ABSTRACT
We identified the carbapenemase gene blaOXA-499, a variant of blaOXA-143, from a clinical isolate of Acinetobacter pittii for the first time. OXA-499 shared 93.1% amino acid identity with OXA-143, and the gene was located on the chromosome. By cloning the OXA-499-encoding gene into the pWH1266 vector and transforming it into susceptible Acinetobacter spp., we were able to show that OXA-499 confers resistance to carbapenems.
KEYWORDS: Acinetobacter pittii, carbapenemase resistant, OXA-143-like, OXA-499
TEXT
Acinetobacter spp. are opportunistic pathogens that cause various nosocomial infections, such as pneumonia, bacteremia, wound infections, and meningitis (1). Over the course of time, these pathogens have acquired resistance genes to nearly all antibiotic classes, including fluoroquinolones, aminoglycosides, and carbapenems, which make them difficult to treat. There has been a rapid increase in carbapenem-resistant Acinetobacter spp. in Korea, which may be due to a significant increase in carbapenem usage (2). Carbapenem resistance in Acinetobacter spp. is associated mainly with carbapenem-hydrolyzing class D β-lactamases (CHDLs), such as OXA-23, OXA-24, OXA-58, OXA-51, OXA-143, and their variants (3–8). Acinetobacter pittii is frequently associated with hospital-associated infections and was the most commonly isolated Acinetobacter sp. in a recent German study (9). The substantial increase in carbapenem resistance in A. pittii needs to be further studied for us to understand its clinical significance (10–12). Here, we report the occurrence of carbapenemase OXA-499, a variant of OXA-143, in carbapenem-resistant Acinetobacter pittii clinical isolate YMC2010/8/T346 belonging to the novel sequence type 1385 (ST1385), recovered from a patient in a university-affiliated hospital in South Korea.
In 2010, a 69-year-old male patient was admitted to the emergency room in a university-affiliated hospital in South Korea with signs of fever and abdominal pain. A week earlier, the patient had undergone an esophagojejunostomy, and peritonitis due to leakage was suspected. The patient's initial white blood cell count was 34,480/μl, and his C-reactive protein and procalcitonin levels were elevated. Empirical therapy with piperacillin-tazobactam (4.5 g intravenously [i.v.] thrice daily [TID]) was administered for 2 days. Due to the unstable vital signs of the patient, the regimen was changed to metronidazole (500 mg i.v. twice daily for 4 days), teicoplanin (400 mg i.v. once daily for 10 days), and meropenem (1 g i.v. TID for 13 days). In the culture of isolates obtained from the peritoneal catheter tip, Enterococcus faecium, coagulase-negative staphylococci, and A. pittii were identified by using the Bruker Biotyper matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) system.
MICs were determined by Etest (bioMérieux, Marcy-l'Étoile, France). A. pittii isolate YMC2010/8/T346 was susceptible to piperacillin (MIC, 32 μg/ml), ceftazidime (MIC, 4 μg/ml), cefepime (MIC, 4 μg/ml), ciprofloxacin (MIC, 0.25 μg/ml), and imipenem (MIC, 2 μg/ml) but resistant to meropenem (MIC, 16 μg/ml) according to CLSI guidelines (13). Genomic DNA was isolated using the Wizard genomic DNA purification kit (Promega, Madison, WI), and a whole-genome library was constructed and sequenced on a 318 chip using the Ion Torrent PGM system and Ion Sequencing 400 kit (Life technologies, CA, USA). Additionally, PacBio single-molecule real-time (SMRT) sequencing and genome assembly were performed to identify the location of the blaOXA-499 allele. Annotation was performed with RAST (http://rast.nmpdr.org) and the NCBI Prokaryotic Genome Annotation Pipeline. Genomic analysis was performed using Geneious pro 8.0 (https://www.geneious.com), and resistance genes were screened using ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/) with manual scrutiny.
