ABSTRACT
Acquisition of vancomycin resistance in Staphylococcus aureus is often accompanied by a reduction in virulence, but the mechanisms underlying this change remain unclear. The present study was undertaken to investigate this process in a clinical heterogeneous vancomycin-intermediate S. aureus (hVISA) strain, 10827; an hVISA reference strain, Mu3; and a VISA reference strain, Mu50, along with their respective series of vancomycin-induced resistant strains. In these strains, increasing MICs of vancomycin were associated with increased expression of the vancomycin resistance-associated regulator gene (vraR) and decreased expression of virulence genes (hla, hlb, and coa) and virulence-regulated genes (RNAIII, agrA, and saeR). These results suggested that VraR might have a direct or indirect effect on virulence in S. aureus. In electrophoretic mobility shift assays, VraR did not bind to promoter sequences of hla, hlb, and coa genes, but it did bind to the agr promoter region. In DNase I footprinting assays, VraR protected a 15-nucleotide (nt) sequence in the intergenic region between the agr P2 and P3 promoters. These results indicated that when S. aureus is subject to induction by vancomycin, expression of vraR is upregulated, and VraR binding inhibits the function of the Agr quorum-sensing system, causing reductions in the virulence of VISA/hVISA strains. Our results suggested that VraR in S. aureus is involved not only in the regulation of vancomycin resistance but also in the regulation of virulence.
KEYWORDS: Staphylococcus aureus, global regulatory networks, two-component regulatory systems, vancomycin resistance, virulence regulation
INTRODUCTION
Staphylococcus aureus is an important pathogen that can be acquired in both community and hospital environments and is responsible for illnesses ranging from superficial skin infections to deep-seated and life-threatening diseases (1, 2). Infections caused by methicillin-resistant S. aureus (MRSA) are associated with mortality and morbidity, an aggressive course, multidrug resistance, and hospital outbreaks (3). Vancomycin is the first-choice drug for treatment of MRSA infection, but its increasing clinical use has led to the emergence of S. aureus strains with reduced vancomycin susceptibility, including vancomycin-resistant S. aureus (VRSA), vancomycin-intermediate S. aureus (VISA), and heterogeneous VISA (hVISA) (4–6). According to the Clinical and Laboratory Standards Institute (CLSI) (7), VRSA is defined by a vancomycin MIC of ≥16 μg/ml, whereas VISA isolates have MICs between 4 and 8 μg/ml. hVISA strains appear to be susceptible to vancomycin (MICs of ≤2 μg/ml) but contain a subpopulation of vancomycin-resistant cells.
Most VRSA isolates carry plasmid-borne copies of transposon Tn1546, which was acquired from vancomycin-resistant Enterococcus species (8). Tn1546-based vancomycin resistance involves alteration of the d-alanyl-d-alanine dipeptide residue in the S. aureus cell wall precursor lipid II to d-alanyl-d-lactate, which has substantially lower affinity for vancomycin (9). Unlike in VRSA, the molecular mechanisms of resistance in VISA/hVISA strains are not well understood. Comparative genomics shows diverse genetic mutations in VISA relative to vancomycin-susceptible S. aureus (VSSA), and only a few of these mutations (such as vraSR, graSR, walKR, stk1/stp1, rpoB, clpP, sigB, and trfAB genes) have been experimentally verified (10–17). Transcriptomic studies have revealed changes in expression levels in two-component regulatory systems (TCRSs), in particular, vraSR, walKR, and graSR (10, 18–20). These TCRSs seem to be involved in cell wall synthesis and thickening, which restricts the access of vancomycin to its target sites. A particularly interesting member of this group is vraSR, which encodes a histidine kinase (VraS) and a response regulator (VraR) that can rapidly sense and transduce cell wall stress (20). The VraSR system is highly expressed in VISA/hVISA strains (18, 21–23). After induction by an inhibitor of cell wall synthesis, VraS and VraR autoactivate the expression of the vra operon and about 46 other unlinked genes known collectively as the cell wall stimulon, which positively regulate synthesis of the cell wall, leading to thickening and subsequent resistance to vancomycin (21–24).
Acquisition of vancomycin resistance in VISA/hVISA strains is often accompanied with a decrease in virulence (25–27). Our previous report has also demonstrated that VISA/hVISA strains have reduced coagulase activity and reduced or no hemolysis (28). Hattangady et al. performed complete genome comparison, along with transcriptomic and metabolomic studies, of two laboratory-selected VISA strains and found that expression of surface-associated virulence determinants was decreased in VISA isolates (29). Majcherczyk et al. and Peleg et al. used a rat model and a Galleria mellonella model, respectively, to show that virulence and infectivity of VISA/hVISA strains attenuate as the vancomycin MIC for the strains increases (26, 27). However, the specific mechanism that underlies the attenuation of virulence in VISA/hVISA strains was still not clear.
In our study, a clinical hVISA strain, 10827; an hVISA reference strain, Mu3; and a VISA reference strain, Mu50, were exposed to increased concentrations of vancomycin and produced their series of vancomycin-induced resistant strains. Quantitative real-time PCR (qRT-PCR) was performed to investigate the expression of vraR (encoding a vancomycin resistance-associated regulator); virulence genes hla (encoding alpha-toxin), hlb (encoding β-toxin), and coa (encoding coagulase); and virulence-regulated genes (RNAIII, agrA, and saeR) in all strains. Electrophoretic mobility shift assays (EMSAs) were conducted to identify the potential of VraR to affect virulence through transcriptional regulation, and the precise location of VraR binding to promoter sequences was determined by DNase I footprinting.
RESULTS
Expression of vraR and virulence-associated genes is altered in VISA strains.
