Abstract
Background
Infliximab shows good efficacy in treating refractory rheumatoid arthritis (RA). However, many patients responded poorly and related studies were inconsistent in predictive biomarkers. This study aimed to identify circulating biomarkers for predicting infliximab response in RA.
Material/Methods
Public databases of Gene Expression Omnibus (GEO) and ArrayExpress were searched for related microarray datasets, focused on the response to infliximab in RA. All peripheral blood samples were collected before infliximab treatment and gene expression profiles were measured using microarray. Differential genes associated with infliximab efficacy were analyzed. The genes recognized by half of the datasets were regarded as candidate biomarkers and validated by prospective datasets.
Results
Eight microarray datasets were identified with 374 blood samples of RA patients, among which 191 (51.1%) were diagnosed as non-responders in the subsequent infliximab treatment. Five genes (FKBP1A, FGF12, ANO1, LRRC31, and AKR1D1) were associated with the efficacy and recognized by half of the datasets. The 5-gene model showed a good predictive power in random- and prospective-designed studies, with AUC (area under receiver operating characteristic [ROC] curve)=0.963 and 1.000, and it was also applicable at the early phase of treatment (at week 2) for predicting the response at week 14 (AUC=1.000). In the placebo group, the model failed to predict the response (AUC=0.697), indicating the model’s specificity in infliximab treatment.
Conclusions
The model of FKBP1A, FGF12, ANO1, LRRC31, and AKR1D1 in peripheral blood is useful for efficiently predicting the response to infliximab treatment in rheumatoid arthritis.
MeSH Keywords: Arthritis, Rheumatoid; Biomarkers, Pharmacological; Genetic Testing
Background
Rheumatoid arthritis (RA) is an autoimmune disease of unknown etiology, characterized by chronic polyarthritis that induces joint damage and functional disability [1]. Tumor necrosis factor (TNF) plays a key role in the RA pathogenesis, and anti-TNF agents have shown good efficacy in treating refractory RA, especially when methotrexate (MTX) and disease-modifying anti-rheumatic drugs (DMARDs) proved ineffective [2]. As the first clinically approved TNF blocker, infliximab has been widely used in RA treatment. However, approximately 30% of RA patients achieved inadequate response [3]. Clinical parameters related with RA diagnosis or prognosis, such as C-reactive protein (CRP) and disease activity score 28 (DAS28), failed to predict clinical response to infliximab [4,5]. Thus, considering the high cost, adverse effects, and prolonged disease severity in non-responders, a search for predictive biomarkers was needed [6].
In recent years, gene expression profiling has been successfully used on tissue samples or blood to identify biomarkers in various disorders [7,8]. Compared to synovial biopsy in RA, peripheral blood samples were easy to obtain and facilitated dynamic monitoring, which could also reflect the inflammatory condition. Thus, several studies tested the gene expression pattern of peripheral blood mononuclear cells (PBMCs) in RA patients to identify genetic markers for predicting subsequent infliximab response. Unfortunately, huge discrepancies between studies restrained the clinical application of those reported biomarkers, and no systematic reviews have concentrated on this. Therefore, we conducted a systematic bioinformatics analysis of microarray data in public databases to identify the most recognized differential genes associated with RA infliximab response, and then test the viability of these genes as predictive biomarkers.
Material and Methods
Search strategy
Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) are international public repositories that archive and freely distribute microarray data submitted by the research community. We searched related microarray data in the databases using key words including “infliximab”, “anti-TNF” or “anti-tumor necrosis factor” in combination with “rheumatoid arthritis” or “RA”.
Data selection and extraction
All clinical information and published articles of the microarray datasets were reviewed independently by 2 researchers. Microarray datasets were included if they satisfied the following criteria: (i) Diagnosis of RA was based on the 1987 revised criteria of the American College of Rheumatology (ACR) for the classification of RA or on the 2010 ACR/European League Against Rheumatism (EULAR) classification criteria [9,10]; (ii) Patients responded inadequately to MTX; (iii) naïve to anti-TNF treatment or had an interval of at least 3 months after the last treatment; (iv) Peripheral blood samples were collected before first administration of infliximab, and total RNA was separated from PBMCs to analyze the global expression pattern of numerous well-annotated genes by microarray; and (v) Response to infliximab was assessed at least 6 weeks later according to certain criteria. For those eligible datasets, the normalized data of gene expression profiling were downloaded from the databases of GEO or ArrayExpress.
