Abstract
The biosynthesis of multiple secondary metabolites in the phytopathogenic ascomycete Fusarium fujikuroi is strongly affected by nitrogen availability. Here, we present the first genome-wide transcriptome and proteome analysis that compared the wild type and deletion mutants of the two major nitrogen regulators AreA and AreB. We show that AreB acts not simply as an antagonist of AreA counteracting the expression of AreA target genes as suggested based on the yeast model. Both GATA transcription factors affect a large and diverse set of common as well as specific target genes and proteins, acting as activators and repressors. We demonstrate that AreA and AreB are not only involved in fungal nitrogen metabolism, but also in the control of several complex cellular processes like carbon metabolism, transport and secondary metabolism. We show that both GATA transcription factors can be considered as master regulators of secondary metabolism as they affect the expression of more than half of the 47 putative secondary metabolite clusters identified in the genome of F. fujikuroi. While AreA acts as a positive regulator of many clusters under nitrogen-limiting conditions, AreB is able to activate and repress gene clusters (e.g. bikaverin) under nitrogen limitation and sufficiency. In addition, ChIP analyses revealed that loss of AreA or AreB causes histone modifications at some of the regulated gene clusters.
Introduction
The ascomcyete Fusarium fujikuroi is a notorious plant pathogen, most famous for causing the ‘bakanae’ or ‘foolish seedling’ disease in rice plants [1]. Infected plants display an abnormal elongation of internodes, which is caused by the fungal production of gibberellins (GA), a class of bioactive diterpenoid plant hormones [2–4]. Besides GA, F. fujikuroi produces a broad spectrum of other secondary metabolites (SM), including the red pigments bikaverin (BIK) and fusarubins (FSR) [5,6], as well as the mycotoxins fusarins (FUS), fusaric acid (FSA), fumonisins (FUM), beauvericin (BEA) and apicidin F (APF), the latter being uniquely produced by F. fujikuroi [7–15].
The recently sequenced genome of F. fujikuroi spans 12 chromosomes containing 14,813 predicted gene models and a total of 47 putative SM gene clusters centered around the following biosynthetic key-enzymes: 14 polyketide synthases (PKS), 15 non-ribosomal peptide synthetases (NRPS), 4 PKS/NRPS hybrid enzymes, 2 dimethylallyl tryptophan synthases (DMATS) and 12 terpene cyclases (TC) [8]. Most of these gene clusters (32 out of 47), including the GA cluster, do not contain cluster-specific transcription factors (TFs) suggesting that they are regulated by global TFs depending on the environmental conditions. One crucial environmental cue affecting the expression of many SM clusters in F. fujikuroi is the nitrogen (N) availability to the fungus. In fact, 30 of the F. fujikuroi SM gene clusters were found to be regulated by N [8]. Some of them are only expressed under N-sufficient conditions (e.g. FUS, FUB (fusaric acid biosynthesis), APF), while others are subject to nitrogen metabolite repression (NMR) and are therefore only expressed under N-limiting conditions (e.g. GA, FUM, BIK, and FSR) [4–6,8–11]. N-repression of SM has also been described for other fungi, e.g. the biosynthesis of penicillin in Penicillium chrysogenum, trichothecenes and fusarielin H in Fusarium graminearum, and cephalosporin in Acremonium chrysogenum [16–19]. The GA biosynthetic genes in F. fujikuroi were the first genes shown to be under the control of the major N metabolism regulator AreA [20].
AreA (NIT2 in Neurospora crassa) is a GATA-family TF with a Cys2Cys2 Zinc finger DNA-binding domain preferentially binding to at least two 5’HGATAR DNA sequence motifs within a distance of 30 bp [21–24]. This key regulator mediates de-repression of genes involved in the utilization of alternative N sources in the absence of preferred N sources like glutamine and ammonium [25]. Alternatively, AreA can control expression of genes synergistically with other TFs. The best studied model is the regulation of the nitrate assimilation system in A. nidulans, which relies on the activities of the two TFs AreA and NirA [26–28]. Here, the activity of AreA opens the chromatin structure at the nitrate utilization gene cluster via histone acetylation thereby enabling binding of the nitrate-activated TF NirA to its target promoters, the nitrate assimilatory genes [28–30]. These findings suggest that AreA can bind directly to promotors of target genes and is able to regulate gene expression by recruitment of other TFs and/or post-translational histone modifications.
Besides AreA, a second GATA TF was shown to be involved in N-dependent gene regulation in A. nidulans and P. chrysogenum termed AreB and NreB, respectively [31–35]. In these two fungi, AreB/NreB is generally regarded as the negative counterpart to AreA, acting as a major repressor of AreA-activated N catabolism genes [32,35]. However, the function of AreB appears to be more complex: both AreA and AreB were shown to repress the arginine catabolism genes agaA and otaA under N-repressing and carbon(C)-limiting conditions [36,37]. Recently, we investigated the role of AreB and its interplay with AreA in F. fujikuroi, where we could show that the deletion of AREB resulted in a general growth reduction on various media, but had no effect on the use of alternative N or C sources [31]. Furthermore, expression analysis of selected N-regulated genes indicated a dynamic regulatory interplay of AreA and AreB: expression of the GA cluster genes was completely lost in ΔAREA as well as ΔAREB, suggesting a synergistic activation regulation by both TFs. Finally, we showed for the first time that AreA and AreB physically interact in the nucleus under N-limiting conditions [31].
In view of these unexpected and intriguing results we have examined the common, as well as the different, effects of AreA and AreB on gene expression on a genome-wide level in F. fujikuroi using microarray analysis. Additionally, we compared the abundance of proteins between the wild type (Wt) and ΔAREA and ΔAREB mutants and investigated the potential effect of AREA and AREB deletion on histone H3 lysine 9 (H3K9) acetylation profiles around SM gene clusters by chromatin immunoprecipitation (ChIP) analysis. These comprehensive studies demonstrate that both GATA TFs are major regulators of many metabolic processes which may coordinately activate or repress common target genes, but that both of them have also specific functions. Besides regulating alternative N assimilation pathways, both AreA and AreB are global regulators of secondary metabolism. While AreA mainly acts as a positive regulator of many SM gene clusters under N-limiting conditions, AreB activates and represses certain SM genes under N-limiting as well as N-sufficient conditions. The combined transcriptome, proteome and ChIP analyses give insights into their mode of regulation and provide a basis for further studies of the complex N regulation network in F. fujikuroi and other filamentous fungal species.
Results
To gain a deeper insight into the regulatory effects of AreA and AreB in F. fujikuroi, an integrative analysis of the transcriptome and proteome under N-limiting (6 mM glutamine) and N-sufficient (60 mM glutamine) conditions was performed in biological duplicates. Differences between the Wt and mutant strains under either condition were studied at the transcriptome and proteome levels by microarray and high performance liquid chromatography coupled to tandem mass spectrometry (HPLC-MS/MS), respectively.
Deletion of AREA and AREB affects the transcription of large gene sets
The genome-wide search for AreA- and AreB-dependent genes was performed by use of high quality 126135 K NimbleGen microarrays that were manufactured based on the present genome annotation of F. fujikuroi IMI58289 [8]. Based on the selection criteria 2-fold change in expression at the 95% confidence interval (False Discovery Rate < 0.05), 4,432 of the 14,813 annotated genes were affected by N availability in the Wt, and about 80% of them are affected in an AreA and/or AreB-dependent manner (Fig 1A, S1 Table). Potential AreA/AreB binding sites (double GATA/TATC sequence elements with a distance of <30 bp) [23,38] were found in the promoters of 73% of all 14821 annotated F. fujikuroi genes, while 80% of the genes affected in the ΔAREA and/or ΔAREB mutant contained at least one pair of these motifs (S1 Table).
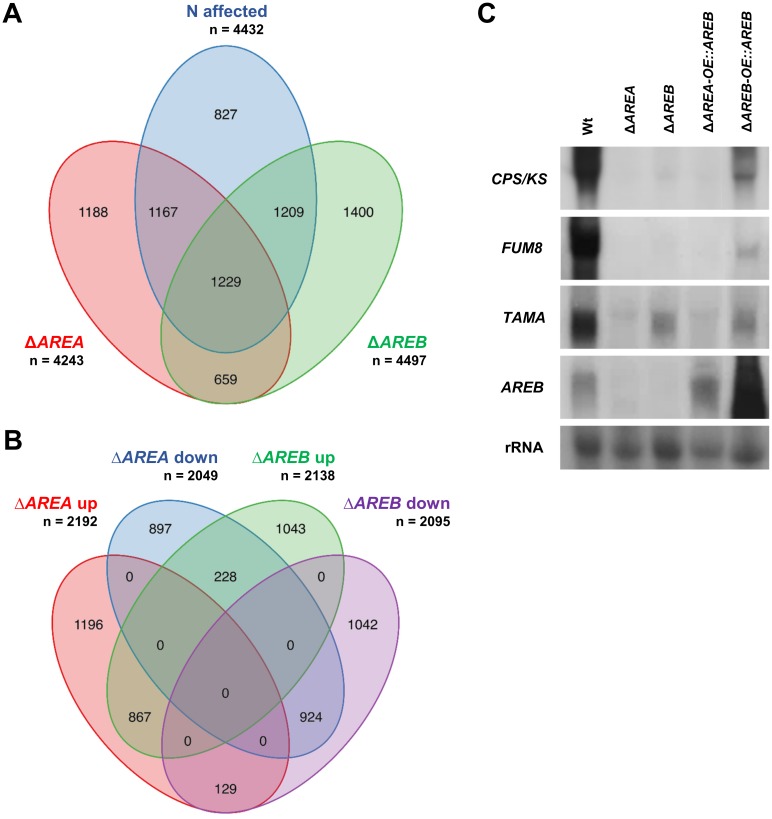
Fig 1. Distribution of genes affected in ΔAREA and ΔAREB, as well as by nitrogen availability.