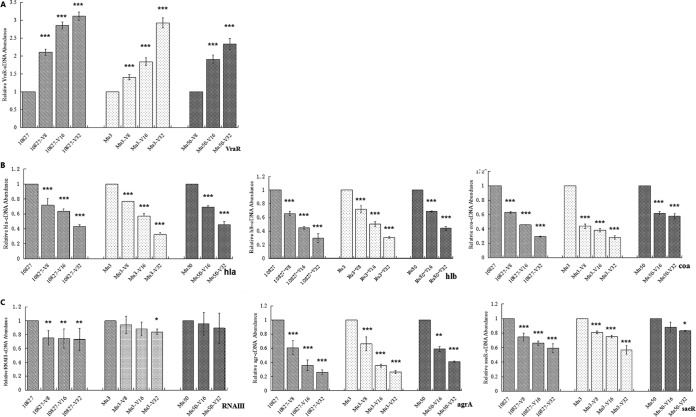
Expression of vraR, hla, hlb, coa, RNAIII, agrA, and saeR was analyzed by qRT-PCR. As shown in Fig. 1A and Table 1, transcription of vraR was upregulated in the vancomycin-resistant strains 10827-V32 (3.11-fold), Mu3-V32 (2.92-fold), and Mu50-V32 (2.33-fold) compared to their parental isolates (P < 0.001). Transcription of hla, hlb, and coa was downregulated in the vancomycin-resistant strains 10827-V32 (0.43-fold, 0.30-fold, and 0.29-fold, respectively), Mu3-V32 (0.33-fold, 0.31-fold, and 0.28-fold, respectively), and Mu50-V32 (0.46-fold, 0.44-fold, and 0.58-fold, respectively) compared to their parental strains (P < 0.001) (Fig. 1B). Transcription of agrA and saeR was downregulated in the vancomycin-resistant strains 10827-V32 (0.26-fold and 0.59-fold, respectively), Mu3-V32 (0.26-fold and 0.57-fold, respectively), and Mu50-V32 (0.41-fold and 0.83-fold, respectively) compared to their parental strains (P < 0.05) (Fig. 1C). Moreover, we also observed a trend toward reduced transcription of RNAIII with higher vancomycin MIC, although this was not statistically significant.
FIG 1.
Quantitative real-time PCR analysis of the expression of resistance-associated and virulence-associated genes in S. aureus strains 10827, Mu3, and Mu50 and their series of vancomycin-resistant induced strains. Results are presented relative to their respective parental strain 10827, Mu3, or Mu50, the value for which has been normalized to 1. (A) Expression of vraR. (B) Expression of hla, hlb, and coa. (C) Expression of RNAIII, agrA, and saeR. Bars represent mean values from three or more independent experiments. Error bars indicate standard deviations. ***, P < 0.001; **, P < 0.01; *, P < 0.05.
TABLE 1.
Quantitative real-time PCR analysis of gene expression in S. aureus strains 10827, Mu3, and Mu50 and their series of vancomycin-resistant induced strains
| Strain | Vancomycin MIC (μg/ml) | Relative cDNA abundance |
||||||
|---|---|---|---|---|---|---|---|---|
| vraR | hla | hlb | coa | RNAШ | agr | sae | ||
| 10827 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 10827-V8 | 8 | 2.10 ± 0.09 | 0.71 ± 0.09 | 0.66 ± 0.03 | 0.63 ± 0.01 | 0.75 ± 0.10 | 0.61 ± 0.10 | 0.75 ± 0.06 |
| 10827-V16 | 16 | 2.85 ± 0.10 | 0.64 ± 0.03 | 0.45 ± 0.02 | 0.46 ± 0.00 | 0.74 ± 0.14 | 0.36 ± 0.08 | 0.66 ± 0.03 |
| 10827-V32 | 32 | 3.11 ± 0.12 | 0.43 ± 0.02 | 0.30 ± 0.06 | 0.2939 ± 0.0081 | 0.73 ± 0.16 | 0.26 ± 0.03 | 0.59 ± 0.06 |
| Mu3 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Mu3-V8 | 8 | 1.40 ± 0.07 | 0.77 ± 0 | 0.72 ± 0.05 | 0.44 ± 0.03 | 0.94 ± 0.13 | 0.66 ± 0.10 | 0.81 ± 0.02 |
| Mu3-V16 | 16 | 1.84 ± 0.11 | 0.57 ± 0.04 | 0.51 ± 0.04 | 0.38 ± 0.02 | 0.88 ± 0.10 | 0.3555 ± 0.0224 | 0.75 ± 0.02 |
| Mu3-V32 | 32 | 2.92 ± 0.14 | 0.33 ± 0.02 | 0.31 ± 0.02 | 0.28 ± 0.02 | 0.84 ± 0.04 | 0.26 ± 0.02 | 0.57 ± 0.60 |
| Mu50 | 8 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Mu50-V16 | 16 | 1.90 ± 0.12 | 0.70 ± 0.01 | 0.69 ± 0.01 | 0.62 ± 0.02 | 0.95 ± 0.17 | 0.59 ± 0.04 | 0.88 ± 0.07 |
| Mu50-V32 | 32 | 2.33 ± 0.16 | 0.46 ± 0.04 | 0.44 ± 0.03 | 0.58 ± 0.04 | 0.89 ± 0.22 | 0.49 ± 0.01 | 0.83 ± 0.01 |
Oligomeric state of VraR in solution.