Identify biomarkers for predicting infliximab response
For each microarray dataset, all samples were divided into different groups according to the therapeutic outcomes. Differential genes associated with infliximab response were analyzed using the “gcRMA” package of R language (version 3.2.3, https://www.r-project.org/). The genes recognized by half of the datasets were regarded as candidate biomarkers for predicting infliximab response. P<0.05 was considered significant, and P values from these analyses were not adjusted for multiple testing.
Candidate biomarkers validation
ACR and EULAR response criteria are commonly used to measure individual response in clinical trials, dependent on the extent of change and the level of disease activity reached [11]. In the included datasets, GSE58795 and GSE42296 were randomly or prospectively designed datasets using the ACR and EULAR response criteria, respectively. Thus, we chose these datasets as validation sets to determine the accuracy of the biomarkers in predicting infliximab response. ROC (receiver operating characteristic) analysis was conducted to assess the predictive power of each biomarker. If no single biomarker showed good predictive power, a logistics regression model was established to evaluate the predictive accuracy of multiple gene models. In GSE42296, blood samples were also collected for microarray testing at week 2 after first infliximab treatment, and the data were used to assess the predictive power of biomarkers at the early phase of treatment. In GSE58795, another 29 patients were randomly divided into control group and received placebo treatment, and 13 patients were categorized as moderate response at week 14. Through comparing the biomarkers’ predictive values between infliximab and placebo groups, the specificity and accuracy of these biomarkers could be assessed indirectly.
Gene ontology analysis
To overview biological aspects of the biomarkers, the online tool ToppGene (https://toppgene.cchmc.org/) was used for a gene ontology analysis of the molecular function, biological processes, and cellular components in the biomarkers.
Results
Basic characteristics
Eight microarray datasets [12–19] were identified with 183 pre-treated blood samples of infliximab responders and 191 non-responders (GEO accession numbers: GSE3592, GSE8350, GSE12051, GSE42296, GSE58795, GSE20690, GSE33377, and GSE78068). Eight different platforms were involved (GPL3064, GPL5460, GPL2507, GPL6244, GPL10379, GPL4133, GPL5175, and GPL6480). The dataset of GSE19821 was excluded for absence of response categories [20]. Among the eligible datasets, most RA patients were diagnosed according to the ACR (1987) criteria. Before infliximab treatment, all patients suffered from moderate to severe disease activity, and responded inadequately to disease-modifying anti-rheumatic drugs (DMARD) and/or MTX. Most datasets used the ACR or EULAR response criteria, and categorized the therapeutic outcomes into response and non-response, while the criteria based on clinical disease activity index (DAI) and serum CRP level were used in GSE78068 and GSE20690, respectively. The characteristics of included microarray datasets are listed in Table 1.
Table 1.
Characteristics of included microarray datasets.