The F. fujikuroi Wt and the ΔAREA and ΔAREB deletion mutants were cultivated for 3 days in ICI liquid cultures with 6 mM glutamine (nitrogen limitation) or 60 mM glutamine (nitrogen sufficiency) as sole nitrogen source. Data is based on microarray analysis. (A) Differentially regulated genes at 60 mM compared to 6 mM glutamine (N affected), as well as in ΔAREA and ΔAREB compared to Wt. (B) Differentially up-regulated (log2FC > 1) and down-regulated (log2FC < -1) genes at 6 mM glutamine in ΔAREA and ΔAREB compared to Wt. (C) Northern blot analysis of SM cluster genes CPS/KS (GA) and FUM8 (FUM) as well as TF genes TAMA and AREB. The F. fujikuroi Wt, the ΔAREA and ΔAREB deletion mutants and two strains expressing AREB ectopically under control of a strong, constitutive promoter (ΔAREA-OE::AREB) were cultivated at 6 mM glutamine for 3 days.
Under N-limiting conditions, 4,241 genes and 4,233 genes were regulated differentially in the ΔAREA and ΔAREB mutant, respectively, either being up- or down-regulated compared to the Wt (see S1 Table for details). Under N sufficiency, only six genes with currently unknown function exhibited differential regulation in the ΔAREA mutant, indicating that AreA does not play a significant role as a transcriptional regulator under these conditions. In contrast, a large set of 4,432 genes was affected in the ΔAREB mutant in the presence of sufficient N (S1 Table). 2,084 of these genes were affected in the ΔAREB mutant irrespective of N availability, suggesting an additional N-independent regulatory role of AreB.
Importantly, by using stricter selection criteria (> 4-fold change in expression at the 95% confidence interval), the deletion of AREA and AREB still affects the expression of large gene sets (S1f Table).
Analyzing the effects of AreA, AreB and nitrogen on individual gene sets using a two-way ANOVA, we found that nitrogen (P = 0.0031) and the interaction of AreA and nitrogen (P = 0.0149) have a significant effect on the expression of secondary metabolism genes [8]. In contrast, AreB significantly influences secondary metabolism gene expression (P = 0.0113) independent of nitrogen. We also found that nitrogen has a significant effect on secreted proteins (P = 0.0048) and that AreB affects gene expression of transcription factors (P = 0.0005).
In summary, our findings show that AreA and AreB are global regulators of large gene sets and that both may act either as activators or as repressors. The genome-wide transcriptome analysis also demonstrates that the function of AreB is much more complex than being a regulatory counterpart of AreA. Although the transcript level of AREB is significantly down-regulated under N-sufficient conditions (S1 Table) confirming our previous data [31], AreB but not AreA performs regulatory functions also under these conditions.
Commonly affected genes in ΔAREA and ΔAREB: Elucidating AreA and AreB target genes
Previously we have shown that the expression of AREB depends on AreA, while AreB does not affect AREA expression [31], which was also reflected by our microarray analysis (S1 Table). This AreA-dependent transcriptional regulation of AREB makes it hard to distinguish between differentially expressed genes that are affected by AreA (referred to as ‘AreA targets’) and those that are affected by AreB (referred to as ‘AreB targets’). A gene that is affected in ΔAREA as well as in ΔAREB (Fig 1B) may be either a common target of both TFs or an AreB target only, which is indirectly affected in ΔAREA due to the drastically reduced AREB transcript levels in this mutant.
Therefore, we sorted the differentially expressed genes in the ΔAREA and ΔAREB mutants at N-limiting conditions into two major groups: (1) genes that are exclusively affected in one mutant and (2) genes that are affected in both mutants (Table 1, S2 Table). For the first group of genes, the differential expression is clearly the result of the lack of the respective TF, and the affected genes can be considered either as AreA or AreB targets, respectively. For the genes in the second group that are equally down- or up-regulated in both mutants, AreA might act only indirectly via down-regulation of AREB-expression (see Table 1 for details).
Table 1. Overview of all differentially expressed genes in ΔAREA and ΔAREB cultivated with 6 mM glutamine (N limitation).
| gene set | No. of diff. regulateda genes | Targets of? | ||
|---|---|---|---|---|
| general overview | ||||
| total | up | down | ||
| Genes affected in ΔAREA | 4241 | 2192 | 2049 | |
| Genes affected in ΔAREB | 4233 | 2138 | 2095 | |
| (1) affected in one mutant | ||||
| total | up | down | ||
| Genes affected in ΔAREA only | 2093 | 1196 | 897 | AreA |
| Genes affected in ΔAREB only | 2085 | 1043 | 1042 | AreB |
| (2) affected in both mutants | ||||
| total | up/down | down/up | ||
| antagonistic directed regulation in ΔAREA/ΔAREB | 357 | 129 | 228 | AreA, AreB |
| total | up/up | down/down | ||
| equally directed regulation in ΔAREA/ΔAREB | 1791 | 867 | 924 | AreB, AreA? |
a selection criteria: 2-fold change (up or down) in expression at the 95% confidence interval (False Discovery Rate < 0.05).
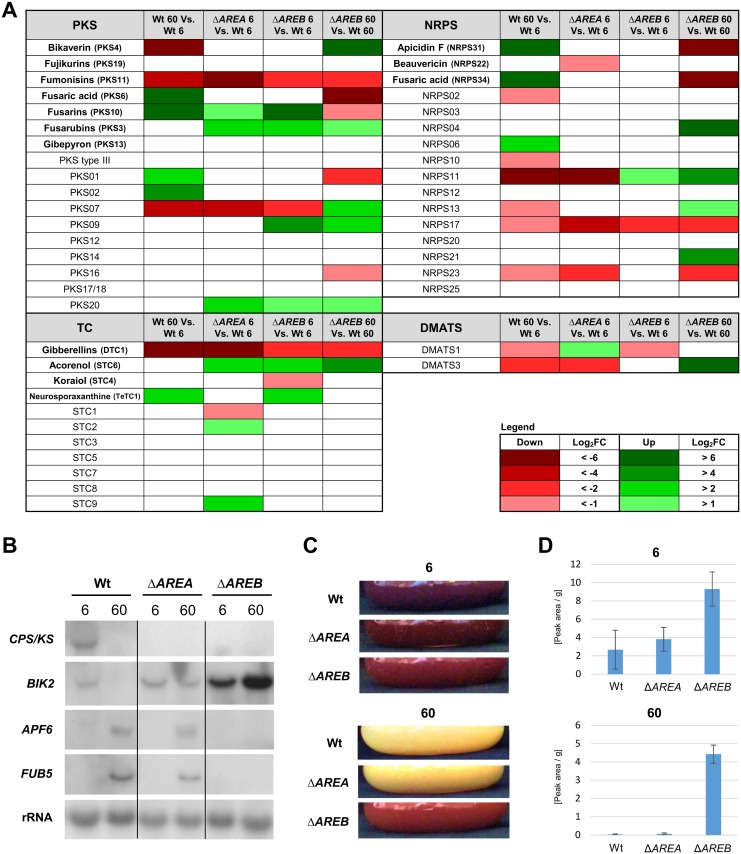
To shed light on the role each of the two TFs plays in regulating the expression of some of these unclear AreA target genes, we constitutively expressed AREB in the ΔAREA background (ΔAREA-OE::AREB), thereby circumventing the AreA-dependent expression of AREB. To verify functionality of the OE::AREB construct we complemented also the ΔAREB mutant with the same vector (ΔAREB-OE::AREB). As an example, we studied the expression of three genes which were strongly down-regulated in both deletion mutants: the key genes for GA (CPS/KS) and FUM (FUM8) biosynthesis, as well as the TF-encoding gene FFUJ_04390, a homologue of the A. nidulans nitrogen regulator gene TAMA [39,40] (Fig 1C). As expected, expression of the SM cluster genes was lost in ΔAREA as well as ΔAREB and restored in ΔAREB-OE::AREB. A similar expression pattern was observed for TAMA, which was significantly down-regulated but not completely lost in ΔAREB. Expression of none of these genes was restored in ΔAREA-OE::AREB indicating the essential role of both TFs for these genes. By this approach we provide for the first time experimental proof, that both AreA and AreB are indeed essential to synergistically activate expression of the GA and FUM gene clusters, going in line with our previous results where the expression of both SM gene clusters was strongly down-regulated in ΔAREA and ΔAREB mutants [13,20].