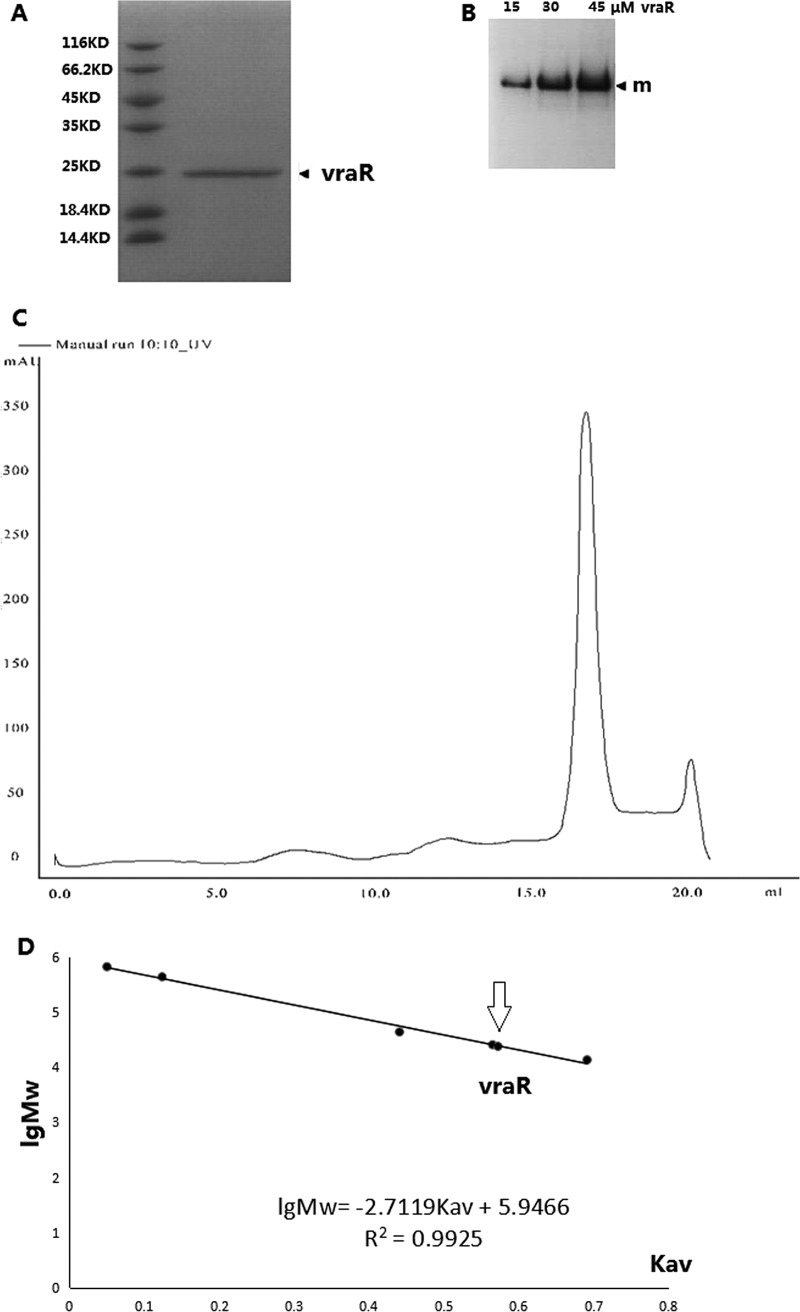
The analysis of purified VraR protein by gel filtration chromatography revealed a single peak with an elution volume of 17.06 ml. On the assumption that the shape and partial specific volume of VraR are similar to those of the standard proteins, the native molecular mass of VraR was estimated to be 24,058 Da, which was calculated from a standard linear regression equation, IgMw = 5.9466 − 2.7119 × Kav (Fig. 2). The native molecular mass is approximately the same as the molecular mass of a VraR monomer (∼23.5 kDa). Native PAGE analysis also indicated that purified VraR protein was present in a single oligomeric state in solution. Thus, we conclude that VraR in solution is a stable monomer.
FIG 2.
Determination of VraR purity and oligomeric state. (A) SDS-PAGE analysis of VraR. (B) Native PAGE analysis of VraR (“m” denotes the monomer). (C) Gel filtration chromatographic analysis of VraR. (D) Molecular size calibration for standard proteins. Molecular size estimated from the Kav value for VraR is indicated by an arrow.
VraR binds specifically to the agr promoter region.
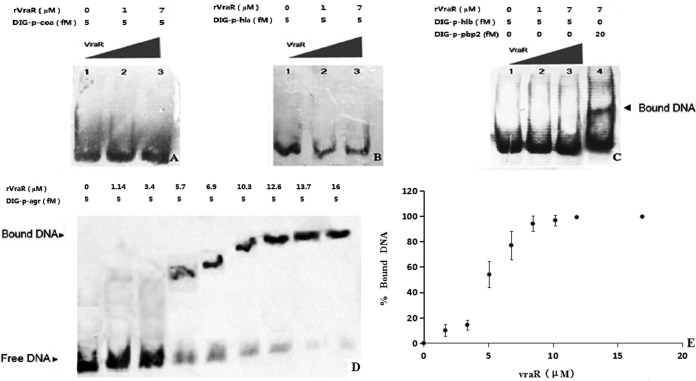
The vraR gene was cloned, and His-tagged VraR protein was expressed and purified to perform EMSA with DNA probes containing the putative promoter sequences of the coa, hla, hlb, and agr target genes. With an increasing concentration of VraR in the assays, the amount of free agr promoter DNA substrate decreased, and the intensity of the shifted band increased (Fig. 3). The VraR concentration that resulted in shifting of half of the agr-promoter probe gave a computed dissociation constant (Kd) of 5.06 ± 0.90 μM. In these assay, VraR did not bind to the promoter regions of coa, hla, and hlb.
FIG 3.
Electrophoretic mobility shift assay (EMSA) for VraR. (A) EMSA with the coa promoter. (B) EMSA with the hla promoter. (C) EMSA with the hlb promoter. (D) Analysis of the DNA-binding affinity of VraR. (E) Plot of the level of bound agr promoter in the EMSA against the VraR concentration in the assay.
Characterization of the VraR binding site in the agr promoter.
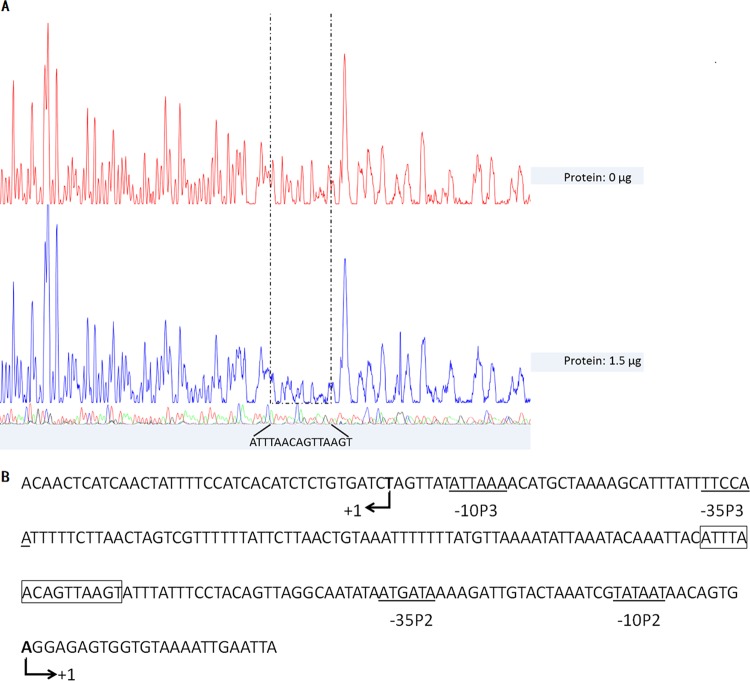
To further investigate the mechanism of regulation of the agr locus by VraR, the location of potential VraR binding sites in the promoter region of the agr locus was investigated by DNase I footprinting analysis. As shown in Fig. 4A, VraR protected a 15-nucleotide (nt) region of the agr promoter (5′-ATTTAACAGTTAAGT-3′ on the agrA coding strand) against DNase I digestion. The protected sequence was located in the intergenic region (IR) between the P2 and P3 promoters (Fig. 4B).
