloxacin | ≥1 | 25 | 0 |
| Trimethoprim-sulfamethoxazole | ≥4/76 | 39 | 14 |
| Tetracycline | ≥16 | 79 | 0 |
| Colistin | >2 | 100 | 100 |
| Azithromycin | ≥16 | 14 | 0 |
“% of resistance” refers to the no. of resistant strains/no. of total strains (n). WT, wild type; TC, transconjugant.
Mechanisms of transmission of mcr-1 among Salmonella isolates.
Conjugation experiments showed that 21 out of the 28 mcr-1-positive Salmonella isolates were able to transfer their colistin resistance phenotype to E. coli J53 (Fig. 1). PFGE using S1 nuclease (S1-PFGE) and Southern-hybridization analysis indicated that all 28 isolates, regardless of their conjugative status, carried only one mcr-1-carrying plasmid. Most of the conjugative plasmids carrying mcr-1 belonged to three major groups, ∼30 kb (n = 3), ∼60 kb (n = 11), and ∼250 kb (n = 7). All nonconjugative plasmids had sizes of ∼140 kb (n = 2), ∼180 kb (n = 4), and ∼240 kb (n = 1) (Fig. 1).
Genetic features of mcr-1-carrying plasmids recovered from Salmonella strains.
Representative plasmids were selected from different strains, including plasmids of ∼30 kb, ∼60 kb, ∼140 kb, ∼180 kb, ∼240 kb, and ∼250 kb, for sequencing using the Illumina platforms. Two of them, ∼60-kb and ∼250-kb plasmids, were further sequenced by the PacBio platform to obtain the complete maps (Table 2). Illumina contigs of the ∼30-kb plasmid harbored by Salmonella strain HSGX1 were obtained and aligned to several previously reported plasmids, including pOW3E1 (GenBank accession number KX129783.1) and pECJP-B65-33 (GenBank accession number KX084392.1). This plasmid from HSGX1 was shown to belong to the IncX4 type and could be aligned very well to pOW3E1 (KX129783.1) (>99% in both identity and coverage) (data not shown).
TABLE 2.
Origin and genetic features of mcr-1-bearing plasmids in Salmonella strains subjected to sequence analysis in this study
| Strain name | Serotype | Yr of isolation | Approx size by S1-PFGE (kb) | Conjugative nature | Plasmid type | Sequence(s) analyzed |
|---|---|---|---|---|---|---|
| HSSH21 | Typhimurium | 2014 | 30 | Conjugative | IncX4 | Contigs |
| HSSH23 | Enteritidis | 2012 | 60 | Conjugative | IncI2 | pHSSH23-MCR1 |
| HXGX4 | Typhimurium | 2013 | 140 | Nonconjugative | IncHI2 | Contigs |
| HXGX3 | Typhimurium | 2015 | 180 | Nonconjugative | IncHI2 | Contigs |
| HSSH4 | Typhimurium | 2015 | 240 | Nonconjugative | IncHI2 | Contigs |
| HSHLJ1 | Typhimurium | 2015 | 250 | Conjugative | IncHI2 | pHSHLJ1-MCR1 |
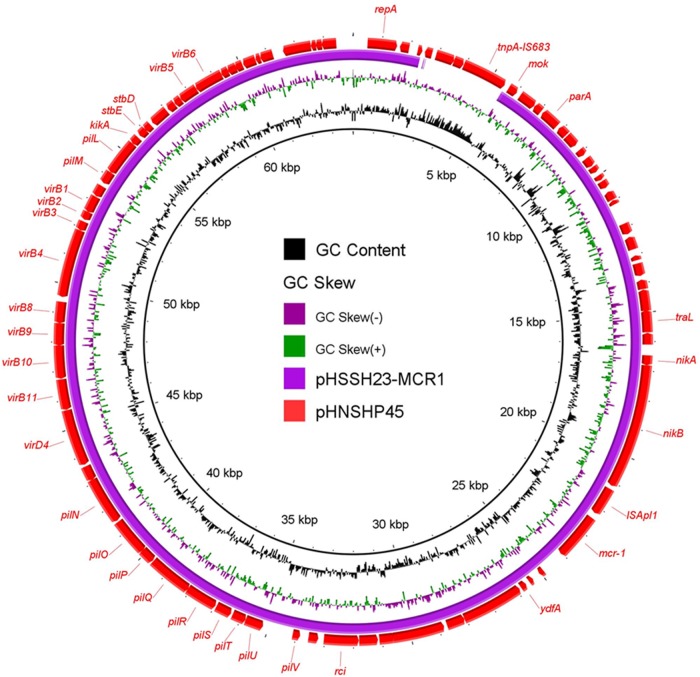
The complete sequence of a ∼60-kb conjugative plasmid, recovered from the S. Enteritidis HSSH23 strain, pHSSH23-MCR1, was obtained. It was found to belong to an IncI2-type plasmid with a backbone structure similar to that of pHNSHP45 (GenBank accession number KP347127) and other plasmids reported previously (8, 9). Compared to plasmid pHNSHP45, pHSSH23-MCR1 with a size of 60,035 bp was found to lack an IS683-mediated fragment (Table 2 and Fig. 2). The two types of IncI2 plasmids differentiated by the presence or absence of ISApl1 and IS683 are known to have been disseminated worldwide and have also been recovered from E. coli and Salmonella spp. strains in the United Kingdom (8).