In conclusion, equally large sets of common but also distinct genes were affected in ΔAREA and ΔAREB. While TF-specific genes, and genes which were regulated by the two TFs in opposite directions, can clearly be assigned to the activity of one of the two TFs (AreA and/or AreB target genes), the impact of AreA on the equally regulated genes (Table 1) remains elusive. To dissect AreA function from that of AreB, the expression of a given gene needs to be studied in the ΔAREA mutant with constitutive, AreA-independent, expression of AREB as shown for the GA and FUM cluster genes and TAMA in this work.
Impact of AREA and AREB deletion on the proteome
Apart from the transcriptional regulation, we examined the impact of AreA and AreB on protein abundance under both N conditions in biological duplicates, employing quantitative MS analysis and phosphoproteomics. Proteins were defined as significantly differentially expressed when the abundance of corresponding peptides was increased or decreased by log2FC 0.5, or when peptides were detected only in the Wt (down-regulated) or the mutant strains (up-regulated), respectively. The absence of peptides can be evaluated with high confidence due to advanced retention time alignment and enhancement as shown previously [41]. Overall, the strict selection criteria allow protein regulations to be identified with high biological significance.
In total 19,658 distinct peptides (posterior error probability (PEP) < = 1%) were identified, which correspond to 1,964 proteins encoded by the genome of F. fujikuroi. This set was subsequently refined to 1,010 proteins, for which at least two unique peptides were identified (S3 Table). Under N-limiting conditions, 446 proteins were differentially regulated in the ΔAREA mutant. Unexpectedly, a set of 138 proteins was affected in ΔAREA under N sufficiency, in contrast to the minor impact of AreA on gene expression under these conditions. In the ΔAREB mutant, 386 proteins were regulated differentially under N-limiting and 446 proteins under N-sufficient conditions (S1 Fig, Table 2, S3 Table). In a similar manner to the microarray analysis, we differentiated between proteins which are specifically up- or down-regulated only in one mutant (1), and others that are commonly affected in both (2) (Table 2).
Table 2. Overview of all differentially expressed proteins in ΔAREA and ΔAREB cultivated with 6 mM glutamine and 60 mM glutamine.
| protein set | No. of diff. regulateda proteins | Targets of? | |
|---|---|---|---|
| general overview | |||
| total 6 | total 60 | ||
| Proteins affected in ΔAREA | 446 | 138 | |
| Proteins affected in ΔAREB | 386 | 446 | |
| (1) exclusively affected in one mutant | |||
| total 6 | total 60 | ||
| Proteins affected in ΔAREA only | 175 | 43 | AreA |
| Proteins affected in ΔAREB only | 115 | 351 | AreB |
| (2) commonly affected in both mutants | |||
| total 6 | total 60 | ||
| antagonistic regulation in ΔAREA and ΔAREB | 38 | 16 | AreA, AreB |
| total 6 | total 60 | ||
| equally directed regulation in ΔAREA and ΔAREB | 233 | 79 | AreB, AreA? |
a selection criteria: 1-fold change (up or down) in protein abundance. Nitrogen-limiting and nitrogen-sufficient conditions are indicated by 6 (6 mM glutamine) and 60 (60 mM glutamine), respectively
Comparison between the proteome and transcriptome datasets revealed that a large set of proteins affected by N-availability or in the ΔAREA and/or ΔAREB mutants were not significantly regulated (log2FC < -1; > 1) on transcriptional level (S3 Table). In particular, more than 80% of the N-regulated proteins (Wt under N limitation compared with Wt under N sufficiency) were exclusively regulated at the protein level (S3 Table), suggesting the involvement of post-transcriptional regulations.
Taken together, the abundance of more than half of the quantified proteins was affected in the ΔAREA and/or the ΔAREB mutants, stressing the strong impact of both TFs on the proteome of F. fujikuroi. We also observed AreA- and AreB-mediated protein-regulations that were not affected at the transcriptional level, indicating the involvement of post-translational regulations.
AreA and AreB regulate a large set of putative downstream regulators
To examine the hierarchical regulation networks, we were especially interested in downstream acting AreA and/or AreB-dependent TFs. From about 980 TFs identified in the genome of F. fujikuroi [8], 240 were differentially regulated in the ΔAREA mutant under N-limiting conditions (S4a Table). In the ΔAREB mutant, 311 and 296 TFs were affected under N-limiting and N-sufficient conditions, respectively, representing about 30% of the TFs in the genome under each condition (S4a Table). Examples of notable and strongly regulated TFs in ΔAREA and ΔAREB are shown in Table 3. Among them are TF-encoding genes which are putatively involved in N metabolism (e.g. TamA), C metabolism (e.g. homologs of Acu-15 involved in acetate metabolism [42]) as well as pathway-specific regulators belonging to certain SM-clusters (e.g. apicidin, beauvericin, bikaverin, DMATS1, fusaric acid, PKS1).
Table 3. Selection of differentially regulated transcription factor genes in ΔAREA and ΔAREB cultivated with 6 mM glutamine and 60 mM glutamine.
| GENOME ID | CLASS | GATA PAIRS | ANNOTATION | FUNCTION/ SM-CLUSTER | WT 60 VS WT 6a | ΔAREA 6 VS WT 6a | ΔAREB 6 VS WT 6a | ΔAREB 60 VS WT 60a |
|---|---|---|---|---|---|---|---|---|
| FFUJ_00012 | bZIP | 2 | Apf2—apicidin cluster transcription factor | apicidin cluster | 9.48 | N/A | 2.07 | -6.04 |
| FFUJ_00054 | Zn2Cys6 | 3 | related to UPC2—regulatory protein involved in control of sterol uptake | sterol metabolism | N/A | 4,82 | 2,37 | N/A |
| FFUJ_02117 | Zn2Cys6 | 4 | Fub 10 –fusaric acid cluster transcription factor | fusaric acid cluster | 9.99 | 1.67 | 2.30 | -6.11 |
| FFUJ_02119 | Zn2Cys6 | 3 | Fub 12 –fusaric acid cluster transcription factor | fusaric acid cluster | 3.26 | N/A | N/A | -2.22 |
| FFUJ_02223 | unknown | 5 | uncharacterized protein | PKS1 cluster | 5.39 | N/A | N/A | -3.60 |
| FFUJ_02504 | bZIP | 2 | related to SRP40—suppressor of mutant AC40 of RNA polymerase I and III | transcription/ splicing | N/A | 4,23 | 3,15 | 3,42 |
| FFUJ_02801 | Cys2His2 | 1 | related to krueppel protein Klp1 | asexual development | N/A | -1.28 | -1.78 | -1.39 |
| FFUJ_03660 | Zn2Cys6 | 10 | related to pathway-specific regulatory protein nit-4 | nitrogen metabolism | -1,21 | -2,79 | -1,45 | -3,62 |
| FFUJ_04390 | Zn2Cys6 | 10 | TamA—transcriptional activator for allantoin and GABA catabolic genes | nitrogen metabolism | -2,36 | -3,42 | -3,29 | -4,41 |
| FFUJ_06522 | Zn2Cys6 | 4 | ARG81—Transcription factor involved in arginine metabolism | arginine metabolism | -4,55 | -3,65 | -1,19 | N/A |
| FFUJ_06723 | Zn2Cys6 | 0 | related to transcriptional activator Mut3p | carbon metabolism | 4,43 | 1,47 | 1,79 | -4,30 |
| FFUJ_08895 | Zn2Cys6 | 2 | related to transcriptional activator acu-15 | carbon metabolism | N/A | -3,21 | N/A | -1,40 |
| FFUJ_09177 | Zn2Cys6 | 0 | uncharacterized transcription factor | DMATS1 cluster | -1.10 | N/A | -1.40 | N/A |
| FFUJ_09190 | Zn2Cys6 | 4 | related to STB5—SIN3 binding protein | carbon metabolism | -8,06 | -8,19 | 1,09 | N/A |
| FFUJ_09298 | Zn2Cys6 | 19 | Bea4—beauvericin cluster transcription factor | beauvericin cluster | -3.42 | -1.75 | N/A | N/A |
| FFUJ_11293 | Cys2His2 | 0 | related to TRI15—putative transcription factor | secondary metabolism | N/A | 4,56 | N/A | 3,47 |
| FFUJ_12023 | Zn2Cys6 | 15 | related to nitrate assimilation regulatory protein nirA | nitrogen metabolism | N/A | N/A | 2,99 | -6,71 |
| FFUJ_12033 | Zn2Cys6 | 4 | related to transcription activator protein acu-15 | carbon metabolism | N/A | N/A | -1,54 | -3,49 |
| FFUJ_12043 | Zn2Cys6 | 1 | related to transcriptional activator Mut3p | carbon metabolism | -1,55 | N/A | -3,33 | -3,39 |
| FFUJ_12646 | Zn2Cys6 | 3 | related to transcriptional activator Mut3p | carbon metabolism | -1,67 | -3,03 | -3,04 | -2,64 |
| FFUJ_12938 | Zn2Cys6 | 8 | related to transcription activator protein acu-15 | carbon metabolism | -3,01 | -3,95 | N/A | -2,03 |
| FFUJ_12947 | Zn2Cys6 | 6 | related to C6 zink-finger protein PRO1A | sexual development | N/A | -2,48 | -2,11 | -4,25 |
| FFUJ_13618 | Zn2Cys6 | 0 | related to thiamine repressible genes regulatory protein thi1 | thiamine synthesis | 1,45 | 1,36 | 3,34 | 1,60 |
| FFUJ_13963 | Zn2Cys6 | 3 | Bik5—bikaverin cluster transcription factor | bikaverin cluster | -5,95 | -3,22 | 1,55 | 1,60 |
| FFUJ_14054 | Zn2Cys6 | 2 | related to transcription activator protein acu-15 | carbon metabolism | N/A | N/A | N/A | -4,69 |
| FFUJ_14818 | Zn2Cys6 | 4 | related to ARG81—transcription factor involved in arginine metabolism | arginine metabolism | -3,66 | -3,39 | -4,71 | -2,69 |
a RNA foldchange (FC) between two experimental conditions or strains in log2 scale (False Discovery Rate < 0.05). N/A indicates no significant foldchange (< 1, > -1) of gene expression under the respective condition. Nitrogen-limiting and nitrogen-sufficient conditions are indicated by 6 (6 mM glutamine) and 60 (60 mM glutamine), respectively.