FIG 4.
Identification of VraR binding sequences. (A) DNase I footprinting analysis of the agr promoter with VraR. (B) agr promoter sequence with a summary of the DNase I footprinting assay results. The −10 and −35 promoter regions are indicated by solid lines below the sequence. VraR-protected regions are in solid boxes. The translational start site is indicated by an angled arrow, and the corresponding nucleotide is in boldface.
DISCUSSION
It has been found that the natural parental strains of VSSA were alterable to hVISA and VISA by serial passage with stepwise increases of vancomycin concentrations (30, 31). A link has also been implied between the acquisition of vancomycin resistance in S. aureus and attenuated virulence (10, 26, 27, 29, 32). In our study, vancomycin resistance was induced in three strains of S. aureus, including Mu3 and Mu50, which both carry the mutation in vraS(I5N) and result in replacement of isoleucine at residue 5 of the protein with asparagine (31, 33). The third strain (10827) was a clinical hVISA isolate, and its genome has not been fully sequenced. We found that the vancomycin MICs of the three strains were increased to 32 μg/ml by incrementally increasing induction concentrations, and the MICs were positively correlated with the expression of vraR and negatively correlated with the expression of virulence-associated genes. These results correspond well with those of previous studies. Whether similar trends also exist in other clinical strains remains to be determined.
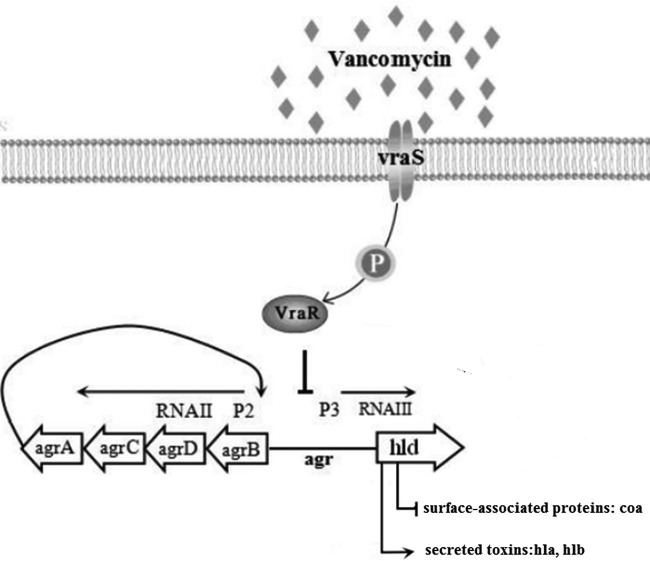
The virulence of S. aureus is largely determined by surface-associated proteins, secreted toxins, and enzymes, expression of which is tightly regulated by regulatory loci, such as agr, saeR, sarA, and sigB, which form a complex and delicate regulatory network that acts to coordinate temporal expression of virulence genes (34). The Agr quorum-sensing system is a global regulatory system which downregulates the expression of many genes encoding surface-associated proteins (such as coa and fnb) and upregulates expression of genes encoding secreted toxins (such as hla and hlb) (35, 36). The SaeSR system regulates expression of many virulence genes, including those encoding surface proteins and toxins, primarily at the transcriptional level (37). Giraudo et al. showed by Northern blot analysis that the SaeSR system can regulate transcription of hla, hlb, and coa independently of the Agr system (37). Another regulatory locus, sarA, encodes the 14.5-kDa DNA-binding proteins that also regulate virulence factor expression. Unlike agr, SarA activates the synthesis of both surface-associated proteins and exoproteins in S. aureus (35, 38). Moreover, the alternative transcription factor sigma B (encoded by sigB) affects the expression of several genes that encode virulence factors and stress response proteins and seems to counterbalance the influence of the agr system on the expression of virulence factors. In our study, when S. aureus was under the induction of vancomycin, the expression of vraR was increased, and expression of hla, hlb, coa, RNAIII, agrA, and saeR was decreased. The results of EMSAs showed that VraR did not bind to the promoter regions of hla, hlb, and coa but it did bind to the agr promoter. This result indicated that VraR was indirectly involved in the regulation of virulence through binding to the agr promoter region. Previous studies have shown that agr activates the transcription of hla and hlb and represses the transcription of coa (35, 36). Therefore, we speculated that VraR inhibits other virulence regulation loci such as saeRS by an indirect path (Fig. 5).
FIG 5.
Proposed model of the molecular events in VraR-mediated gene regulation. Direct repression and activation of genes are shown by the solid bars and arrows, respectively. Based on experimental data from this study and others, we propose that vraS responds to vancomycin-elicited cell wall stress and results in transphosphorylation of VraR, which is followed by directly targeting and binding the agr promoter region to inhibit the expression of virulence factors.
The agr locus is composed of two divergent transcriptional units, the transcription of which is activated from divergent promoters, P2 and P3 (39). The P2 promoter drives the transcription of the agrBDCA operon (40). The P3 promoter drives the synthesis of the RNAIII molecule (41). Previous research has shown that the intracellular signaling molecules that bind the agr promoter are AgrA, SarA, and SarR (42, 43). AgrA can bind as a dimer to each of two 9-bp direct repeats in the IR between the P2 and P3 promoters to modulate agr transcription (44). The SarA binding site on the agr promoter covered a 29-bp region between the P2 and P3 promoters devoid of any direct repeats (45). SarR is a dimer that recognizes and binds to an overlapping site between two AgrA binding direct repeats (45, 46). Reyes et al. reported that AgrA activates agr P2 and P3 promoters, whereas SarA activates and SarR represses P2 transcription (47). In our study, DNase I footprinting showed that VraR protects a 15-nt sequence in the P2 and P3 interpromoter region of agr and that this sequence contains overlapping AgrA, AarA, and SarR binding sites.