Whole-genome analysis indicated the presence of blaOXA-506, a variant of the A. pittii intrinsic blaOXA-213-like and blaADC-41 alleles, which were not associated with insertion sequence (IS) elements, as well as the acquired blaOXA-499 gene. Since previous studies indicated that the intrinsic blaOXA from A. pittii has thus far not been shown to confer resistance, we considered blaOXA-499 as the probable candidate conferring carbapenem resistance (14). Further analysis of the blaOXA-499 gene revealed that this enzyme is a variant of OXA-143, first described in A. pittii isolated in Germany, which has been deposited under GenBank accession number NG_049775. However, the related data on its carbapenemase characteristics have not been reported to date. OXA-143-like variants have been identified worldwide. OXA-143 and OXA-231 were identified in Brazil, OXA-253 in Honduras and Brazil, OXA-182 in South Korea, and OXA-255 in the United States (8). OXA-499 shared amino acid identities of 93.1%, 92.7%, 96.0%, 92.7%, and 98.9% with OXA-143, OXA-231, OXA-253, OXA-182, and OXA-255, respectively (see Table S1 in the supplemental material). As a result, all of these were grouped together as an OXA-143-like clade in the amino acid phylogeny of OXA-carbapenemase genes (see Fig. S1 in the supplemental material). The deduced protein sequence showed 3 amino acid differences between OXA-499 and OXA-255, namely, Thr22-Ser, Lys29-Asn, and Ser158-Asn. The amino acids conserved in residues STFK (position 81–84), FGN (position 154–156), and KSG (position 218–220) were the same as those in other class D β-lactamases and CHDLs (see Fig. S2 in the supplemental material).
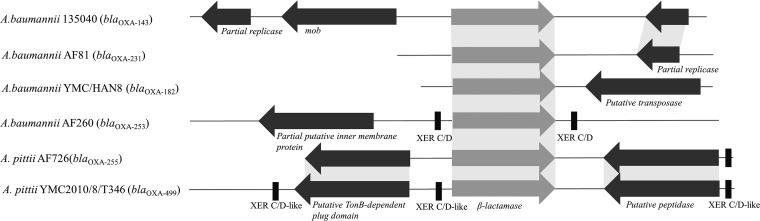
The open reading frame of the blaOXA-499 gene was amplified using primers OXA-499A (5′-ATGAAAAAATTTATACTTCCTATCTTCAGC-3′) and OXA-499B (5′-TTATATAATCCCTAAATTTTCTAATG-3′) and ligated into the PstI-digested shuttle vector pWH1266 before transforming it into electrocompetent carbapenem-susceptible Acinetobacter baumannii ATCC 19606 and A. pittii YMC2013/1/R3000 as described previously (15). This led to increases in the MICs of imipenem, meropenem, and ertapenem from 0.38, 0.75, and 4 μg/ml to 4, 8, and ≥32 μg/ml, respectively; i.e., an 8- to 10-fold increase in carbapenem resistance was observed. Additionally, cloning encompassing the endogenous promoter of blaOXA-499, as described by Zander et al. (16), was performed, which increased the MICs of all three carbapenems used in this study to ≥32 μg/ml (Table 1). In addition, positive results were seen for the three-dimensional extract and modified-Hodge tests (17) with carbapenem discs when the blaOXA-499 gene was cloned into pET28a(+) and transformed into Escherichia coli BL21(DE3) (see Fig. S3 in the supplemental material). Despite repeated transformation of the blaOXA-499 gene using a plasmid preparation from A. pittii YMC2010/8/T346 to either A. baumannii or A. pittii, the gene was not transferable. PacBio SMRT sequencing indicated the presence of the blaOXA-499 gene in the chromosome. Genomic sequence comparisons of various Acinetobacter spp. (see Fig. S4 in the supplemental material) indicated the insertion of a 4.5-kb fragment consisting of a plasmid-associated putative peptidase gene present upstream and a TonB-dependent receptor plug domain present downstream of the blaOXA-499 gene (Fig. 1). The Xer C/D-like sites were located 40 bp upstream (5′-ATTTAATATAATACGCCTTATACGAAAT-3′) of the putative peptidase, 79 bp upstream (5′-ATTTAACATAATGGGCGTTATGTTAAGT-3′) of blaOXA-499, and 44 bp downstream (5′-TTACGCATAAGCCGTATTATGTTAATT-3′) of the TonB-dependent receptor plug domain, which indicates the mobility and probable recombination of this fragment into the chromosome. The putative peptidase gene showed 77% similarity to a peptidase encoded on A. baumannii plasmid p3ABSDF (GenBank accession number CU468233), and the TonB-dependent receptor plug domain showed 55% amino acid identity with a hypothetical protein in Acinetobacter spp. NIPH1867 (locus tag WP_005210788), which was similar to findings reported previously (16). In addition, the regions surrounding this 4.5-kb fragment insertion showed similarity with the plasmids identified in Acinetobacter spp. in a BLAST search. The above findings implied that the initial location of blaOXA-499 was in a plasmid, which subsequently integrated into the chromosome.
