ABSTRACT
The blaVIM-2-carrying In58 integron has been linked to a chromosomal location in different bacterial species, including Pseudomonas aeruginosa. This work reports the first fully sequenced In58-harboring plasmid, which is significantly different from the two previously identified blaVIM-2-carrying plasmids in P. aeruginosa. blaVIM-2 might have been acquired by transposition of Tn6352, a novel transposon composed of the In58 and ISPa17 elements. The recognition of similar inverted repeat (IR) sites by ISPa17 reveals a common mobilization process associated with acquisition of the blaVIM-2 and blaVIM-1 genes.
KEYWORDS: Pseudomonas aeruginosa, carbapenemase, plasmid-mediated resistance, transposon
TEXT
Carbapenemase-producing Pseudomonas aeruginosa strains have been increasingly documented, a particular worrisome situation due to associated resistance to several β-lactams (1, 2). The most widespread carbapenemases produced by P. aeruginosa are metallo-β-lactamases (MBLs), particularly the VIM and IMP types (1, 2). VIM-2 represents the MBL most frequently found in P. aeruginosa, and evidence of endemic spread throughout southeast Asia and southern European countries has been documented (1–3).
In P. aeruginosa, blaVIM-2 is most often associated with class 1 integrons containing additional antibiotic resistance genes (3–11). The blaVIM-2 gene has been commonly associated with chromosomally located Tn402-like class 1 integrons (3–8). This gene also has been less frequently linked to plasmids, usually smaller than 100 kb, and cannot be transferred by conjugation, at least not to Escherichia coli (1, 9).
Until now, only two complete blaVIM-2-carrying plasmid sequences in P. aeruginosa, pNOR-2000 and pDCPR1, have been described (10, 11). In order to obtain new insight about the mobile elements contributing to the dissemination of this gene, we report here the complete sequence of pJB12, a blaVIM-2-carrying plasmid from a P. aeruginosa clinical isolate, and we explore the genetic background involved in acquisition of the blaVIM-2-harboring In58 integron.
Bacterial isolate.
In the context of a regular surveillance of several Portuguese hospitals conducted by our lab for the screening of MBL producers among P. aeruginosa strains, four isolates belonging to the high-risk sequence type 175 (ST175) clone were recovered in different years from inpatients of two geographically distinct hospitals. These isolates presented the same pulsed-field gel electrophoresis (PFGE) pattern and carried the blaVIM-2 gene in a ca. 30-kb plasmid (S1 nuclease PFGE-based sizing). The pJB12 plasmid was extracted from one of these isolates, FFUP_PS_12, which was obtained in 2006 at the Centro Hospitalar do Porto (Portugal). FFUP_PS_12 was initially identified by Vitek-2 (bioMérieux), and the species was confirmed by multilocus sequence analysis (rpoD, gyrB, and 16S rRNA genes) (12). Antimicrobial susceptibility testing was conducted by standard disc diffusion, Etest (carbapenems), and broth microdilution (colistin) methods according to EUCAST guidelines (http://www.eucast.org/).
Plasmid analysis.
Extraction of plasmid DNA from FFUP_PS_12 was performed with a plasmid midikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Sequencing was accomplished with a MiSeq (Illumina) sequencer with an average median depth of coverage of 35×. The quality of the high-throughput sequence data was assessed with FastQC. De novo assembly of the paired-end reads was performed with SPAdes 3.9.0 (13). Evaluation of the genome assembly was performed by QUAST (http://quast.bioinf.spbau.ru/). ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/) was used to identify acquired antimicrobial resistance genes. Annotation and plasmid visualization were performed by Geneious 9.1.6 (Biomatters, USA). Annotations were manually curated using BLASTn and BLASTp (https://blast.ncbi.nlm.nih.gov/Blast.cgi) similarity searches of each predicted open reading frame (ORF).
Conjugation was performed as previously described (5); briefly, a spontaneous rifampin-resistant mutant of P. aeruginosa PAO1 was used as the recipient strain, and transconjugant selection was performed in Mueller-Hinton (MH) agar plates containing rifampin (100 μg/ml) and imipenem (4 μg/ml). E. coli DH5α- and P. aeruginosa PAO1-competent cells were prepared and transformed by electroporation as described previously (14). Transformants were selected in MH agar plates containing imipenem (0.5 μg/ml for E. coli DH5α and 4 μg/ml for P. aeruginosa PAO1) and confirmed by PCR for blaVIM and aacA7 genes and antimicrobial susceptibility testing.