FIG 2.
Alignment of one mcr-1-bearing IncI2 plasmid against pHNSHP45. The reference plasmid, the previously reported pHNSHP45, is indicated by red arrows. pHSSH23-MCR1, labeled in blue, was aligned to the reference plasmid using BLAST Ring Image Generator (BRIG) software. The gaps in pHSSH23-MCR1 represent missing sequences compared to the sequence of the reference plasmid.
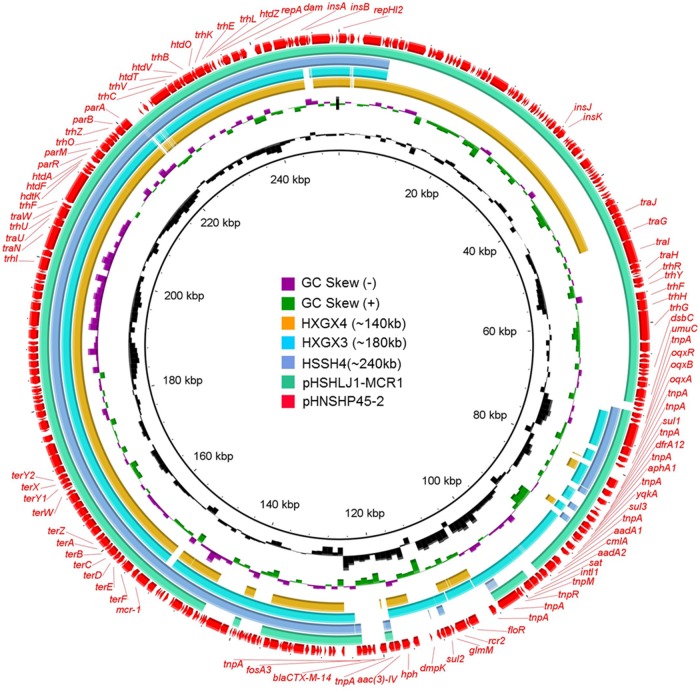
The complete sequence of a conjugative ∼250-kb plasmid recovered from S. Typhimurium HSHLJ1, plasmid pHSHLJ1-MCR1 (238,539 bp), was obtained and compared to that of pHNSHP45-2; results showed that this conjugative plasmid displayed a high degree of sequence homology to pHNSHP45-2 (Fig. 3). Antimicrobial resistance gene analysis showed that pHSHLJ1-MCR1 contained some additional insertion sequences (ISs) and one mph(A) gene (Fig. 4). Interestingly, alignment of Illumina contigs of the nonconjugative ∼240-kb plasmid recovered from the S. Typhimurium strain HSSH4 to pHNSHP45-2 revealed that this plasmid aligned very well with pHNSHP45-2 except that a region containing the tra genes, which were responsible for plasmid conjugation, was absent in the plasmid of strain HSSH4 (Fig. 3). Antimicrobial resistance gene analysis showed that plasmids from S. Typhimurium strain HSSH4 also lacked the mobile elements carrying blaCTX-M14, oqxAB, fosA3, and other antimicrobial resistance genes but gained other mobile elements carrying other antimicrobial resistance genes and transposase genes (Fig. 4). The genetic differences resulting from the absence of the tra region may explain why the ∼240-kb plasmid of S. Typhimurium strain HSSH4 was nonconjugative.
FIG 3.