TABLE 1.
Carbapenem susceptibility of A. pittii YMC2010/8/T346, A. pittii YMC2013/1/R3000, A. baumannii ATCC19606, and transformants harboring blaOXA-499- encoding recombinant pWH1266
| Strain and vector | MIC (μg/ml) and interpretation ofa: |
||
|---|---|---|---|
| Meropenem | Imipenem | Ertapenemb | |
| A. pittii YMC2010/8/T346 | 16, R | 1.5, S | ≥32 |
| A. baumannii ATCC19606 | 0.75, S | 0.38, S | 4 |
| A. baumannii ATCC19606 + pWH1266 | 1, S | 0.38, S | 6 |
| A. baumannii ATCC19606 + pWH1266::Oxa499__Pc | ≥32, R | ≥32, R | ≥32 |
| A. baumannii ATCC19606 + pWH1266::Oxa499 | 8, I | 4, S | ≥32 |
| A. pittii YMC2013/1/R3000 | 0.75, S | 0.38, S | 4 |
| A. pittii YMC2013/1/R3000 + pWH1266 | 0.75, S | 0.38, S | 8 |
| A. pittii YMC2013/1/R3000 + pWH1266::Oxa499__Pa | ≥32, R | ≥32, R | ≥32 |
| A. pittii YMC2013/1/R3000 + pWH1266::Oxa499 | 8, I | 4, S | ≥32 |
R, resistant strains; I, intermediate strains; S, susceptible strains.
CLSI did not provide MIC interpretation guidelines for ertapenem in Acinetobacter spp.
Oxa499_P indicates that the cloning was performed including the natural promoter of the blaOXA-499 gene.
FIG 1.
Schematic drawing of the blaOXA-143-like flanking regions in A. baumannii 135040 (GenBank accession no. NG_049441), A. baumannii AF81 (NG_049527), A. baumannii HAN8 (NG_049483), A. baumannii AF260 (NG_049548), A. pittii AF726 (NG_049550), and A. pittii YMC2010/8/T346 (KX828713). Shaded regions indicate the similarities between the isolates.
Initially described in A. baumannii isolated from Brazil (18), OXA-143 variants are now being detected all around the globe in Acinetobacter spp. The detection of OXA-499 in a novel sequence type of A. pittii, including the previously detected OXA-182, in South Korea is a cause of great concern and indicates the possibility that more variants of the carbapenemase gene blaOXA-143 exist.
Nucleotide sequence accession number(s).
The nucleotide sequence of the blaOXA-499 gene and the whole-genome sequences of A. pittii isolate YMC2010/8/T346 generated from Ion Torrent PGM and PacBio SMRT sequencing are available under the GenBank accession numbers KX828713, MKHF00000000, and CP017938, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the BioNano Health-Guard Research Center, funded by the Ministry of Science, ICT and Future Planning (MSIP) of Korea as a Global Frontier Project (grant H-GUARD_ 2014M3A6B2060509); by a grant from the Brain Korea 21 PLUS Project for Medical Science, Yonsei University; and by the Bio & Medical Technology Development Program of the NRF funded by the Korean government (grant 2014M3A9E5073818).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02676-16.