Screening and Sanger sequencing for the pJB12-like repA gene (with the primers rep_JB12_Fw [CTCCTTGGAGCGATACGACC] and rep_JB12_Rv [GGACTCATACAGGCTCACGG]) were performed for the remaining isolates belonging to ST175 and with the blaVIM-2 gene located in a ca. 30-kb plasmid.
General features of FFUP_PS_12 and pJB12.
FFUP_PS_12 displayed an extensively drug-resistant (XDR) phenotype, including resistance to imipenem (MICs, ≥32 mg/liter), meropenem (MICs, ≥32 mg/liter), ceftazidime, cefepime, piperacillin-tazobactam, gentamicin, tobramycin, amikacin, netilmicin, and ciprofloxacin and susceptibility to aztreonam and colistin (MICs, 1 mg/liter) (15). The isolate belonged to the high-risk clone ST175, which has been associated mainly with XDR and multidrug-resistant (MDR) P. aeruginosa isolates throughout several European countries and in Japan and was responsible for the dissemination of VIM-2, IMP-1, and IMP-22 carbapenemases (1).
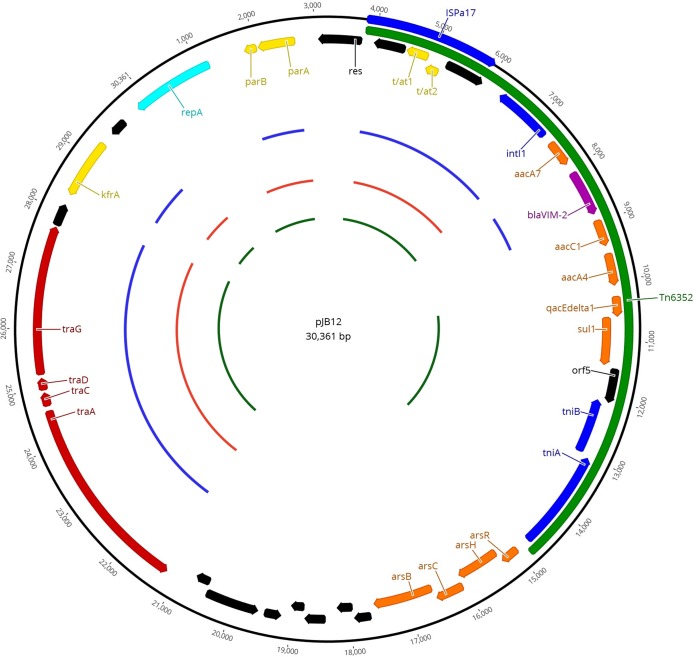
The complete plasmid sequence of pJB12 was identified in a single 30,465-bp contig, which is in agreement with the result obtained from S1 nuclease PFGE-based sizing (ca. 30 kb). Since extremities overlapped, suggesting a circular sequence, a 30,361-bp plasmid was obtained. With a GC content of 62.6%, pJB12 comprised 36 ORFs, including genes associated with a transposon structure harboring the blaVIM-2-carrying In58 integron (Fig. 1; see also Table S1 in the supplemental material).
FIG 1.
Schematic representation of the pJB12 plasmid. Annotated coding sequences are shown as arrows and are colored depending on gene function. Yellow arrows represent genes involved in plasmid housekeeping functions such as partition and maintenance. Red arrows point out the genes involved in conjugation transfer. Light-blue arrows indicate the replication gene. Dark-blue arrows point out genes responsible for DNA rearrangements (such as integrases and transposases). Orange arrows represent resistance genes. The purple arrow points out the blaVIM-2 gene. Tn6352 is presented in dark green. Genes of unknown function are shown in black. The inner circles illustrate homology regions between plasmid pJB12 and those characterized previously. The inner dark-green circle corresponds to plasmid pAX22 from Achromobacter xylosoxidans (GenBank accession number NC_022242.1), and the two outer circles represent plasmids pKLC102 (red circle) and pNOR-2000 (blue circle), both from P. aeruginosa (GenBank accession numbers AY257538.1 and KC189475.1, respectively).
In silico analysis and comparison of the pJB12 scaffold with related plasmids.
Plasmid pJB12 displays two plasmid stabilization systems (ParA-ParB and T/AT1-T/AT2) which ensure the maintenance and inheritance of the plasmid (Fig. 1; see also Table S1) (10). An ars operon, which mediates arsenic resistance, was also present (16). The identified genes (arsR, arsH, arsC, and arsB) encode for proteins that display high similarity to the ones identified in pAB3, a plasmid identified in Acinetobacter baumannii (GenBank accession number CP012005 ) (Fig. 1; Table S1). Contamination with arsenic compounds (e.g., arsenic-contaminated drinking water, arsenic compounds used in animal husbandry and as rodenticides) may lead to the selection of plasmids carrying different antimicrobial genes (16).