| GEO accession | Contributors (submission date) | RA diagnosis criteria | Patient included criteria | Infliximab infusion | Efficacy evaluation (date) | Response categories |
|---|---|---|---|---|---|---|
| GSE3592 | Lequerre et al. (2005) | ACR (1987) | MTX treatment; DAS28=5.1; resistance to at least one DMARD (MTX included) | 3 mg/kg at weeks 0, 2, 6, and every 8th week thereafter | EULAR criteria (at week 14) | Good; moderate (categorized as non-respond); none |
| GSE8350 | Sekiguchi et al. (2007) | ACR (1987) | At least 18 yrs of age; an ACR functional class of I–III; receiving MTX ≥6 mg/week for a minimum of 3 months and a stable dose for at least 6 weeks at the time of study enrolment | 3 mg/kg at weeks 0, 2, 6, and every other 8 weeks thereafter | ACR criteria (at week 22) | Respond (ACR50%); non-respond |
| GSE12051 | Julia et al. (2008) | ACR (1987) | Active disease (DAS28 >3.2); naive to anti-TNF alpha treatment; receiving concomitant MTX treatment of ≤20 mg/wk or maximum tolerable; concomitant therapy with prednisolone (GC, dose ≤10 mg/day or equivalent) and NSAID; having stable MTX, GC and NSAID doses during the previous 4 weeks to the inclusion in the study; having discontinued previous DMARDs at least 4 weeks prior to the inclusion | At weeks 0, 2 and 14 | EULAR criteria (at week 14) | Good; moderate (categorized as respond); none |
| GSE42296 | Mesko et al. (2012) | EULAR/ACR (2010) | Age between 20 and 60 years; failure to respond to at least two DMARDs; active disease (DAS28 >3.2); anti-TNFα therapy-naive patients or previous anti-TNFα use at least 3 months prior to blood sampling | At weeks 0 and 2 | ACR criteria (at week 6 or 14) | Respond (ACR0% or ACR20%); non-respond (ACR50% or ACR70%) |
| GSE58795 | MacIsaac et al. (2014) | ACR (1987) | At least 6 tender and 6 swollen joints, CRP ≥1.0 mg/L; on a stable dose of MTX; naive to anti-TNF biologics; RAMRIS synovitis score ≥1 in the radio-carpal or intercarpal joints of one hand | 3 mg/kg at weeks 0, 2, 6 and 14 | EULAR criteria (at week 14) | Good; moderate; none |
| GSE20690 | Tanino et al. (2010) | NA | Resistant to standard MTX treatment | At week 0, and NA | Serum CRP level (at week 14) | No inflammation (CRP ≤0.3 mg/dl); residual inflammation |
| GSE33377 | Toonen et al. (2011) | ACR (1987) | First course of a TNF-blocking agent; DAS28 >3.2; previous failure on at least two DMARDs (MTX included) | At week 0, and NA | EULAR criteria (at week 14) | Good; moderate (excluded); none |
| GSE78068 | Nakamura et al. (2016) | ACR (1987) or EULAR/ACR (2010) | Responded inadequately to MTX (≥6 mg/week) | At week 0, and NA | CDAI (at month 6) | Achieving remission (CDAI ≤2.8); not achieving remission |
GEO – Gene Expression Omnibus; RA – rheumatoid arthritis; ACR – American College of Rheumatology; MTX – methotrexate; DAS28 – disease activity score 28; EULAR – European League Against Rheumatism; TNF – tumor necrosis factor; GC – glucocorticoid; NSAID – non-steroidal anti-inflammatory drugs; DMARD – disease-modifying anti-rheumatic drugs; RAMRIS – RA MRI Score; CRP – C-reactive protein; CDAI – clinical disease activity index; NA – not available.
Differential genes associated with infliximab response
The differential genes in GSE33377 were obtained from the supplementary material of published studies due to the poor annotated files in the microarray dataset. The distribution of differential genes was heterogeneous across the datasets. Only 5 genes were recognized by at least half of the datasets: FKBP1A (FK506 binding protein 1A), FGF12 (fibroblast growth factor 12), ANO1 (anoctamin 1, calcium activated chloride channel), LRRC31 (leucine rich repeat containing 31), and AKR1D1 (aldo-keto reductase family 1, member D1) (Figure 1). Gene ontology analysis indicated that these 5 genes were significantly associated with the regulation of transmembrane transport (Table 2).
Figure 1.
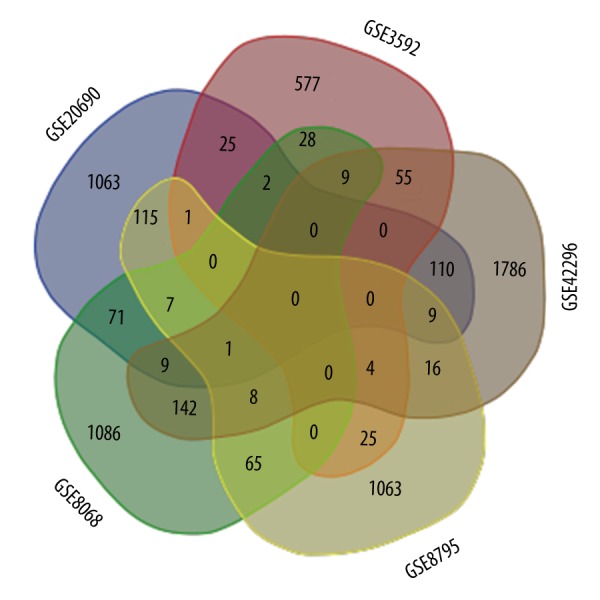
The distribution of differential genes between studies (only list the top 5 of the differential genes).