The agr locus has been shown to be polymorphic and can be divided into four distinct genetic groups (48, 49). Previous studies have shown that induction of the VISA phenotype is more likely in agr group II strains than in other groups (50). However, Sirichoat et al. reported that the S. aureus strains from agr groups I to IV all developed to become VISA strains, and when hVISA or VISA developed from VSSA, a reduction in levels of agr expression occurred in the resistant isolates (51). Sakoulas et al. examined Agr function in VISA/hVISA and found that all VISA strains were defective in Agr function (52). Harigaya et al. and Sirichoat et al. have also confirmed that VISA/hVISA strains have been detected with reduced or absent Agr activity (51, 53). As yet, the molecular mechanisms of agr dysfunction in VISA/hVISA strains have not been elucidated. In this study, we found that with the increase of vraR expression, the agr expression gradually decreased and VraR did bind to the agr promoter. This observation may partly explain the finding that reduced agr function in VISA/hVISA strains was frequently detected. However, there have been reports that loss of Agr function directly contributes to the development of vancomycin resistance in S. aureus. Tsuji et al. used an in vitro pharmacodynamic model to evaluate the role of Agr in wild-type and knockout S. aureus strains and found that strains with a disruption in the agr locus were more likely to develop intermediate resistance to vancomycin (54). Therefore, whether a direct or indirect feedback-regulation mechanism exists would be determined by further study.
Conclusion.
It has been reported that VISA/hVISA strains with attenuated virulence may represent a “stealth” strategy to evade host immune surveillance and promote clinical persistence and chronic infections (10). The VraSR system is known as vancomycin resistance associated. Our results suggest that the VraSR system, as the central regulation factor, is not only involved in the regulation of vancomycin resistance but is also involved in the regulation of virulence. This finding indicates that the resistance and virulence regulatory network of S. aureus is more complex than we previously recognized, which is worth further attention and exploration.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. aureus strains 10827 (vancomycin MIC, 1 μg/ml), Mu3 (vancomycin MIC, 2 μg/ml), and Mu50 (vancomycin MIC, 8 μg/ml) were induced by continuous passage through medium containing increasing concentrations of vancomycin. Briefly, overnight cultures of the strains were diluted to an optical density at 600 nm (OD600) of 0.5 in fresh brain heart infusion (BHI) broth, and then 10 μl of each suspension was streaked onto vancomycin-supplemented BHI plates (starting concentration of 2 μg/ml), which were incubated overnight at 35°C. Individual colonies were picked and grown in BHI broth containing the same concentration of vancomycin. This procedure was repeated with increasing concentrations of vancomycin (up to 32 μg/ml) until the isolate grew stably in the presence of vancomycin. These homogenous series of vancomycin-induced resistant strains were named according to the MIC values: 10827-V8, 10827-V16, and 10827-V32; Mu3-V8, Mu3-V16, and Mu3-V32; and Mu50-V16 and Mu50-V32.
RNA preparation and qRT-PCR assays.
RNA preparations were made as described previously (55). Briefly, the parental strains 10827 and Mu3 was grown to exponential phase in BHI broth at 35°C, and the viability of bacteria was determined via spectrophotometry (OD600 of 0.7). A homogenous series of vancomycin-induced resistant strains was harvested until they reached an OD600 of 0.7 in BHI broth, and then the cells were collected by centrifugation for 10 min at 5,000 × g and used for total RNA extraction using TRIzol (Invitrogen). cDNA was synthesized and labeled according to the manufacturer's recommendations for S. aureus antisense genome arrays (Affymetrix Inc.). Transcript levels of vraR, hla, hlb, coa, RNAIII, agrA, and saeR were quantified by quantitative real-time PCR (qRT-PCR) using a Kapa SYBR qPCR kit (Kapa Biosystems) in a LightCycler (LC-32; Roche, USA). The 16S rRNA gene was used as an internal control as described previously (56). All qRT-PCR assays were repeated three times. The primer sets for expression analysis of all genes are described in Table 2.
TABLE 2.
Primers used in this study
| Name | Primer sequence (5′–3′) |
|---|---|
| 16sRNA-1 | CGTGCTACAATGGACAATACAAA |
| 16sRNA-2 | ATCTACGATTACTAGCGATTCCA |
| vraR-1 | AAGACTAAACACCAACAAAACAGAG |
| vraR-2 | GAAAAGTTACTTACGCCAATCACA |
| coa-1 | GAGATACAGACAATCCACATAA |
| coa-2 | CTACCTTCAAGACCTTCTAAAA |
| hla-1 | GTAAGTCGTATTAGAACTAAAGCGG |
| hla-2 | GCACGCAAGAATCTTGTAGTTC |
| hlb-1 | AATCAATTTTGCATCTATTTTGTTG |
| hlb-2 | CAAAACGGTCGATAACATATAAACG |
| RNAIII-1 | TTCACTGTGTCGATAATCCA |
| RNAIII-2 | GGAAGGAGTGATTTCAATGG |
| agrA-1 | ATGGTATCGAGAATCTTAAAGTACG |
| agrA-2 | TACTTACTTCATCGGGTATTTCG |
| saeR-1 | CGCCTTAACTTTAGGTGCAGATGAC |
| saeR-2 | ACGCATAGGGACTTCGTGACCATT |
| coa-promoter-1 | GTGTTGTCATGCTTTGTTACTCC |
| coa-promoter-2 | GCGCCTAGCGAAATTATTTGC |
| hla-promoter-1 | TTTTCATCATCCTTCTATTT |
| hla-promoter-2 | CTAACCCTCGAAATTGAAAT |
| hlb-promoter-1 | TACTCAAAAAACATTTACTTAAAAATATAAATTCGAT |
| hlb-promoter-2 | TTTTATATAGCTTACAACAAAATAGATGCAAAATTG |
| agr-promoter-1 | ATCAACTATTTTCCATCACATCT |
| agr-promoter-2 | TTACACCACTCTCCTCACT |
Cloning, expression, and purification of VraR.