The putative replicase protein from pJB12 shared 83% homology with that of pMATVIM-7, a blaVIM-7-carrying plasmid identified in a P. aeruginosa isolate recovered from sputum of a cancer patient diagnosed with pneumonia at a hospital in Houston, Texas (17). However, pJB12 presented a backbone different than that of pMATVIM-7 (GenBank accession number AM778842), suggesting independent evolutionary routes for the emergence of these enzymes. Interestingly, the remaining blaVIM-2-carrying 30-kb plasmids in our collection presented the same nucleotide sequence for the replicase gene of plasmid pJB12. A putative origin of replication was found upstream of the repA gene. An AT-rich region, a 9-mer DnaA-box (5′-TTTTACACA-3′), and iterons, which are typical functional elements of plasmid replication origins, were identified (18). Considering the replicase proteins associated with the two blaVIM-2-harboring plasmids previously described in P. aeruginosa (pNOR-2000 and pDCPR1, GenBank accession numbers KC189475.1 and KJ577613.1, respectively) (10, 11), we found no detectable homology compared with the one found in plasmid pJB12, suggesting a separate origin for the replication regions. Nevertheless, for parts of pNOR-2000 and two other plasmids (pAX22, a blaVIM-1-carrying plasmid from Achromobacter denitrificans, and pKLC102, an aadB-harboring plasmid from P. aeruginosa) (19, 20), the pJB12 plasmid backbone shared high nucleotide homology with the highest amino acid sequence identity being verified for plasmid pNOR-2000 (from 80% to 97% for the maintenance system and from 87% to 94% for the transfer region) (Table S1). This homology included the partitioning and stable inheritance system genes (parA, parB, and kfrA) and transfer genes (traA, traC, traD, and traG) (Fig. 1).
The pJB12 plasmid carried a relaxase gene (traA), a putative oriT site upstream of the traA gene, and a type IV coupling protein (T4CP) gene (traG) (10, 21). However, the machinery for self-conjugation is incomplete, since the type IV secretion system (T4SS) is absent (10, 21). Even so, pJB12 might be mobilizable in the presence of a helper plasmid (10, 21). The incomplete transfer operon we observed justifies the failure of the conjugation experiments. Electrotransformation of the pJB12 plasmid to P. aeruginosa PAO1 was successful, with transformants acquiring resistance to β-lactams (MICs, ≥32 mg/liter against imipenem) and aminoglycosides.
In58 mobilization in plasmid pJB12 resembles that observed in blaVIM-1-carrying In70 from plasmid pAX22.
In this study, the blaVIM-2 gene was associated with the In58 integron, a genetic platform that also carries aminoglycoside resistance genes (aacA7, aacA4, and aacC1), which was previously identified among P. aeruginosa, Pseudomonas putida, and Citrobacter freundii isolates in different countries (6–8). This structure was found to be chromosomally located (6, 8), and plasmid pJB12 represents the only reported plasmidic support for In58. This integron was frequently found in Portuguese P. aeruginosa clinical isolates carrying the blaVIM-2 gene associated with different high-risk clones (J. Botelho, F. Grosso, and L. Peixe, unpublished data).
In58 was associated with a defective Tn402-like transposon, with ISPa17 upstream of Tn402 inverted repeat initial (IRi) in a head-to-tail orientation (Fig. 1). ISPa17 represents an atypical structure initially described in plasmid Rms149 (22). ISPa17 encodes a putative transposase, a toxin/antitoxin system (A/AT1 and A/AT2), and a putative resolvase (Fig. 1). ISPa17 may represent a role in the mobilization of defective transposons carrying class 1 integrons, as demonstrated for the blaVIM-1-harboring In70 integron (19, 23). Interestingly, the IRs flanking ISPa17 were similar to those surrounding the Tn402-like transposon, suggesting that these IRs can be targeted by the transposases of both structures. Direct repeats (DRs) of the 5′-AATTG sequence were found flanking the inverted repeat left (IRL) of ISPa17 and the inverted repeat terminal (IRt) of the Tn402-like transposon, suggesting that a transposition event mediated the en bloc insertion of these structures (19). This new complex transposable element was inserted next to the plasmid resolvase gene and designated Tn6352 (http://www.ucl.ac.uk/eastman/research/departments/microbial-diseases/tn) according to the criteria proposed by Roberts et al. (24). Future studies are warranted to investigate the mobility of Tn6352 and to address if the transposition event was mediated by the ISPa17 transposase or by a complete trans-acting tni module of another Tn402-like transposon.