Furthermore, we studied the impact of both GATA TFs on the expression of genes encoding potential histone-modifying enzymes (HMEs), such as histone methyltransferases and demethylases, as well as histone acetyltransferases (HAT) and deacetylases. Of 144 genes putatively encoding HME [8], about 35 and 20 were up- or down-regulated in the ΔAREA and ΔAREB mutant, respectively, under N-limiting conditions (S4b Table). Under N-sufficient conditions, AreB regulates 29 HME-encoding genes (S4b Table). Among the affected genes are FFUJ_13544, which is a homolog of SPT10, a yeast HAT [43] and the gene FFUJ_08441, which is a homolog to the major DNA methyltransferase Dim-2 of N. crassa [44].
The transcriptomics and proteomics data also indicated a regulatory function of both GATA TFs on putative protein kinases and phosphatases both on transcript and protein level (S1 and S3 Tables). We therefore searched for proteins that are phosphorylated in an AreA- and/or AreB-dependent manner by a phosphoproteomics approach. Among the 1,010 identified proteins, 277 were found to contain phosphorylated amino acids (S3 Table). However, we did not observe any differentially phosphorylated proteins in the ΔAREA and/or ΔAREB mutants compared to the Wt, indicating that AreA and AreB have no significant impact on the phosphorylation of the identified proteins.
These results suggest that both GATA TFs are on top of regulatory circuits, as they control the expression of a large set of genes encoding TFs and putative HMEs.
AreA and AreB affect genes and proteins involved in several cellular processes
To gain a deeper insight into the most prominent functions of genes affected in ΔAREA and ΔAREB, we performed a functional enrichment analysis using the MIPS ‘FunCat’ tool [45]. This analysis indicated an over-representation of genes likely involved in amino acid metabolism and disease, cellular transport, virulence and defense, and secondary metabolism (S5 Table). In contrast to AreA, AreB regulates many genes also at N sufficiency. This includes a significant number of genes involved in secondary metabolism, but most of the AreB target genes at high N are not yet characterized (S5 Table).
Similar to the transcriptome analysis, the majority of the proteins affected by the loss of AreA and/or AreB belong to the functional categories of secondary metabolism, but also amino acid and carbohydrate metabolism, including enzymes of the TCA and glyoxylate cycle (e.g. isocitrate-, malate-, and succinate dehydrogenases, isocitrate lyase) and the main enzymes of the glutamate/glutamine cycling (S2 Fig, S3 Table).
AreA and AreB are major regulators of secondary metabolism
The functional enrichment analysis revealed a major impact of both AreA and AreB on secondary metabolism in F. fujikuroi. Therefore, we studied the differential expression of the 47 predicted gene clusters under N-limiting as well as N-sufficient conditions in more detail (Fig 2A). The depicted expression of PKS-, TC-, NRPS- or DMATS-encoding genes is representative for the whole gene clusters. In the Wt, 12 SM gene clusters (BIK, FUM, PKS07, GA, NRPS02, NRPS10, NRPS11, NRPS13, NRPS17, NRPS23, DMATS1, DMATS3), were down-regulated, while the expression of eight SM gene clusters (FUB, FUS, PKS01, PKS02, neurosporaxanthine, APF, NRPS06) was elevated under N sufficiency. Of the 12 N-repressed SM clusters, seven were down-regulated in ΔAREA under inducing N-limiting conditions including the GA and FUM gene clusters. In addition, several cryptic gene clusters with yet unknown function are also significantly down-regulated in ΔAREA, e.g. PKS07, NRPS11, NRPS17, NRPS23 and DMATS3 (Fig 2A).
Fig 2. Influence of AreA and AreB on secondary metabolite cluster gene expression.
The F. fujikuroi Wt and the ΔAREA and ΔAREB deletion mutants were cultivated for 3 (A, B, C) or 7 (D) days in ICI liquid cultures with 6 mM glutamine (6) or 60 mM glutamine (60) as sole nitrogen source. (A) Differentially regulated secondary metabolite gene clusters, represented by PKS-, NRPS-, TC- and DMATS-encoding genes. Data are based on microarray analysis. (B) Northern blot analysis of secondary metabolite cluster gene expression: CPS/KS (copalyl diphosphate/kaurene-synthase, GA-cluster), BIK2 (monooxygenase, BIK-cluster), APF6 (o-methyltransferase, APF-cluster), FUB5 (homoserine o-acyltransferase, FUB-cluster). (C) Variation in pigmentation of the strains in ICI liquid culture. (D) Variation in bikaverin production. Culture supernatant was analyzed by HPLC-DAD. The peaks of bikaverin and norbikaverin were integrated at wavelength of 510 nm and depicted in relation to the total cell dry mass of the cultures.
Compared to AreA, the impact of AreB on SM-regulation appears to be more diverse. Some of the N-repressed gene clusters, whose expression was significantly down-regulated in ΔAREA, e.g. GA, FUM, PKS07 and NRPS17, were also down-regulated in ΔAREB. For the GA and the FUM cluster the essential function of both GATA TFs regarding transcriptional activation has been demonstrated by constitutive expression of AREB in the ΔAREA background (Fig 1C), while the involvement of AreA in regulating PKS07 and NRPS17 still needs to be determined.
Interestingly, AreB appears to be essential also for the activation of the N-induced APF, PKS1 and FUB (PKS6; NRPS34) gene clusters, which were all down-regulated in ΔAREB (Fig 2A). In addition, AreB also acts as a transcriptional repressor of certain SM clusters. The PKS09, PKS20, α-acorenol and the neurosporaxanthine clusters, which are induced or unaffected by N in the Wt, are up-regulated under both conditions in ΔAREB. Furthermore, the N-repressed BIK, NRPS11 and DMATS3 clusters, were also significantly up-regulated in ΔAREB at N sufficiency, indicating a de-regulation of these SM under normally repressing conditions (Fig 2A).
To verify the microarray data, northern blot analyses under identical conditions were done using one of the co-regulated genes of the GA (CPS/KS), BIK (BIK2), APF (APF6) and FUB (FUB5) clusters as probes. The expression pattern of the tested clusters fully confirmed our findings from the microarray analysis: we recorded a complete loss of GA expression in both mutants, of APF and FUB gene expression in ΔAREB (Fig 2B), and an elevated expression of the BIK genes in the ΔAREB mutant under N sufficiency.
The most significant role of AreB as a repressor was observed for the bikaverin biosynthesis. Concomitant with elevated BIK transcription, the mycelium of the ΔAREB mutant was deeply red colored in liquid cultures under both N conditions (Fig 2C), indicating a de-regulation of BIK production under normally repressing N-sufficient conditions. HPLC-DAD analysis confirmed the expression data and revealed significantly elevated amounts of BIK under both N limitation and sufficiency in the ΔAREB mutant, while no significant differences have been observed between the Wt and the ΔAREA mutant (Fig 2D).