Alignment of conjugative and nonconjugative IncHI2 plasmids/contigs against pHNSHP45-2. The circular map was created using BLAST Ring Image Generator (BRIG) tools; the linear map was generated by EasyFig. Genes in the reference plasmid, pHNSHP45-2, which was reported previously, are labeled by red arrows. Plasmid pHSHLJ1-MCR1 and contigs of other plasmids labeled with different colors were aligned to sequence of the reference plasmid. The gaps in the plasmid sequences represent the missing sequences compared to the sequence of the reference plasmid.
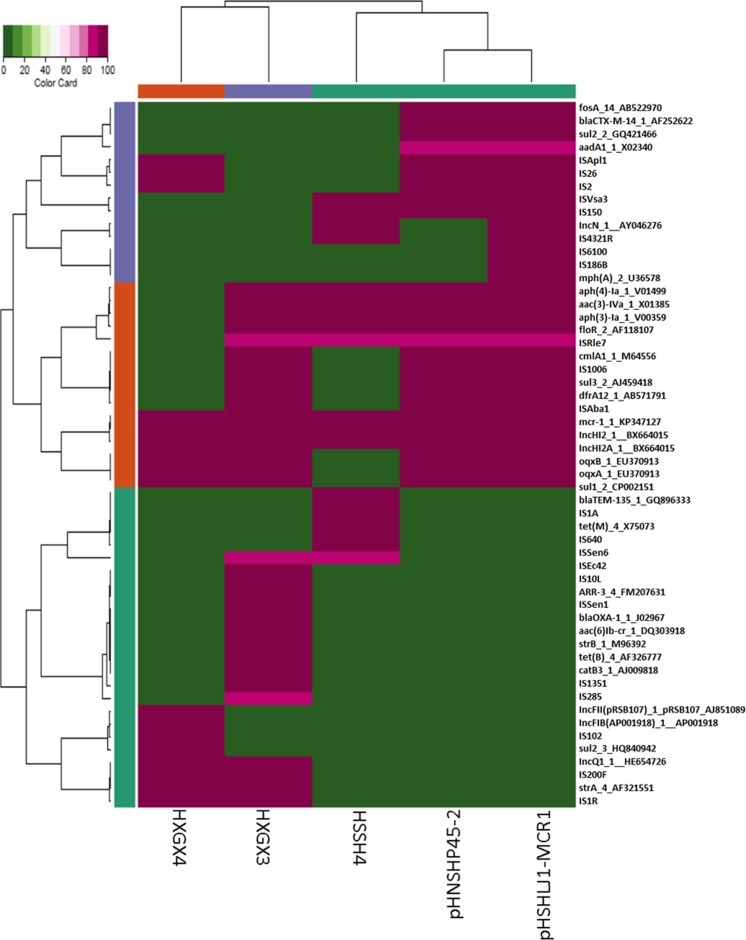
FIG 4.
Antimicrobial resistance-related gene analysis for all IncHI2 plasmids of various sizes. Plasmid pHNSHP45-2 was reported previously; plasmid pHSHLJ1-MCR1, with a size of 238,539 bp, was isolated from S. Typhimurium strain HSHLJ1. Illumina sequencing was performed for contigs of nonconjugative plasmids of ∼240 kb, ∼180 kb, and ∼140 kb recovered from S. Typhimurium strains HSSH4, HXGX3, and HXGX4, respectively.
Alignment of Illumina contigs of the nonconjugative ∼180-kb plasmid recovered from S. Typhimurium strain HXGX3 to pHNSHP45-2 revealed that this plasmid aligned very well with pHNSHP45-2 except that a large region of ca. 70 kb harboring the tra region was missing compared to the sequence of pHNSHP45-2. In addition, this ∼180-kb plasmid also lacked the mobile element carrying blaCTX-M14 and fosA3 but gained a mobile element carrying blaOXA-1, aac(6′)-lb-cr, and catB3, according to antimicrobial resistance gene analysis (Fig. 3). These different genetic features, detectable among the two plasmids, depicted the evolution routes of the IncHI2 class of mcr-1-carrying plasmids and indicated that the loss of the ca. 70-kb tra region in plasmid from strain HXGX3 could contribute to the loss of transferability of this plasmid. Illumina reads of the ∼140-kb plasmid recovered from S. Typhimurium strain HXGX4 were aligned to pHNSHP45-2. This alignment of this plasmid was very similar as the alignment of the ∼180-kb plasmid from HXGX3 to pHNSHP45-2 except for the further deletions in the MDR region, but the plasmid also gained some more antimicrobial resistance genes that were not detected in other IncHI1 plasmids reported in this study (Fig. 3).