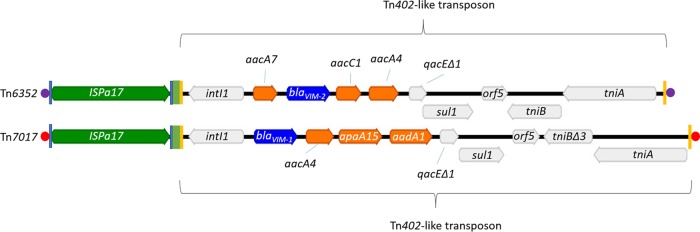
The 113-bp nucleotide sequence found between the inverted repeat right of ISPa17 and the IRi of the Tn402-like transposon (Fig. 2) was identical to the one found in plasmid pAX22 and the ParA-TraC-encoding region of several IncP-1ε plasmids, such as pHH128, pHH3414, and pHH3408 (25). This suggests that Tn6352 was assembled among these plasmids after independent insertion of ISPa17 and the Tn402-like transposon (19). In fact, IncP-1ε plasmids are frequently found in environmental settings and seem to be fundamental vectors for the spread of antibiotic resistance genes (25).
FIG 2.
Comparison of putative transposons Tn6352 and Tn7017. Both structures are composed of ISPa17- and Tn402-like transposons in a head-to-tail orientation. Carbapenemase-encoding genes are represented by dark-blue arrows. Orange arrows symbolize aminoglycoside resistance genes. ISPa17 is represented by a green arrow. The remaining genes identified in the Tn402-like transposons are shown in light gray. Light-blue rectangles represent the inverted repeats (IRs) identified in ISPa17. The IRs of the Tn402-like transposons are illustrated by yellow rectangles. The 113-bp nucleotide sequence found between the IRR of ISPa17 and the IRi of Tn402-like transposons is represented by green rectangles. Direct repeats (DRs) flanking the putative transposon Tn6352 (5′-AATTG-3′) and Tn7017 (5′-GTGGC-3′) are illustrated by purple and red circles, respectively.
In summary, this work presents the sequence of pJB12, the first complete nucleotide sequence of an In58-harboring plasmid. The blaVIM-2 gene was most likely acquired by plasmid pJB12 by transposition of Tn6352, a novel putative transposon comprising an In58 integron and an ISPa17 in a head-to-tail orientation. The recognition of DNA regions with similar IR sites by ISPa17 demonstrates a common mobilization process associated with acquisition of the blaVIM-2 and blaVIM-1 genes.
Accession number(s).
The complete sequence of plasmid pJB12 has been submitted to GenBank under the accession number KX889311.
Supplementary Material
ACKNOWLEDGMENTS
The isolate was kindly provided by the Microbiology Departments of Centro Hospitalar do Porto, Portugal.
This work had financial support from FCT/MEC through national funds and cofinanced by FEDER, under the Partnership Agreement PT2020 (grant no. UID/Multi/04378/2013). João Botelho and Filipa Grosso were supported by grants from Fundação para a Ciência e a Tecnologia (SFRH/BD/104095/2014 and SFRH/BPD/95556/2013, respectively).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02532-16.
REFERENCES
- 1.Oliver A, Mulet X, Lopez-Causape C, Juan C. 2015. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21–22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Potron A, Poirel L, Nordmann P. 2015. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int J Antimicrob Agents 45:568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Hong DJ, Bae IK, Jang IH, Jeong SH, Kang HK, Lee K. 2015. Epidemiology and characteristics of metallo-beta-lactamase-producing Pseudomonas aeruginosa. Infect Chemother 47:81–97. doi: 10.3947/ic.2015.47.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moyo S, Haldorsen B, Aboud S, Blomberg B, Maselle SY, Sundsfjord A, Langeland N, Samuelsen O. 2015. Identification of VIM-2-producing Pseudomonas aeruginosa from Tanzania is associated with sequence types 244 and 640 and the location of blaVIM-2 in a TniC integron. Antimicrob Agents Chemother 59:682–685. doi: 10.1128/AAC.01436-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quinteira S, Sousa JC, Peixe L. 2005. Characterization of In100, a new integron carrying a metallo-β-lactamase and a carbenicillinase, from Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:451–453. doi: 10.1128/AAC.49.1.451-453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinteira S, Peixe L. 2006. Multiniche screening reveals the clinically relevant metallo-beta-lactamase VIM-2 in Pseudomonas aeruginosa far from the hospital setting: an ongoing dispersion process? Appl Environ Microbiol 72:3743–3745. doi: 10.1128/AEM.72.5.3743-3745.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos C, Caetano T, Ferreira S, Mendo S. 2010. Tn5090-like class 1 integron carrying blaVIM-2 in a Pseudomonas putida strain from Portugal. Clin Microbiol Infect 16:1558–1561. doi: 10.1111/j.1469-0691.2010.03165.x. [DOI] [PubMed] [Google Scholar]