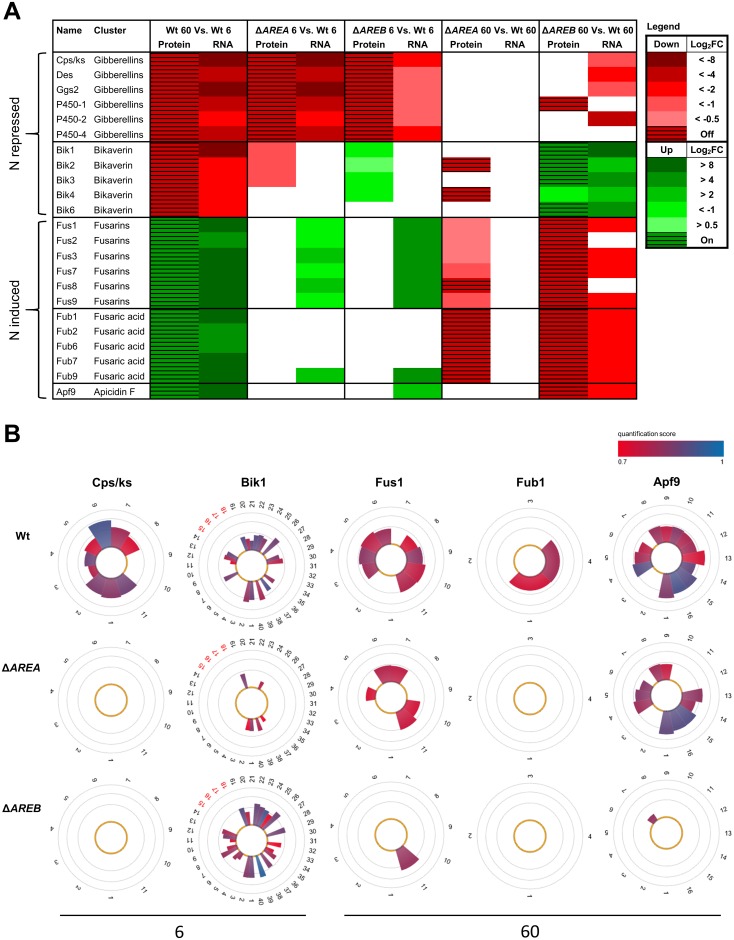
The observed different types of regulation of the GA, BIK, FUS, FUB and APF cluster genes in ΔAREA and ΔAREB correspond very well with the regulation of the respective proteins (Fig 3). These SM cluster genes are among the strongest regulated genes in both deletion mutants. High expression levels were shown to be a pre-condition for a good correlation between transcript and proteome data [46,47].
Fig 3. Comparison of AreA- and AreB-mediated regulation on secondary metabolite proteins and genes.
The F. fujikuroi Wt and the ΔAREA and ΔAREB deletion mutants were cultivated for 3 days in ICI liquid cultures with 6 mM glutamine (6) or 60 mM glutamine (60) as sole nitrogen source. (A) Differentially regulated secondary metabolite proteins (Protein) and genes (RNA). Data is based on proteome and microarray analysis, respectively. (B) MS-based label free quantitation (pyQms) of peptides (numbers) detected during proteome analysis, corresponding to Cps/ks, Bik1, Fus1, Fub1 and Apf9 at 6 mM or 60 mM glutamine, respectively. A pie slice is used to represent each peptide; red numbering indicates phosphopeptides. The area of a pie slice correlates with the amount of the corresponding peptide, colors represent the quality of the pyQms quantification event. Results stemming from non-enriched samples of two independent experiments are depicted in log10 scale.
In conclusion, the transcriptomics and proteomics data demonstrate that AreA and AreB act as master regulators of secondary metabolism in F. fujikuroi. Both GATA TFs may function as transcriptional regulators of common, but also distinct, target SM clusters. While AreA acts exclusively as a strong positive regulator of N-repressed gene clusters (e.g. GA, FUM, PKS07 and NRPS17 genes) under N limitation, AreB can act as an activator (e.g. of APF, PKS1 and FUB genes) and repressor (e.g. of BIK, PKS09, NRPS04 and NRPS11 cluster genes) under both N conditions.
Histone acetylation at some SM gene clusters is affected in ΔAREA and ΔAREB
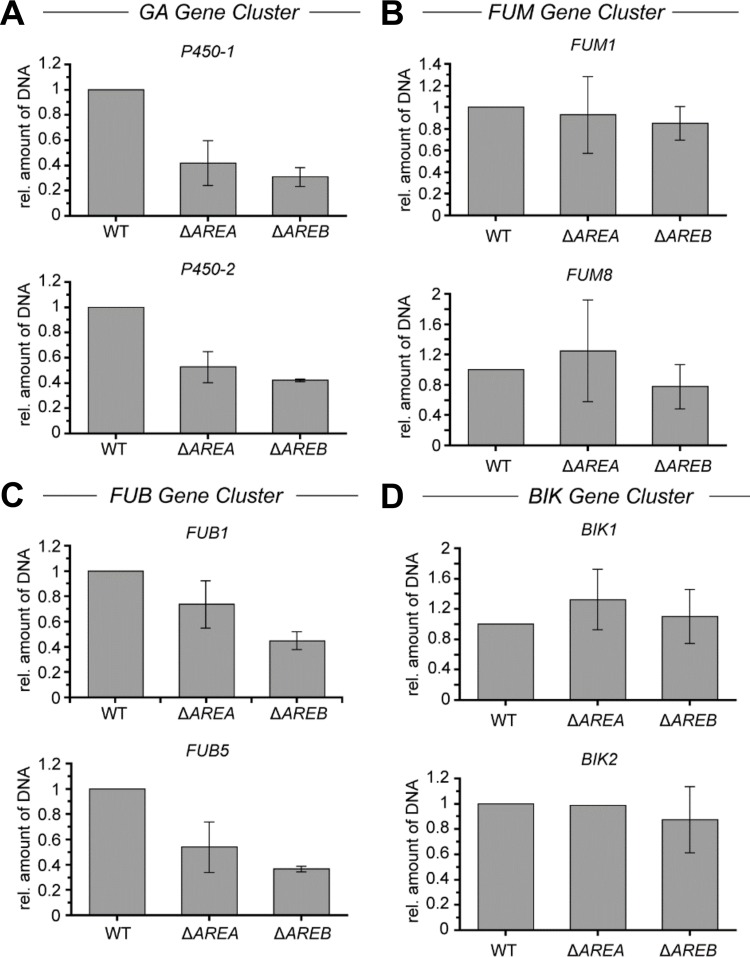
Previously we have shown that the expression of several SM gene clusters in F. fujikuroi correlates with high levels of H3K9ac [6,8,9]. Histone acetylation is mediated by HAT complexes that are generally targeted to specific promoters through their physical interaction with sequence-specific TFs [48]. To study the possible role of AreA and AreB in the recruitment of HAT complexes in F. fujikuroi, we performed chromatin immunoprecipitation (ChIP) coupled to quantitative PCR using an anti-H3K9ac-specific antibody and measured the amount of precipitated DNA at four SM gene clusters that showed differential gene regulation in ΔAREA and/or ΔAREB. We chose the GA and FUM clusters (positively regulated by both TFs), FUB (positively regulated by AreB and only slightly affected in ΔAREA) and BIK (negatively regulated by AreB).
As expected, the GA cluster genes P450-1 and P450-2 encoding P450 monooxygenases [49] showed significantly reduced H3K9ac levels upon deletion of AREA and AREB, suggesting that at least AreB is involved in HAT-mediated histone acetylation at the GA gene cluster (Fig 4A). Contrary to this finding, no significant differences in H3K9ac were observed at the FUM cluster genes FUM1 and FUM8 encoding the PKS [50] and an aminotransferase [51], respectively, albeit showing a similar reduced gene expression in ΔAREA and ΔAREB as the GA cluster genes (Fig 4B).
Fig 4. H3K9 acetylation is affected in ΔAREA and ΔAREB at the GA and FUB gene cluster.
The F. fujikuroi Wt and the ΔAREA and ΔAREB deletion mutants were cultivated for 3 days in ICI liquid cultures with 6 mM (A, B, D) or 60 mM (C) glutamine as sole nitrogen source. The mycelium was subsequently cross-linked and used for chromatin immunoprecipitation (ChIP) experiments using an anti-H3K9ac antibody (AM39137). The precipitated amount of DNA was quantified at (A) GA (P450-1 and P450-2), (B) FUM (FUM1 and FUM8), (C) FA (FUB1 and FUB5) and (D) BIK (BIK1 and BIK2) cluster genes by qPCR. In each case, the amount of DNA in the Wt was arbitrarily set to 1. Mean values and standard deviations are shown. Experiments were done in technical and biological replicates.
In a manner similar to GA cluster genes, H3K9ac levels were significantly reduced in the ΔAREB mutant also at the FUB genes FUB1 and FUB5 encoding the PKS and a putative homoserine-O-acyltransferase, respectively, in agreement with a significant decrease in FSA production [11]. Deletion of AREA led to only slightly reduced FSA biosynthesis and only slightly reduced H3K9ac levels at FUB1 and FUB5 (Fig 4C). H3K9ac levels remained almost unchanged at BIK1 and BIK5 upon deletion of AREB and AREA, under inducing (Fig 4D) as well as under repressing conditions (data not shown), albeit significantly enhanced BIK production in the AREB-deficient strain under both conditions, suggesting that additional trans-acting factors are involved in de-repression of BIK upon deletion of AREB.