DISCUSSION
Similar to the situation in other clinical Enterobacteriaceae isolates, the prevalence rate of mcr-1 in clinical Salmonella isolates was found to remain very low (1.4%). Interestingly, our data showed that most of the Salmonella strains that harbored mcr-1-bearing plasmids were S. Typhimurium, suggesting that a specific genetic background is required for acquisition and maintenance of mcr-1-bearing mobile elements. The close association between S. Typhimurium and mcr-1-bearing plasmids requires further investigation (8). These human clinical isolates exhibited more diverse genetic backgrounds, suggesting that clonal spread is not the major mechanism of mcr-1 transmission in Salmonella.
Clinical Salmonella isolates also carried various conjugative plasmids reported in other Enterobacteriaceae so far, including a ∼30-kb IncX4, ∼60-kb IncI1, and ∼250-kb IncHI2. Surprisingly, we detected several nonconjugative plasmids with sizes of ∼140 kb, ∼180 kb, and ∼240 kb in these clinical Salmonella isolates. These plasmids all belonged to IncHI2 and were very similar to IncHI2 plasmids reported earlier. Sequence analysis showed that the inability of ∼240-kb plasmids to undergo conjugation was attributed to deletion of part of the tra region, which is present in the conjugative ∼250-kb plasmids. The ∼140-kb and ∼180-kb plasmids showed further deletions of the tra region and MDR regions while they gained some other MDR regions which were different from those of pHNSHP45-2. In fact, the IncHI2 type of plasmids without mcr-1 have not been reported in other bacterial species so far. The backbone of the IncHI2 plasmid lacking the mcr-1-carrying mobile element, namely, pHXY0908 (GenBank accession number KM877269.1), has been reported only in Salmonella. The presence of nonconjugative plasmids of ∼140 kb, ∼180 kb, and ∼240 kb in these Salmonella isolates together with the presence of a prototype of these IncHI2 plasmids in Salmonella may suggest that insertion of an mcr-1-carrying mobile element into the backbone of this type of plasmid might be responsible for one of the modes of mcr-1 transmission in Salmonella.
In conclusion, results of this nationwide surveillance of the mcr-1 carriage rate and vector structure in clinical Salmonella isolates provide a comprehensive understanding of the features and mechanisms of transmission of mcr-1 in Salmonella and, hence, lay the foundation for future development of strategies to control the transmission of this colistin resistance determinant among Gram-negative bacterial pathogens.
MATERIALS AND METHODS
Salmonella strains.
Salmonella strains were collected from human clinical samples nationwide in China by Shanghai Municipal Center for Disease Control and Prevention, Shanghai and National Institute for Communicable Disease Control and Prevention (ICDC), and Chinese Center for Disease Control and Prevention, Beijing. All test strains were isolated in CHROMagar Salmonella agar (CHROMagar Company, Paris, France) and XLT4 agar (Oxoid). Suspected Salmonella colonies were selected for biochemical confirmation using an API 20E system (bioMérieux, Marcy l' Etoile, France), as well as via molecular identification by PCR assay targeting the invA gene, followed by sequencing. Salmonella serotyping was conducted by performing a slide agglutination test, using Salmonella antiserum (S&A Reagents Lab, Ltd., Bangkok, Thailand) according to the Kaufman-White scheme.
Screening of the mcr-1 gene in Salmonella.
Salmonella genomic DNA was prepared using the boiling method. PCR was performed using primers targeting mcr-1 as reported previously (6). The genetic identity of all amplification products was confirmed by nucleotide sequencing.
Antimicrobial susceptibility tests.
All Salmonella isolates were subjected to antimicrobial susceptibility tests by the standard agar dilution method as described by the Clinical and Laboratory Standards Institute (15, 16). Sixteen antimicrobials, as shown in Table 1, were tested. Escherichia coli strain ATCC 25922 was used as the quality control.
Conjugation, PFGE, S1-PFGE, and Southern hybridization.
Conjugation experiments were carried out using the mixed broth method as previously described (17). PFGE, S1-PFGE, and Southern hybridization were performed as previously described (18).
Plasmid sequencing.
Representative plasmids with sizes of ∼30 kb, ∼60 kb, ∼140 kb ∼180 kb, ∼240 kb, and ∼250 kb recovered from Salmonella parental strains and transconjugants were subjected to plasmid sequencing using Illumina and PacBio platforms and analyzed as previously described (19).
Accession number(s).