Cloning and expression of VraR proteins were performed as described previously (20). Briefly, the vraR gene was amplified using the primers 5′-GAGGATCCATGACGATTAAAGTATTGTTTG-3′ (forward) and 5′-GCCTCGAGCTATTGAATTAAATTATGTTGG-3′ (reverse), containing BamHI (italicized) and XhoI (underlined) sites, respectively. The amplified product was cloned into pGEM-T vector (Promega) using the TA cloning procedure and transformed into Escherichia coli XL1-Blue cells (Invitrogen). vraR was subcloned into the expression vector pET28a (Invitrogen), and the recombinant plasmid was transformed into E. coli BL21(DE3). The transformants were checked for the insert by using colony PCR and DNA sequencing. The clones were incubated in Luria-Bertani (LB) broth with shaking at 35°C. The overnight cultures were diluted 1:50 in LB broth and incubated with shaking again at 35°C until the OD600 reached 0.3. The cultures were then induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated at 25°C for 12 h. Cells were harvested by centrifugation and resuspended in 50 ml of lysis buffer (20 mM Tris-HCl, pH 8.0, and 0.5 M NaCl). Then, cells were lysed by sonication and centrifuged at 12,000 × g for 30 min at 4°C. The supernatant was loaded onto a nickel affinity column as recommended by the manufacturer (His-select nickel affinity gel; Sigma), and the protein was eluted using an elution buffer composed of 50 mM Tris (pH 8.0), 300 mM NaCl, and 250 mM imidazole. The purity of the eluted protein was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (SDS-PAGE). The protein concentration was measured using a Bradford assay, with bovine serum albumin as a standard.
Determination of the oligomerization state of His-tagged VraR protein.
The oligomerization state of His-VraR protein was analyzed by gel filtration chromatography and native polyacrylamide gel electrophoresis (native PAGE; Tris-glycine system). The methods were performed as described previously (20). Briefly, gel filtration chromatography was carried out by the Äkta fast protein liquid chromatography (FPLC) system (Amersham Biosciences). In brief, a purified VraR sample (1 mg/ml) in 500 mM Tris buffer, pH 7.0, 0.5 mM MgCl2, was applied to a Superdex 200 HR 10/300 column (GE Healthcare) equilibrated with the same buffer. The column was operated at a flow rate of 0.36 ml/min, and the proteins were detected at 280 nm. The column was calibrated with proteins of known molecular mass: thyroglobulin (669 kDa), ferritin (440 kDa), ovalbumin (44 kDa), chymotrypsinogen A (25.7 kDa), and RNase A (13.7 kDa). The Kav values for the standard proteins and VraR were calculated from the equation Kav = (Ve − V0)/(Vc − V0), where V0 is the column void volume, Ve is the elution volume, and Vc is the geometric column volume. The retention time of VraR was interpolated to obtain an approximate mass of VraR protein. The purified His-VraR protein also was examined by native PAGE to verify the major oligomeric forms. Concentrations of VraR sample analyzed by native PAGE were 15, 30, and 45 μM.
EMSA.
DNA fragments containing the coa, hla, hlb, and agr promoters were amplified from the S. aureus Mu50 chromosome. The primers are described in Table 2. The resulting PCR products were labeled using a digoxigenin gel shift kit (Roche) according to the manufacturer's instructions. The labeled fragments were then incubated at 25°C for 15 min with various amounts of purified VraR protein (1 to 16 μM) in 10 μl of incubation buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 3 mM magnesium acetate, 0.1 mM EDTA, 0.1 mM dithiothreitol). Following incubation, the mixtures were separated by electrophoresis in a 4.5% native polyacrylamide gel in 0.5× Tris-borate-EDTA buffer. Band shifts were detected and analyzed by a Universal Hood 2 electrophoresis imager (Bio-Rad). The quantitative analysis of the bands was carried out using NIH ImageJ (version 1.3). Determination of the dissociation constants was based on the results obtained from three independent experiments.
DNase I footprinting assay.
For preparation of fluorescent 6-carboxyfluorescein (FAM)-labeled probes, the promoter region of agr was amplified from plasmid pEASY-Blunt Simple-agr using Dpx DNA polymerase (Tolo Biotech) and primers Agr-p-1 (5′-ATCAACTATTTTCCATCACATCT-3′; FAM labeled) and Agr-p-2 (5′-TTACACCACTCTCCTCACT-3′). The amplified 235-bp probes consist of the full lengths of the P2 and P3 promoters. The FAM-labeled probes were purified using the Wizard SV gel and PCR cleanup system (Promega) and quantified using a NanoDrop 2000C spectrophotometer (Thermo). DNase I footprinting assays were performed according to the method of Wang et al. (57). Briefly, 400 ng of probe was incubated with different amounts of protein (0, 0.5, 1, 1.5, 2.0, 2.5, 3.0, 3.5, and 4.0 μg) in a total volume of 40 μl, and 1.5 μg VraR protein was chosen to carry out the experiment. Following incubation for 30 min at 25°C, 10 μl of solution containing ∼0.015 U of DNase I (Promega) and 100 nmol of freshly prepared CaCl2 was added to each mixture and then further incubated for 1 min at 25°C. The reaction was stopped by the addition of 140 μl of DNase I stop solution (200 mM unbuffered sodium acetate, 30 mM EDTA, and 0.15% SDS). Samples were first extracted with phenol-chloroform and then precipitated with ethanol, and the resulting pellets were dissolved in 30 μl of Milli-Q ultrapure water (Millipore). Then, we added 200 ng/μl poly(dI-dC) in binding buffer to exclude the possibility of nonspecific binding. Preparation of the DNA ladder, electrophoresis of the reaction products, and data analysis were performed as described previously (57), except for use of the GeneScan-LIZ500 size standard (Applied Biosystems).
Statistical analysis.