Table 2.
Gene ontology (GO) analysis of biological processes in FKBP1A, FGF12, ANO1, LRRC31 and AKR1D1 (gene limits ≥3).
| ID | Name | P Value | FDR B&H | Genes |
|---|---|---|---|---|
| GO:0034765 | Regulation of ion transmembrane transport | 0.0001361 | 0.01028 | 3 |
| GO:0034762 | Regulation of transmembrane transport | 0.0001528 | 0.01028 | 3 |
| GO:0032844 | Regulation of homeostatic process | 0.0001759 | 0.01028 | 3 |
| GO:0043269 | Regulation of ion transport | 0.0004262 | 0.01585 | 3 |
| GO:0098655 | Cation transmembrane transport | 0.0005691 | 0.01771 | 3 |
| GO:0098660 | Inorganic ion transmembrane transport | 0.0006062 | 0.01771 | 3 |
| GO:0034220 | Ion transmembrane transport | 0.001662 | 0.02615 | 3 |
| GO:0006812 | Cation transport | 0.00188 | 0.02848 | 3 |
| GO:0048878 | Chemical homeostasis | 0.002022 | 0.02953 | 3 |
| GO:0055085 | Transmembrane transport | 0.00336 | 0.03926 | 3 |
Candidate biomarkers validation
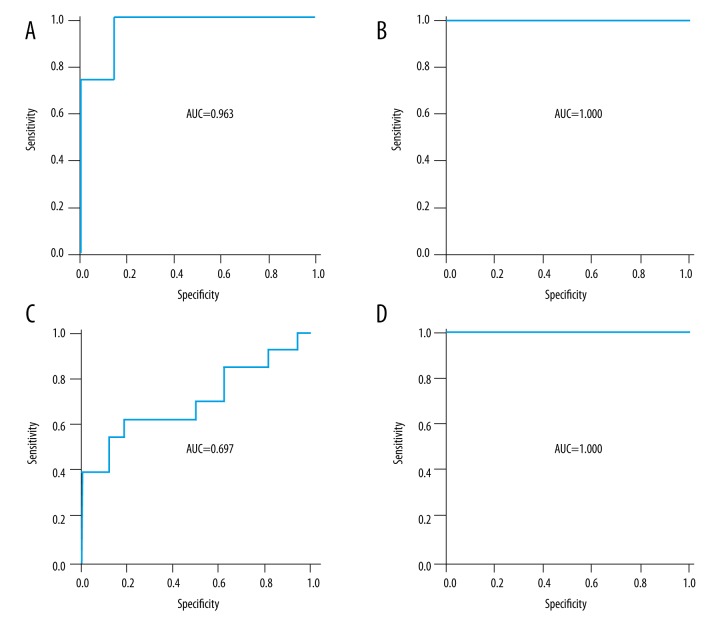
The 5 most recognized genes were regarded as candidate biomarkers, while GSE58795 and GSE42296 were chose as validation sets. No single gene showed good power in predicting infliximab response (AUC <0.85). However, the logistics regression model of these 5 genes showed good accuracy in predicting infliximab response (AUC=0.963 in GSE58795 and 1.000 in GSE42296) (Figure 2A, 2B). At week 2 after the first infliximab infusion, the 5-gene model in peripheral blood samples also showed a good accuracy in predicting the infliximab response at week 14 (AUC=1.000 in GSE42296) (Figure 2D). In the placebo treatment group, the 5-gene model failed to predict the response to placebo (AUC=0.697) (Figure 2C), indirectly indicating the specificity of the 5-gene model in predicting infliximab response.
Figure 2.
Area under receiver operating characteristic (ROC) curve (AUC) of 5-gene model in predicting infliximab response in rheumatoid arthritis patients. The 5-gene model for predicting infliximab response in GSE58795 (A) and GSE42296 (B), and predicting placebo response (C). Five-gene model at the early phrase of infliximab treatment (week 2) to predict long-term response (week 14) (D).