Taken together, deletion of AREA and AREB results in reduced H3K9ac levels at GA and FUB cluster genes, which is in line with an abolished and reduced biosynthesis of GA and FUB, respectively. On the contrary, no correlation was observed between gene expression and H3K9ac levels in case of BIK and FUM clusters upon deletion of either AREA or AREB.
Discussion
In this work, we studied the genome-wide regulatory impact of the two major N regulators, the GATA TFs AreA and AreB, in F. fujikuroi by quantitative transcriptomics and proteomics. By combining these techniques, we gained a deeper insight into the regulatory interplay of both TFs, their involvement in common but also different biological processes and their role in histone acetylation, which will be summarized and discussed in the following paragraphs.
AreA and AreB are crucial regulators of nitrogen-dependent genes
AreA is considered as the major regulator of a large number of genes involved in uptake and metabolism of non-favored N sources in fungi [21,52,53]. Therefore, it is not surprising that AreA affects many genes and proteins that are involved in N metabolism and transport, including many putative N permeases, such as amino acid/peptide, ammonium, allantoin and GABA transporters. Our results show that most of these genes are activated by both GATA TFs under N-limiting conditions in F. fujikuroi. Much less is known about the function of the second N-responsive GATA TF, AreB. Recently, we have shown that AreB in F. fujikuroi is involved in regulating several processes of fungal life besides nitrogen metabolism, e.g. transport and secondary metabolism [31].
In this study, we provide the first comprehensive insight into the central role both TFs play in regulating metabolism using a genome-wide approach. About 80% of all N-regulated genes were affected by one or both TFs (Fig 1A). Interestingly, more than 4,000 genes and a large set of proteins were found to be up- or down-regulated by AreB at high N. In contrast to the gene expression data, we found that there is also a substantial number of AreA target proteins under these conditions (S3 Table, S1 and S2 Figs). This is an unexpected result because the expression of both GATA TF-encoding genes was shown to be repressed under N-sufficient conditions [31]. The most plausible explanation for this discrepancy might be that small quantities of AreA and AreB are probably still present under high N, which are sufficient to mediate the observed regulations. Furthermore, AreB regulates different sets of genes and proteins under N-limiting and N-sufficient conditions, and there are even examples of targets that are either activated or repressed by AreB in relation to the N availability. These different regulatory functions of AreB are most likely due to varying interaction partners and/or the three different transcript (AREB-a, AREB-b, AREB-c) and protein sizes which are formed depending on the N availability to the fungus [31]. While the shortest transcript (AREB-c) is the predominant form under N-limiting conditions, all three transcripts are present in equal proportions under N sufficiency. Furthermore, only the product of the largest transcript (AreB-a) has been detected inside the nucleus under N-sufficient conditions [31], indicating that the different regulatory functions of AreB at changing N conditions are probably be mediated by the products of the three different transcripts.
Surprisingly, AreA and AreB also regulate a large set of genes and proteins that are not affected by N availability, indicating that both GATA transcription factors play additional roles as regulators of N-independent processes. These data provide novel insights into additional roles of AreA besides being the major regulator of nitrogen metabolism and a master regulator of secondary metabolism in F. fujikuroi. Pleiotropic effects have also been described for AreA and AreB homologs in other filamentous fungi. In A. nidulans, AreB is involved in vegetative growth and asexual development [35] and plays a role in PacC-mediated pH regulation and virulence in Colletotrichum gloeosporioides [54]. In Magnaporthe oryzae, the GATA-type transcription factor Asd4 with high homology to AreB in F. fujikuroi is essential for sporulation, optimal growth on complete media and appressorium formation besides regulating genes involved in nitrogen assimilation [55]. AreA was shown to be essential for full virulence of the human pathogen Aspergillus fumigatus [56].
Mode of action: Novel insights into the AreA-/AreB-dependent regulation
In A. nidulans and P. chrysogenum it was shown that AreA/NreA and AreB/NreB, can act as regulatory counterparts of shared target genes [32,35], which mimics the interplay of the activating AreA-orthologs Gln3p/Nil1p and the repressing AreB-orthologs Dal80p/Nil2p in yeast [57,58]. However, later it was exemplarily demonstrated in A. nidulans that both TFs act as synergistic repressors of arginine biosynthesis genes [36,37], and that AreB can be involved in the regulation of AreA-independent processes [35]. In this work, we have demonstrated for the first time on a genome-wide scale that the role of AreB is much more complex than simply being an antagonist of AreA: (1) AreB regulates a large set of specific target genes not affected by deletion of AREA, (2) AreB can act as positive as well as negative regulator, (3) the majority of genes and proteins affected in both mutants are regulated in the same way (either positively or negatively), (4) AreB, but not AreA, regulates a large set of genes also under N-sufficient conditions, (5) AreA and AreB have an impact on many genes whose expression is not affected by N availability.
For the genes and proteins that are activated or repressed by both TFs, it is hard to distinguish if alterations on transcript and protein levels are caused by the loss of AreA and/or AreB, or if they are indirectly affected by the down-regulation of AREB in the ΔAREA background. The expression of these potential targets should be further elucidated with the help of a constitutively expressed AREB in the ΔAREA deletion background, as we did for the GA and FUM cluster genes. These genes are among the most strongly down-regulated genes in ΔAREA and ΔAREB. Here we provide, for the first time, evidence that their expression clearly depends on the simultaneous presence of both GATA TFs, and that the strong impact of AreA is not due to the down-regulation of AREB in the ΔAREA mutant (Fig 1C). Recently, we have shown that AreA and AreB physically interact in the nucleus under N-limiting conditions. Such physical interaction is one possible explanation for the synergistic regulation of shared target genes and proteins [31]. We are currently investigating potential interaction partners of both GATA TFs by co-immunoprecipitation to better understand their mode of action.
AreA- and AreB-mediated regulation of secondary metabolism
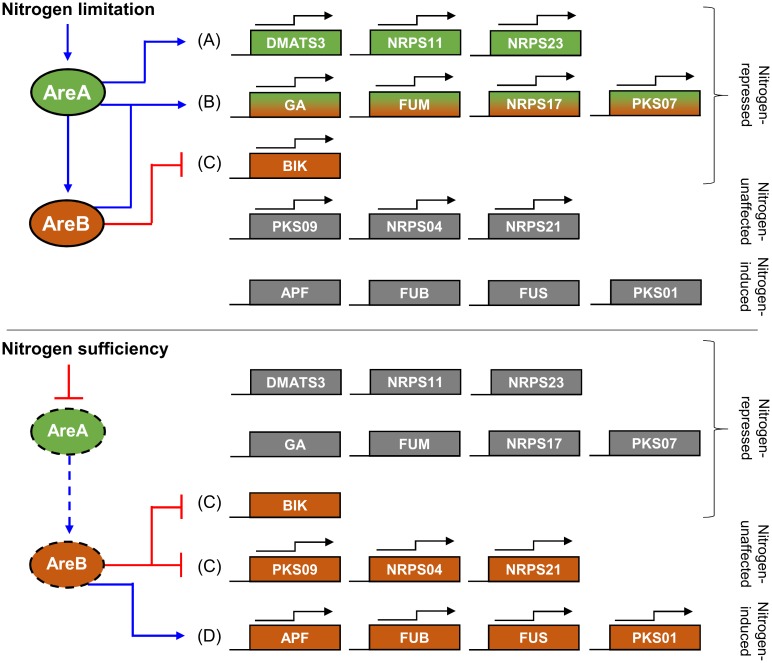
Previously we have shown that AreA and/or AreB have an impact on the regulation of certain aspects of SM in F. fujikuroi, including GA, FSA, APF and FUM [10,11,13,20,31]. Here we show that AreA and/or AreB affect more than half of the genome’s SM biosynthetic pathways, which is summarized in Fig 5. While AreA mainly acts as a positive regulator of N-repressed clusters under N-limiting conditions (Fig 5A), the regulatory function of AreB appears to be more diverse because it regulates multiple genes independently of nitrogen availability as confirmed by two-way ANOVA analysis. AreB was shown to be a synergistic activator that is essential for expression of certain AreA target clusters at N-limiting conditions (Fig 5B). This was demonstrated for the GA and FUM clusters (Fig 1C) and is most likely true for the regulation of the NRPS17 cluster. However, AreB also regulates AreA-independent clusters and can act as a repressor (Fig 5C) as well as an activator (Fig 5D) under both N conditions. One of the most intriguing results is the high expression of BIK genes and proteins under otherwise repressing N-sufficient conditions in the ΔAREB mutant. Accordingly, AreB acts as a strong repressor of BIK biosynthesis, both under inducing low and repressing high N conditions. Besides BIK, AreB also plays a role as a repressor of the silent PKS09, NRPS04 and NRPS21 clusters (Fig 5C). Therefore, the ΔAREB deletion mutant could be a powerful tool to activate those gene clusters and to identify their products.