All statistical analyses were performed using SPSS Statistics 16.0 (SPSS Inc., Chicago, IL). Statistical differences between each group were determined by one-way analysis of variance (ANOVA). A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Baolin Sun and Xu Zhang (University of Science and Technology of China, Hefei, Anhui, China) for assistance with technical assistance. Language editing services were provided by Edanz Group Ltd.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Kim HK, Thammavongsa V, Schneewind O, Missiakas D. 2012. Recurrent infections and immune evasion strategies of Staphylococcus aureus. Curr Opin Microbiol 15:92–99. doi: 10.1016/j.mib.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. 2006. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 4.Sievert DM, Rudrik JT, Patel JB, McDonald LC, Wilkins MJ, Hageman JC. 2008. Vancomycin-resistant Staphylococcus aureus in the United States, 2002–2006. Clin Infect Dis 46:668–674. doi: 10.1086/527392. [DOI] [PubMed] [Google Scholar]
- 5.Sieradzki K, Roberts RB, Haber SW, Tomasz A. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N Engl J Med 340:517–523. doi: 10.1056/NEJM199902183400704. [DOI] [PubMed] [Google Scholar]
- 6.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother 40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing: 22nd informational supplement M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 8.Kos VN, Desjardins CA, Griggs A, Cerqueira G, Van Tonder A, Holden MT, Godfrey P, Palmer KL, Bodi K, Mongodin EF, Wortman J, Feldgarden M, Lawley T, Gill SR, Haas BJ, Birren B, Gilmore MS. 2012. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with methicillin-resistant S. aureus hospital-acquired infection in the United States. mBio 3:e00112-12. doi: 10.1128/mBio.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Severin A, Wu SW, Tabei K, Tomasz A. 2004. Penicillin-binding protein 2 is essential for expression of high-level vancomycin resistance and cell wall synthesis in vancomycin-resistant Staphylococcus aureus carrying the enterococcal vanA gene complex. Antimicrob Agents Chemother 48:4566–4573. doi: 10.1128/AAC.48.12.4566-4573.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardete S, Kim C, Hartmann BM, Mwangi M, Roux CM, Dunman PM, Chambers HF, Tomasz A. 2012. Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus MRSA strain of clonal type USA300. PLoS Pathog 8:e1002505. doi: 10.1371/journal.ppat.1002505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato Y, Suzuki T, Ida T, Maebashi K. 2010. Genetic changes associated with glycopeptide resistance in Staphylococcus aureus: predominance of amino acid substitutions in YvqF/VraSR. J Antimicrob Chemother 65:37–45. doi: 10.1093/jac/dkp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hafer C, Lin Y, Kornblum J, Lowy FD, Uhlemann AC. 2012. Contribution of selected gene mutations to resistance in clinical isolates of vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 56:5845–5851. doi: 10.1128/AAC.01139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoji M, Cui L, Iizuka R, Komoto A, Neoh HM, Watanabe Y, Hishinuma T, Hiramatsu K. 2011. walK and clpP mutations confer reduced vancomycin susceptibility in Staphylococcus aureus. Antimicrob Agents Chemother 55:3870–3881. doi: 10.1128/AAC.01563-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howden BP, Stinear TP, Allen DL, Johnson PD, Ward PB, Davies JK. 2008. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob Agents Chemother 52:3755–3762. doi: 10.1128/AAC.01613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cameron DR, Ward DV, Kostoulias X, Howden BP, Moellering RC Jr, Eliopoulos GM, Peleg AY. 2012. Serine/threonine phosphatase Stp1 contributes to reduced susceptibility to vancomycin and virulence in Staphylococcus aureus. J Infect Dis 205:1677–1687. doi: 10.1093/infdis/jis252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh VK, Schmidt JL, Jayaswal RK, Wilkinson BJ. 2003. Impact of sigB mutation on Staphylococcus aureus oxacillin and vancomycin resistance varies with parental background and method of assessment. Int J Antimicrob Agents 21:256–261. doi: 10.1016/S0924-8579(02)00359-X. [DOI] [PubMed] [Google Scholar]
- 17.Renzoni A, Kelley WL, Barras C, Monod A, Huggler E, François P, Schrenzel J, Studer R, Vaudaux P, Lew DP. 2009. Identification by genomic and genetic analysis of two new genes playing a key role in intermediate glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother 53:903–911. doi: 10.1128/AAC.01287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAleese F, Wu SW, Sieradzki K, Dunman P, Murphy E, Projan S, Tomasz A. 2006. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J Bacteriol 188:1120–1133. doi: 10.1128/JB.188.3.1120-1133.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui L, Lian JQ, Neoh HM, Reyes E, Hiramatsu K. 2005. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother 49:3404–3413. doi: 10.1128/AAC.49.8.3404-3413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belcheva A, Golemi-Kotra D. 2008. A close-up view of the VraSR two-component system. A mediator of Staphylococcus aureus response to cell wall damage. J Biol Chem 283:12354–12364. doi: 10.1074/jbc.M710010200. [DOI] [PubMed] [Google Scholar]
- 21.Howden BP, Smith DJ, Mansell A, Johnson PD, Ward PB, Stinear TP, Davies JK. 2008. Different bacterial gene expression patterns and attenuated host immune responses are associated with the evolution of low-level vancomycin resistance during persistent methicillin-resistant Staphylococcus aureus bacteraemia. BMC Microbiol 8:39. doi: 10.1186/1471-2180-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuroda M, Kuwahara-Arai K, Hiramatsu K. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem Biophys Res Commun 269:485–490. doi: 10.1006/bbrc.2000.2277. [DOI] [PubMed] [Google Scholar]
- 23.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49:807–821. doi: 10.1046/j.1365-2958.2003.03599.x. [DOI] [PubMed] [Google Scholar]
- 24.McCallum N, Meier PS, Heusser R, Berger-Bächi B. 2011. Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus. Antimicrob Agents Chemother 55:1391–1402. doi: 10.1128/AAC.01213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 26.Majcherczyk PA, Barblan JL, Moreillon P, Entenza JM. 2008. Development of glycopeptide-intermediate resistance by Staphylococcus aureus leads to attenuated infectivity in a rat model of endocarditis. Microb Pathog 45:408–414. doi: 10.1016/j.micpath.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Peleg AY, Monga D, Pillai S, Mylonakis E, Moellering RC Jr, Eliopoulos GM. 2009. Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J Infect Dis 199:532–536. doi: 10.1086/596511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai Y, Zhou X, Ma X, Lu H, Li H. 2012. Misidentification of vancomycin-resistant Staphylococcus aureus as coagulase-negative staphylococcus. J Med Microbiol 61:1454–1458. doi: 10.1099/jmm.0.045518-0. [DOI] [PubMed] [Google Scholar]