Discussion
Response prediction has significant health and economic benefits, but the prediction based on clinical parameters and disease activity scores could not satisfy the need for accuracy. With the development of high-throughput gene detection technology, gene expression profiling has been used in identifying genes associated with disease diagnosis, prognosis, and treatment. Several studies tested the mRNA expression profiles in peripheral blood samples from RA patients before infliximab treatment, and detected gene panels discriminating between future responders and non-responders. Although synovial biopsy samples could reflect the degree of inflammation at a specific joint and the response to infliximab more directly, it was difficult to obtain such samples, particularly from multiple sites and at different time points. Thus, we focused on the use of peripheral blood samples.
The most common approach to searching for biomarkers is to identify individual genes showing statistically significant differences between responders and non-responders. After systematic analyses, we found that differential genes identified in these studies were not consistent with each other, and different gene sets were reported to distinguish between responders and non-responders. Variability in microarray data arose from many sources. First, most studies were limited in sample size, and the response criteria varied from studies (ACR, EULAR, serum CRP, or DAI). The discrepancies between regions and ethnicities were also inevitable. In the experimental aspects, 8 studies used 8 different kinds of microarray platforms. Different platforms contained different gene-oriented probes, and not all platforms were well-annotated. Sampling, RNA amplification and labeling, and the hybridization conditions might also affect the results.
Variation between these studies made it a huge challenge to integrate the findings directly. Thus, we analyzed the differential genes associated with the subsequent infliximab efficacy according to the criteria of each study. Considering that there was no overlap of differential genes across all 8 studies, the genes recognized by at least half of the studies were chosen as candidate biomarkers. Then, 2 random-prospective-designed datasets were used to test the predictive power of candidate biomarkers. These 2 datasets also used the widely recognized response criteria of ACR and EULAR. Five genes were identified for the validation (FKBP1A, FGF12, ANO1, LRRC31, and AKR1D1). Gene ontology analysis indicated that the 5 genes were significantly associated with the regulation of transmembrane transport. However, no single gene showed good predictive power, but the 5-gene model of logistics regression had an accurate prediction at baseline, as well as at the early phrase of treatment. Currently, most, if not all, therapeutic effect prediction studies focused on the predictive values of single genes rather than multiple genes. Our study indicated an excellent performance of joint prediction by multiple genes, which could also illuminate the low predictive values of differential genes in eligible studies.
This study had some strengths and limitations. First, we believe this is the first systematic study to identify the circulating biomarkers for predicting RA infliximab response through bioinformatics analysis of the microarray data from public databases. Second, a 5-gene model was established and showed good predictive power, and it was also applicable at the early phrase of treatment. A limitation of the study is that the included datasets were heterogeneous. Large-scale prospective studies are needed to corroborate our results.
Conclusions
The models of FKBP1A, FGF12, ANO1, LRRC31, and AKR1D1 in peripheral blood is useful for efficiently predicting the response to infliximab treatment in rheumatoid arthritis patients.
Footnotes
Conflict of interest
The authors have no conflicts of interest.