Fig 5. Overview of the main AreA- and AreB-mediated regulations of secondary metabolite cluster expression at nitrogen-limiting (6 mM glutamine) and nitrogen-sufficient (60 mM glutamine) conditions.
Blue arrows indicate positive regulations, red lines indicate repressing effects. Black arrows indicate expression of clusters in the Wt under the respective nitrogen condition. (A) Clusters activated by AreA only; (B) clusters synergistically activated by AreA and AreB; (C) clusters repressed by AreB only; (D) clusters activated by AreB only.
AreA and AreB could facilitate their regulatory impact on the SM-clusters by one of the following ways or combinations of them: (1) binding to the promoters of the single cluster genes, (2) binding to the promoter of a cluster-specific TF (if present) that activates expression of the remaining cluster genes and (3) activating or recruiting another global regulator that binds to the promoter of the cluster genes or mediates promoter accessibility by chromatin rearrangement. Previous work has indicated that AreB affects transcription of the APF [10] and FUS [11,14] clusters most likely via regulating the cluster-specific TF, which was confirmed by our microarray analysis (Table 3). Furthermore, we showed that AreB represses the TF-encoding gene BIK5, which indicates a similar regulation of the BIK cluster. For the GA cluster, which lacks a specific TF, it was experimentally shown that AreA directly binds to promoter sequences of the single cluster genes [20].
Besides direct binding to the promoters of target genes, AreB and possibly AreA also mediate histone acetylation at some regulated SM gene clusters. Acetylation of histones directs chromatin structural transitions from hetero- to euchromatin and thus directly affects the accessibility of the transcriptional machinery to the underlying DNA. There are several examples demonstrating a role of GATA-type TFs in the recruitment of HAT complexes. In mammalian systems, GATA-1 was shown to recruit histone H3 and H4 acetylation complexes during gene activation [59], and AreA has been linked to histone acetylation in A. nidulans. In the latter, H3K9 and H3K14 are acetylated at the nitrate gene cluster in a strictly AreA-dependent manner [28]. AreA also contributes to chromatin accessibility and expression of two velvet-regulated gene clusters, encoding biosynthesis of the mycotoxin beauvericin and of the siderophore ferricrocin in F. oxysporum [60]. However, a direct interaction between AreA and HAT complexes still awaits proof.
Here, our ChIP analysis revealed that the presence of AreB is required for acetylation of H3K9 at the GA gene cluster. A significant decrease in H3K9ac was also observed at the promoters of FUB genes. Reduction in H3K9ac was more pronounced in AREB- than in AREA-deficient strains, thereby mirroring production levels of both SM in both mutants. Therefore, AreB and possibly also AreA are required for proper H3K9ac and consequently GA and FUB gene expression in F. fujikuroi, in a similar manner to GATA-1 in mammals [59] and AreA in A. nidulans [28].
Contrary to GA and FUB genes, we detected no alteration in H3K9ac at the FUM cluster genes in ΔAREA and ΔAREB strains, albeit there was a complete loss of FUM biosynthesis [13]. However, recent data demonstrated that overexpression of the cluster-specific TF gene FUM21 in the ΔAREA and ΔAREB background leads to only partial restoration of FUM gene expression and supports our suggestion that AreA and AreB are involved in chromatin accessibility [13]. Similar to the FUM genes, no AreA/AreB-dependent changes in H3K9ac were observed at BIK genes, albeit higher gene expression in ΔAREB, suggesting that AreA and/or AreB do not facilitate H3K9ac at this SM gene cluster. Other histone modifications could be involved in the activation of the BIK and FUM clusters.
In conclusion, in addition to their role as major players mediating NMR, AreA and AreB can be considered as master regulators of secondary metabolism in F. fujikuroi. Both GATA factors are involved in the transcriptional regulation of more than half of the 47 SM clusters in the genome. While AreA mainly acts as a strong positive regulator of N-repressed clusters, the regulatory function of AreB appears to be more diverse, including repression of N-induced clusters. In addition, we could prove an involvement of AreB in H3K9 acetylation of some, but not all, AreA and/or AreB-regulated SM gene clusters.
Materials and methods
Fungal strains and culture conditions
In this study the following F. fujikuroi strains were used: wild-type (Wt) strain IMI58289 (Commonwealth Mycological Institute, Kew, UK), ΔAREA-T19 [100] and ΔAREB-T2.1 [39]. Strains were maintained on solid CM [61] and cultivated at 28°C in darkness. Submersive cultivation was performed as described in [62], with synthetic ICI (Imperial Chemical Industries, UK) minimal medium [63] supplemented with 6 mM or 60 mM glutamine as N source in biological duplicates. After cultivation for 72 h at 28°C, cultures were harvested and mycelium was used for extraction of total RNA or proteins and for ChIP analyses.
For yeast recombination cloning, S. cerevisiae strain FGSC9721/FY834 (MATa his3Δ200 ura3-52 leu2Δ1 lys2Δ202 trplΔ63) [64] was cultivated in 5 ml liquid YPD (pH 5.8, 10 g/l yeast extract, 20 g/l Bacto-Trypton (Difco), 20 g/l glucose) medium overnight at 200 rpm and 30°C. The culture was used to inoculate 50 ml liquid YPD and incubated at 200 rpm and 30°C for 4 to 6 h until an OD 600 nm of ~1 was reached. The harvested yeast cells were also used for yeast recombination cloning [65,66].
Bacterial strains and plasmid construction
Escherichia coli strain Top10F’ (Invitrogen, Groningen, The Netherlands), cultivated in Lysogeny Broth medium [67] was used for plasmid propagation and amplification. For generating the pOE::AreB-GFP fusion vector, clones of the shortest AREB transcript of 1305 bp length (AREB-c [31]) were amplified using the primer AreB-GFP-F / AreB-GFP-R, which contain overlapping sequences homologous to the vector pNAH-OGG [66]. This vector contains a hygromycin resistance cassette, and a codon-optimized eGFP [68,69] under control of the constitutive Aspergillus nidulans oliC promoter and the Botrytis cinerea gluc terminator, respectively. The AREB PCR products and the NcoI-digested plasmid pNAH-OGG were co-transformed into S. cerevisiae yielding pOE::AreB-GFP by yeast recombination cloning (see above).
Fungal transformations
Preparation of protoplasts of F. fujikuroi was carried out as described [70]. The ΔAREA and ΔAREB mutant strains were transformed with 20 μg of the pOE::AreB::GFP vector, yielding strains ΔAREA-OE::AREB and ΔAREB-OE::AREB. Transformed protoplasts were regenerated at 28°C in a complete regeneration agar (0.7 M sucrose, 0.5 g/l yeast extract) with 100 μg/ml nourseothricin (Werner Agents, Jena, Germany) or 100 μg/ml hygromycin (Sigma-Aldrich, Taufkirchen, Germany) for 4 to 7 days as specified above. Genomic integration of the overexpression-constructs was checked by diagnostic PCR with primers ogfp-seqR1 / AreB-seq3.
Isolation of total RNA and northern blot
For northern blot analysis, total F. fujikuroi RNA was extracted from freeze-dried mycelium, using the RNAgents total RNA isolation kit (Promega, Mannheim, Germany). 20 μg of RNA were separated in a 1% agarose gel containing formaldehyde [67] and transferred to Hybond-N + membranes. Northern blot hybridizations were done by the method of Church and Gilbert [71]. For expression analysis of SM genes, corresponding probes were generated by PCR and 32P labeled using the random oligomer-primer method [67]. The following probes were used and amplified with the following primers: APF6 (00008_aps6_F / 00008_aps6_R), AREB (areB-for-neu / areB-rev-neu), BIK2 (bik2-F / bik2-R), CPS/KS (cps/ks-RT-for / cps/ks-RT-rev), FUB5 (FF02109-Wt-F / FF02109-Wt-R), FUM8 (fum8_F / fum8_R), TAMA (Tam1-F / Tam1-R).
Microarray analysis
The F. fujikuroi microarray was designed by Roche NimbleGen Systems (Madison, WI) as described previously [8]. Microarray-hybridizations were performed at Arrows Biomedical (Münster, Germany) and RNA quality was checked using Agilent Bioanalyzer 2100 and RNA Nano 6000 Lab-Chip Lab-Chip Kit (Agilent Technologies).
Expression data were analyzed as described before [8]. Genes with an absolute log2-fold change above one or below minus one and an adjusted P-value (FDR) below 0.05 based on biological duplicates were regarded as significantly differentially expressed. The Munich Information Center for Protein Sequences Functional Catalogue (FunCat) [45] was used to identify biological processes. We applied Fisher’s exact test [72] to determine statistically overrepresented functional categories in differentially expressed gene sets. The retained P-values were adjusted using Bonferroni procedure. Tested categories with an adjusted P-value below 0.05 were regarded as significantly overrepresented in the gene set. The influence of nitrogen and genotype factor on gene sets was tested using a two-way Type-III ANOVA [73] at a significance level of P = 0.05. The analyzed gene sets were retrieved from [8].