The complete nucleotide sequences of the ∼60-kb (pHSSH23-MCR1) and ∼250-kb (pHSHLJ1-MCR1) plasmids were submitted to GenBank under accession numbers KX856068 and KX856066, respectively.
ACKNOWLEDGMENTS
This work was supported by the Chinese National Key Basic Research and Development Program (2013CB127200) and Hong Kong Research Grant Council Collaborative Research Fund (C7038-15G).
We have no conflicts of interest to declare.
REFERENCES
- 1.Biswas S, Brunel JM, Dubus JC, Reynaud-Gaubert M, Rolain JM. 2012. Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther 10:917–934. doi: 10.1586/eri.12.78. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 3.Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 4.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother 55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, Hoiby N, Moskowitz SM. 2011. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 55:5761–5769. doi: 10.1128/AAC.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 7.Tse H, Yuen KY. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:145–146. doi: 10.1016/S1473-3099(15)00532-0. [DOI] [PubMed] [Google Scholar]
- 8.Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, Day M, Muller-Pebody B, Ellington MJ, de Pinna E, Johnson AP, Hopkins KL, Woodford N. 2016. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother 71:2300–2305. doi: 10.1093/jac/dkw093. [DOI] [PubMed] [Google Scholar]
- 9.Anjum MF, Duggett NA, AbuOun M, Randall L, Nunez-Garcia J, Ellis RJ, Rogers J, Horton R, Brena C, Williamson S, Martelli F, Davies R, Teale C. 2016. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J Antimicrob Chemother 71:2306–2313. doi: 10.1093/jac/dkw149. [DOI] [PubMed] [Google Scholar]
- 10.Campos J, Cristino L, Peixe L, Antunes P. 2016. MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:− and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill 21(26):pii=30270 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=22516. [DOI] [PubMed] [Google Scholar]
- 11.Yang YQ, Zhang AY, Ma SZ, Kong LH, Li YX, Liu JX, Davis MA, Guo XY, Liu BH, Lei CW, Wang HN. 2016. Co-occurrence of mcr-1 and ESBL on a single plasmid in Salmonella enterica. J Antimicrob Chemother 71:2336–2338. doi: 10.1093/jac/dkw243. [DOI] [PubMed] [Google Scholar]
- 12.Figueiredo R, Card RM, Nunez J, Pomba C, Mendonca N, Anjum MF, Da Silva GJ. 2016. Detection of an mcr-1-encoding plasmid mediating colistin resistance in Salmonella enterica from retail meat in Portugal. J Antimicrob Chemother 71:2338–2340. doi: 10.1093/jac/dkw240. [DOI] [PubMed] [Google Scholar]
- 13.Quesada A, Ugarte-Ruiz M, Iglesias MR, Porrero MC, Martinez R, Florez-Cuadrado D, Campos MJ, Garcia M, Piriz S, Saez JL, Dominguez L. 2016. Detection of plasmid mediated colistin resistance (MCR-1) in Escherichia coli and Salmonella enterica isolated from poultry and swine in Spain. Res Vet Sci 105:134–135. doi: 10.1016/j.rvsc.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Vinueza-Burgos C, Cevallos M, Ron-Garrido L, Bertrand S, De Zutter L. 2016. Prevalence and diversity of Salmonella serotypes in Ecuadorian broilers at slaughter age. PLoS One 11:e0159567. doi: 10.1371/journal.pone.0159567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CLSI. 2015. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. CLSI document M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.CLSI. 2016. Performance standards for antimicrobial susceptibility testing; twenty-sixth informational supplement. CLSI document M100-S26. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 17.Borgia S, Lastovetska O, Richardson D, Eshaghi A, Xiong J, Chung C, Baqi M, McGeer A, Ricci G, Sawicki R, Pantelidis R, Low DE, Patel SN, Melano RG. 2012. Outbreak of carbapenem-resistant Enterobacteriaceae containing blaNDM-1, Ontario, Canada. Clin Infect Dis 55:e109–e117. doi: 10.1093/cid/cis737. [DOI] [PubMed] [Google Scholar]
- 18.Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. 2015. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother 60:709–711. doi: 10.1128/AAC.02173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye L, Li R, Lin D, Zhou Y, Fu A, Ding Q, Chan EW, Yao W, Chen S. 2016. Characterization of an IncA/C multidrug resistance plasmid in Vibrio alginolyticus. Antimicrob Agents Chemother 60:3232–3235. doi: 10.1128/AAC.00300-16. [DOI] [PMC free article] [PubMed] [Google Scholar]