REFERENCES
- 1.Bergogne-Berezin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev 9:148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shibayama K, Lee H, Kim S. 2015. Comparison of antibiotic resistance rate of medically important microorganisms between Japan and Korea. Ann Clin Microbiol 18:111–118. doi: 10.5145/ACM.2015.18.4.111. [DOI] [Google Scholar]
- 3.Girlich D, Poirel L, Nordmann P. 2010. First isolation of the blaOXA-23 carbapenemase gene from an environmental Acinetobacter baumannii isolate. Antimicrob Agents Chemother 54:578–579. doi: 10.1128/AAC.00861-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bou G, Oliver A, Martinez-Beltran J. 2000. OXA-24, a novel class D beta-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob Agents Chemother 44:1556–1561. doi: 10.1128/AAC.44.6.1556-1561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown S, Young HK, Amyes SG. 2005. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin Microbiol Infect 11:15–23. doi: 10.1111/j.1469-0691.2004.01016.x. [DOI] [PubMed] [Google Scholar]
- 6.Tsakris A, Ikonomidis A, Pournaras S, Spanakis N, Markogiannakis A. 2006. Carriage of OXA-58 but not of OXA-51 beta-lactamase gene correlates with carbapenem resistance in Acinetobacter baumannii. J Antimicrob Chemother 58:1097–1099. doi: 10.1093/jac/dkl365. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, D'Souza R, Yong D, Lee K. 2016. Prediction of putative resistance islands in a carbapenem-resistant Acinetobacter baumannii global clone 2 clinical isolate. Ann Lab Med 36:320–324. doi: 10.3343/alm.2016.36.4.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans BA, Amyes SG. 2014. OXA beta-lactamases. Clin Microbiol Rev 27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schleicher X, Higgins PG, Wisplinghoff H, Korber-Irrgang B, Kresken M, Seifert H. 2013. Molecular epidemiology of Acinetobacter baumannii and Acinetobacter nosocomialis in Germany over a 5-year period (2005-2009). Clin Microbiol Infect 19:737–742. doi: 10.1111/1469-0691.12026. [DOI] [PubMed] [Google Scholar]
- 10.Boo TW, Walsh F, Crowley B. 2006. First report of OXA-23 carbapenemase in clinical isolates of Acinetobacter species in the Irish Republic. J Antimicrob Chemother 58:1101–1102. doi: 10.1093/jac/dkl345. [DOI] [PubMed] [Google Scholar]
- 11.Park YK, Jung SI, Park KH, Kim SH, Ko KS. 2012. Characteristics of carbapenem-resistant Acinetobacter spp. other than Acinetobacter baumannii in South Korea. Int J Antimicrob Agents 39:81–85. doi: 10.1016/j.ijantimicag.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Chusri S, Chongsuvivatwong V, Rivera JI, Silpapojakul K, Singkhamanan K, McNeil E, Doi Y. 2014. Clinical outcomes of hospital-acquired infection with Acinetobacter nosocomialis and Acinetobacter pittii. Antimicrob Agents Chemother 58:4172–4179. doi: 10.1128/AAC.02992-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 14.Perichon B, Goussard S, Walewski V, Krizova L, Cerqueira G, Murphy C, Feldgarden M, Wortman J, Clermont D, Nemec A, Courvalin P. 2014. Identification of 50 class D beta-lactamases and 65 Acinetobacter-derived cephalosporinases in Acinetobacter spp. Antimicrob Agents Chemother 58:936–949. doi: 10.1128/AAC.01261-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Zander E, Bonnin RA, Seifert H, Higgins PG. 2014. Characterization of blaOXA-143 variants in Acinetobacter baumannii and Acinetobacter pittii. Antimicrob Agents Chemother 58:2704–2708. doi: 10.1128/AAC.02618-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manchanda V, Singh NP. 2003. Occurrence and detection of AmpC beta-lactamases among Gram-negative clinical isolates using a modified three-dimensional test at Guru Tegh Bahadur Hospital, Delhi, India. J Antimicrob Chemother 51:415–418. [DOI] [PubMed] [Google Scholar]
- 18.Gionco B, Pelayo JS, Venancio EJ, Cayo R, Gales AC, Carrara-Marroni FE. 2012. Detection of OXA-231, a new variant of blaOXA-143, in Acinetobacter baumannii from Brazil: a case report. J Antimicrob Chemother 67:2531–2532. doi: 10.1093/jac/dks223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.