- 29.Hattangady DS, Singh AK, Muthaiyan A, Jayaswal RK, Gustafson JE, Ulanov AV, Li Z, Wilkinson BJ, Pfeltz RF. 2015. Genomic, transcriptomic and metabolomic studies of two well-characterized, laboratory-derived vancomycin-intermediate Staphylococcus aureus strains derived from the same parent strain. Antibiotics (Basel) 4:76–112. doi: 10.3390/antibiotics4010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wongthong S, Dutchanutouch K, Namsaengkang V, Chanawong A, Wilailuckana C, Lulitanond A. 2015. Performance of vancomycin and teicoplanin disk diffusion test in isogenic vancomycin non-susceptible Staphylococcus aureus. J Infect Dev Ctries 9:157–164. doi: 10.3855/jidc.5059. [DOI] [PubMed] [Google Scholar]
- 31.Katayama Y, Murakami-Kuroda H, Cui L, Hiramatsu K. 2009. Selection of heterogeneous vancomycin-intermediate Staphylococcus aureus by imipenem. Antimicrob Agents Chemother 53:3190–3196. doi: 10.1128/AAC.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao W, Cameron DR, Davies JK, Kostoulias X, Stepnell J, Tuck KL, Yeaman MR, Peleg AY, Stinear TP, Howden BP. 2013. The RpoB H481Y rifampicin resistance mutation and an active stringent response reduce virulence and increase resistance to innate immune responses in Staphylococcus aureus. J Infect Dis 207:929–939. doi: 10.1093/infdis/jis772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui L, Neoh HM, Shoji M, Hiramatsu K. 2009. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 53:1231–1234. doi: 10.1128/AAC.01173-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bronner S, Monteil H, Prévost G. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol Rev 28:183–200. doi: 10.1016/j.femsre.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol 183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Recsei P, Kreiswirth B, O'Reilly M, Schlievert P, Gruss A, Novick RP. 1986. Regulation of exoprotein gene expression by agr. Mol Gen Genet 202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 37.Giraudo AT, Cheung AL, Nagel R. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch Microbiol 168:53–58. doi: 10.1007/s002030050469. [DOI] [PubMed] [Google Scholar]
- 38.Chien YT, Manna AC, Projan SJ, Cheung AL. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem 274:37169–37176. doi: 10.1074/jbc.274.52.37169. [DOI] [PubMed] [Google Scholar]
- 39.Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. 1988. Cloning, characterization, and sequencing of an accessory gene regulator agr in Staphylococcus aureus. J Bacteriol 170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, Kreiwirth B, Vandenesch F, Moghazeh S. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 41.Janzon L, Lofdahl S, Arvidson S. 1989. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator agr of Staphylococcus aureus. Mol Gen Genet 219:480–485. doi: 10.1007/BF00259623. [DOI] [PubMed] [Google Scholar]
- 42.Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 43.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 44.Koenig RL, Ray JL, Maleki SJ, Smeltzer MS, Hurlburt BK. 2004. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J Bacteriol 186:7549–7555. doi: 10.1128/JB.186.22.7549-7555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chien Y, Cheung AL. 1998. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J Biol Chem 237:2645–2652. doi: 10.1074/jbc.273.5.2645. [DOI] [PubMed] [Google Scholar]
- 46.Manna AC, Cheung AL. 2006. Transcriptional regulation of the agr locus and the identification of DNA binding residues of the global regulatory protein SarR in Staphylococcus aureus. Mol Microbiol 60:1289–1301. doi: 10.1111/j.1365-2958.2006.05171.x. [DOI] [PubMed] [Google Scholar]
- 47.Reyes D, Andrey DO, Monod A, Kelley WL, Zhang G, Cheung AL. 2011. Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J Bacteriol 193:6020–6031. doi: 10.1128/JB.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jarraud S, Lyon GJ, Figueiredo AM, Lina G, Vandenesch F, Etienne J, Muir TW, Novick RP. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J Bacteriol 182:6517–6522. doi: 10.1128/JB.182.22.6517-6522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ji G, Beavis R, Novick RP. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 50.Sakoulas G, Eliopoulos GM, Moellering RC Jr, Novick RP, Venkataraman L, Wennersten C, DeGirolami PC, Schwaber MJ, Gold HS. 2003. Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? J Infect Dis 187:929–938. doi: 10.1086/368128. [DOI] [PubMed] [Google Scholar]
- 51.Sirichoat A, Wongthong S, Kanyota R, Tavichakorntrakool R, Chanawong A, Welbat JU, Lulitanond A. 2016. Phenotypic characteristics of vancomycin-non-susceptible Staphylococcus aureus. Jundishapur J Microbiol 9:e26069. doi: 10.5812/jjm.26069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakoulas G, Eliopoulos GM, Moellering RC Jr, Wennersten C, Venkataraman L, Novick RP, Gold HS. 2002. Accessory gene regulator agr locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob Agents Chemother 46:1492–1502. doi: 10.1128/AAC.46.5.1492-1502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harigaya Y, Ngo D, Lesse AJ, Huang V, Tsuji BT. 2011. Characterization of heterogeneous vancomycin-intermediate resistance, MIC and accessory gene regulator agr dysfunction among clinical bloodstream isolates of Staphylococcus aureus. BMC Infect Dis 11:287. doi: 10.1186/1471-2334-11-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsuji BT, Rybak MJ, Lau KL, Sakoulas G. 2007. Evaluation of accessory gene regulator agr group and function in the proclivity towards vancomycin intermediate resistance in Staphylococcus aureus. Antimicrob Agents Chemother 51:1089–1091. doi: 10.1128/AAC.00671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beaume M, Hernandez D, Farinelli L, Deluen C, Linder P, Gaspin C, Romby P, Schrenzel J, Francois P. 2010. Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS One 5:e10725. doi: 10.1371/journal.pone.0010725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shang W, Davies TA, Flamm RK, Bush K. 2010. Effects of ceftobiprole and oxacillin on mecA expression in methicillin-resistant Staphylococcus aureus clinical isolates. Antimicrob Agents Chemother 54:956–959. doi: 10.1128/AAC.01024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Cen XF, Zhao GP, Wang J. 2012. Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J Bacteriol 194:5237–5244. doi: 10.1128/JB.00989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]