Source of support: Departmental sources
References
- 1.Siebert S, Lyall DM, Mackay DF, et al. Characteristics of rheumatoid arthritis and its association with major comorbid conditions: Cross-sectional study of 502649 UK Biobank participants. RMD Open. 2016;2:e000267. doi: 10.1136/rmdopen-2016-000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adami G, Orsolini G, Adami S, et al. Effects of TNF inhibitors on parathyroid hormone and Wnt signaling antagonists in rheumatoid arthritis. Calcif Tissue Int. 2016;99:360–64. doi: 10.1007/s00223-016-0161-3. [DOI] [PubMed] [Google Scholar]
- 3.Kievit W, Adang EM, Fransen J, et al. The effectiveness and medication costs of three anti-tumour necrosis factor alpha agents in the treatment of rheumatoid arthritis form prospective clinical practice data. Ann Rheum Dis. 2008;67:1229–34. doi: 10.1136/ard.2007.083675. [DOI] [PubMed] [Google Scholar]
- 4.Miyoshi F, Honne K, Minota S, et al. A novel method predicting clinical response using only background clinical data in RA patients before treatment with infliximab. Mod Rheumatol. 2016;5:1–4. doi: 10.3109/14397595.2016.1168536. [DOI] [PubMed] [Google Scholar]
- 5.Sakthiswary R, Shaharir SS, Mohd Said MS, et al. IgA rheumatoid factor as a serological predictor of poor response to tumour necrosis factorαinhibitors in rheumatoid arthritis. Int J Rheum Dis. 2014;17:872–77. doi: 10.1111/1756-185X.12443. [DOI] [PubMed] [Google Scholar]
- 6.Gavrila BI, Ciofu C, Stoica V. Biomarkers in rheumatoid arthritis, what is new? J Med Life. 2016;9:144–48. [PMC free article] [PubMed] [Google Scholar]
- 7.Likhitrattanapisal S, Tipanee J, Janvilisri T. Meta-analysis of gene expression profiles identifies differential biomarkers for hepatocellular carcinoma and cholangiocarcinoma. Tumour Biol. :2016. doi: 10.1007/s13277-016-5186-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 8.Zou J, Li J, Ji C, et al. Expression profile of plasma microRNAs in nonsyndromic cleft lip and their clinical significance as biomarkers. Biomed Pharmacother. 2016;82:459–66. doi: 10.1016/j.biopha.2016.05.033. [DOI] [PubMed] [Google Scholar]
- 9.Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 10.Aletaha D, Neogi T, Silman AJ, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatoid/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62:2569–81. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 11.Fransen J, van Riel PL. The disease activity score and the EULAR response criteria. Rheum Dis Clin North Am. 2009;35:745–57. doi: 10.1016/j.rdc.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Lequerre T, Gauthier-Jauneau AC, Bansard C, et al. Gene profiling in white blood cells predicts infliximab responsiveness in rheumatoid arthritis. Arthritis Res Ther. 2006;8:R105. doi: 10.1186/ar1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekiguchi N, Kawauchi S, Furuya T, et al. Messenger ribonucleic acid expression profile in peripheral blood cells from RA patients from RA patients following treatment with anti-TNF-alpha monoclonal antibody, infliximab. Rheumatology (Oxford) 2008;47:780–88. doi: 10.1093/rheumatology/ken083. [DOI] [PubMed] [Google Scholar]
- 14.Julia A, Erra A, Palacio C, et al. An eight-gene blood expression profile predicts the response to infliximab in rheumatoid arthritis. PLoS One. 2009;4:e7556. doi: 10.1371/journal.pone.0007556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mesko B, Poliska S, Vancsa A, et al. Peripheral blood derived gene panels predict response to infliximab in rheumatoid arthritis and Crohn’s disease. Genome Med. 2013;5:59. doi: 10.1186/gm463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maclsaac KD, Baumgartner R, Kang J, et al. Pre-treatment whole blood gene expression is associated with 14-week response assessed by dynamic contrast enhanced magnetic resonance imaging in infliximab-treated rheumatoid arthritis patients. PLoS One. 2014;9:e113937. doi: 10.1371/journal.pone.0113937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanino M, Matoba R, Nakamura S, et al. Prediction of efficacy of anti-TNF biologic agent, infliximab, for rheumatoid arthritis patients using a comprehensive transcriptome analysis of white blood cells. Biochem Biophys Res Commun. 2009;387:261–65. doi: 10.1016/j.bbrc.2009.06.149. [DOI] [PubMed] [Google Scholar]
- 18.Toonen EJ, Gilissen C, Franke B, et al. Validation study of existing gene expression signatures for anti-TNF treatment in patients with rheumatoid arthritis. PLoS One. 2012;7:e33199. doi: 10.1371/journal.pone.0033199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura S, Suzuki K, Iijima H, et al. Identification of baseline gene expression signatures predicting therapeutic responses to three biologic agents in rheumatoid arthritis: A retrospective observational study. Arthritis Res Ther. 2016;18:159. doi: 10.1186/s13075-016-1052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Baarsen LG, Wijbrandts CA, Rustenburg F, et al. Regulation of IFN response gene activity during infliximab treatment in rheumatoid arthritis is associated with clinical response to treatment. Arthritis Res Ther. 2010;12:R11. doi: 10.1186/ar2912. [DOI] [PMC free article] [PubMed] [Google Scholar]