- 8.Poirel L, Lambert T, Turkoglu S, Ronco E, Gaillard J, Nordmann P. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing beta-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob Agents Chemother 45:546–552. doi: 10.1128/AAC.45.2.546-552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dortet L, Flonta M, Boudehen YM, Creton E, Bernabeu S, Vogel A, Naas T. 2015. Dissemination of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa in Romania. Antimicrob Agents Chemother 59:7100–7103. doi: 10.1128/AAC.01512-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnin RA, Poirel L, Nordmann P, Eikmeyer FG, Wibberg D, Puhler A, Schluter A. 2013. Complete sequence of broad-host-range plasmid pNOR-2000 harbouring the metallo-beta-lactamase gene blaVIM-2 from Pseudomonas aeruginosa. J Antimicrob Chemother 68:1060–1065. doi: 10.1093/jac/dks526. [DOI] [PubMed] [Google Scholar]
- 11.Vilacoba E, Quiroga C, Pistorio M, Famiglietti A, Rodriguez H, Kovensky J, Deraspe M, Raymond F, Roy PH, Centron D. 2014. A blaVIM-2 plasmid disseminating in extensively drug-resistant clinical Pseudomonas aeruginosa and Serratia marcescens isolates. Antimicrob Agents Chemother 58:7017–7018. doi: 10.1128/AAC.02934-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulet M, Lalucat J, Garcia-Valdes E. 2010. DNA sequence-based analysis of the Pseudomonas species. Environ Microbiol 12:1513–1530. doi: 10.1111/j.1462-2920.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- 13.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 16.Hobman JL, Crossman LC. 2015. Bacterial antimicrobial metal ion resistance. J Med Microbiol 64:471–497. doi: 10.1099/jmm.0.023036-0. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Toleman MA, Bennett PM, Jones RN, Walsh TR. 2008. Complete sequence of p07-406, a 24,179-base-pair plasmid harboring the blaVIM-7 metallo-beta-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob Agents Chemother 52:3099–3105. doi: 10.1128/AAC.01093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajewska M, Wegrzyn K, Konieczny I. 2012. AT-rich region and repeated sequences—the essential elements of replication origins of bacterial replicons. FEMS Microbiol Rev 36:408–434. doi: 10.1111/j.1574-6976.2011.00300.x. [DOI] [PubMed] [Google Scholar]
- 19.Di Pilato V, Pollini S, Rossolini GM. 2014. Characterization of plasmid pAX22, encoding VIM-1 metallo-β-lactamase, reveals a new putative mechanism of In70 integron mobilization. J Antimicrob Chemother 69:67–71. doi: 10.1093/jac/dkt311. [DOI] [PubMed] [Google Scholar]
- 20.Klockgether J, Reva O, Larbig K, Tummler B. 2004. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J Bacteriol 186:518–534. doi: 10.1128/JB.186.2.518-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. 2010. Mobility of plasmids. Microbiol Mol Biol Rev 74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haines AS, Jones K, Cheung M, Thomas CM. 2005. The IncP-6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. J Bacteriol 187:4728–4738. doi: 10.1128/JB.187.14.4728-4738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.San Millan A, Toll-Riera M, Escudero JA, Cantón R, Coque TM, MacLean RC. 2015. Sequencing of plasmids pAMBL1 and pAMBL2 from Pseudomonas aeruginosa reveals a blaVIM-1 amplification causing high-level carbapenem resistance. J Antimicrob Chemother 70:3000–3003. doi: 10.1093/jac/dkv222. [DOI] [PubMed] [Google Scholar]
- 24.Roberts AP, Chandler M, Courvalin P, Guedon G, Mullany P, Pembroke T, Rood JI, Smith CJ, Summers AO, Tsuda M, Berg DE. 2008. Revised nomenclature for transposable genetic elements. Plasmid 60:167–173. doi: 10.1016/j.plasmid.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heuer H, Binh CT, Jechalke S, Kopmann C, Zimmerling U, Krogerrecklenfort E, Ledger T, Gonzalez B, Top E, Smalla K. 2012. IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: diversification driven by class 1 integron gene cassettes. Front Microbiol 3:2. doi: 10.3389/fmicb.2012.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.