Analysis of bikaverin production by HPLC-DAD
Biological triplicates of strains were incubated for 7 days in ICI liquid cultures. Mycelium was harvested and culture supernatant was used for measurement of bikaverin and norbikaverin production by high-performance liquid chromatography coupled to a diode array detector (HPLC-DAD) at 510 nm wavelength. Samples were separated on a LiChrospher 100 RP-18 column (5 μm, 250 mm x 4 mm; Merck KGaA, Darmstadt, Germany) and measurement was performed on a Merck-Hitachi Chromatography System (Merck KGaA) fitted with an autosampler (L-7200), DAD (L-245) and gradient pump (L-7100). Chromatography conditions: solvent A ACN with 1% formic acid (v/v), solvent B 1% formic acid (v/v) with a flow rate of 1 ml/min. A gradient from 30% A to 45% A in 10 min, then for 15 min up to 50% A and followed by column flushing for 5 min at 100% A was used. Equilibration at the starting condition of 30% A was carried out for 3 min. Sample injection volume was 40 μl. Data were processed and analyzed using EZChrom Elite Version 3.3.2 SP1 (Scientific Software, Inc.)
ChIP-coupled quantitative PCR
Mycelia of biological duplicates were crosslinked with 1% formaldehyde, incubated for 15 min at room temperature and 90 rpm, and quenched with 125 mM glycine. Mycelium was filtered over Miracloth, ground in liquid N and stored at -80°C until further use. ChIP was carried out with an anti H3K9ac-specific antibody (AM39137; Active Motif, CA 92008, United States) as described [74]. Precipitation of the protein-antibody conjugate was performed with Dynabeads® Protein A (Novex®, Life Technologies). Precipitated DNA was quantified by qPCR according to protocol (Bio-Rad) using iQ™ SYBR® Green Supermix (Bio-Rad) and normalized to input DNA. Primers used for qPCR are listed in S6 Table. Primer efficiencies were kept between 90 and 110% and experiments were done in technical and biological replicates. Sequences of all F. fujikuroi ORFs were extracted from the publicly available genome sequence of F. fujikuroi [8].
Proteome analysis
Total protein was extracted from freeze-dried mycelium as described before [75]. 200 μg of total protein from the F. fujikuroi Wt, the ΔAREA and ΔAREB mutants (biological duplicates) were digested with trypsin according to the FASP method [76] followed by TiO2 phosphopeptide enrichment as already described [77].
LC-MS/MS measurement
Chromatographic separation of peptides was performed using an Ultimate 3000 RSLCnano System (Dionex, part of Thermo Fisher Scientific). The mobile phases for the loading pump consisted of 0.05% (v/v) TFA in ultrapure water (A) and 80% ACN/0.05% TFA in ultrapure water (B). The sample (1 μl) was loaded on a trapping column (C18 PepMap 100, 300 μM x 5 mm, 5 μm particle size, 100 Å pore size; Thermo Scientific) and desalted for 5 min using eluent A at a flow rate of 20 μl/min. Then the trap column was switched online with the separation column (Acclaim PepMap100 C18, 75 μm x 15 cm, 2 μM particle size, 100 Å pore size, Thermo Scientific). The mobile phases for elution of the peptides from the column consisted of 0.1% (v/v) formic acid in ultrapure water (A*) and 80% ACN/0.08% formic acid in ultrapure water (B*). Peptides were eluted at a flow rate of 300 nl/min and employing the following gradient profile: 0–40% B* over 90 min, 40–100% B* over 5 min, 100% B* for 10 min. Afterwards the column was re-equilibrated with 99% A* for 20 min.
The LC system was coupled via a nanospray source to an LTQ Orbitrap XL mass spectrometer (Thermo Finnigan). The general mass spectrometric conditions were: spray voltage, 1.5 kV; no sheath and auxiliary gas flow; ion transfer tube temperature: 200°C. The mass spectrometer was operated in positive ion mode and a data-dependent automatic switch was employed between MS and MS/MS acquisition modes. MS full scans (m/z 375–1600) were acquired in positive ion mode by FT-MS in the Orbitrap at a resolution of 60,000 (FWHM) with internal lock mass calibration on m/z 445.12003. The 12 most intense ions were fragmented in the linear ion trap by CID (35% normalized collision energy).
For standard samples, automatic gain control (AGC) was enabled with target values of 5 x 105 and 5 x 104 for MS full scans and MS/MS, respectively. One microscan was acquired per MS/MS spectrum and maximum ion trap fill time was 100 ms [78]. Ion selection thresholds were: 500 counts for MS2. An activation q = 0.25 and activation time of 30 ms were applied in MS2 acquisitions. Dynamic exclusion was enabled with an exclusion duration of 90s, repeat count of 1, list size of 500 and exclusion mass width of +/- 5 ppm. Unassigned charge states and charged state 1 were rejected.
To improve the fragmentation of phosphopeptides, multi-stage activation (MSA) in the Xcalibur software was enabled for each MS/MS spectrum. The 5 most intense ions were fragmented in the linear ion trap by CID (35% normalized collision energy). For the Multi Stage Activation (MSA), an MSA for further fragmention of the ions was triggered if in the 5 most intense peaks of a MS2 event a neutral loss peak at -98, -49 or -32.7 Da was observed. Automatic gain control (AGC) was enabled with target values of 5 x 105 and 5 x 104 for MS full scans and MS/MS, respectively. Two microscans were acquired per MS/MS spectrum and maximum ion trap fill time was 150 ms. Ion selection thresholds were: 500 counts for MS2. An activation q = 0.25 and activation time of 30 ms were applied in MS2 acquisitions. Dynamic exclusion was enabled with an exclusion duration of 120 s, repeat count of 1, list size of 500 and exclusion mass width of +/- 5 ppm. Unassigned charge states and charged state 1 were rejected. Additionally, Phosphopeptide-enriched samples (FT, E1 and E2) were measured with the following adjustments: Automatic gain control (AGC) was enabled with target values of 5 x 105 and 5 x 104 for MS full scans and MS/MS, respectively. Two microscans were acquired per MS/MS spectrum and maximum ion trap fill time was 150 ms.
Data analysis
Data analysis was performed as described previously [77]. LC-MS/MS data was converted from RAW format (Thermo scientific) to the open mzML format [79,80] using msconvert (Proteowizard version 2.0.1885 [81]). Then, mzML files were converted to the mascot generic format (mgf) if required using pymzML [82]. Peptide spectrum matches (PSMs) and statistical post processing was performed using Ursgal [83]. The following peptide database search algorithms were employed: OMSSA (version 2.1.9) [84], X! Tandem (version sledgehammer) [85], and MS-GF+ (version 9979 [86] using default values for most parameters (see http://ursgal.readthedocs.io/). A shuffled-peptide based target-decoy database was generated as described previously [87] based on Fusarium fujikuroi database (p3_i2_t5127_Fus_fujik_v21.prot version 21, [8]) and the contaminant database (cRAP, http://www.thegpm.org/crap/). Variable modifications were set as follows: oxidation of methionine (+15.9949 Da), acetylation of the N-terminus (+42.0106 Da), phosphorylation of serine, tyrosine and threonine (+79.966331). Additionally, X! Tandem considers the loss of water (-18.0106 Da) and deamidation (-17.0265 Da) by default. Carbamidomethylation of cysteine was set as fixed modification. Two missed cleavage sites were permitted. Statistical post-processing and the estimation of posterior error probabilities (PEP) for individual PSMs was performed using Percolator [88,89]. All PSMs were filtered on database search engine level with a cutoff ≤ 1% PEP. A total of 19658 unique peptides were identified (PEP ≤ 1%). Quantification was performed using pyQms with default parameters [87,90]. Retention time was aligned as described previously [87,90]. Intensity alignment and ratio calculation was performed as described previously [65]. Illustrations of logical relations between sets of regulated genes and proteins (Venn diagrams) were created using Ursgal [83]. Linear fits and coefficients of determination between gene ratios stemming from microarray data and protein ratios stemming from proteomics data was calculated using the linregress function of Scipy (https://www.scipy.org/).
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
The F. fujikuroi Wt and the ΔAREA and ΔAREB deletion mutants were cultivated for 3 days in ICI liquid cultures with 6 mM (Nitrogen limitation) or 60 mM (nitrogen sufficiency) glutamine as sole nitrogen source. Data is based on proteome analysis. Shown are differentially up-regulated and down-regulated proteins at nitrogen limitation (6) and nitrogen sufficiency (60) in ΔAREA and ΔAREB compared to Wt.
(TIF)
The F. fujikuroi Wt and the ΔAREA and ΔAREB deletion mutants were cultivated for 3 days in ICI liquid cultures with 6 mM (-N) or 60 mM (+N) gln as sole nitrogen source. Data is based on proteome analysis. Differentially regulated proteins in the mutants compared to Wt under respective nitrogen conditions were functionally classified according to their most prominent FunCat category. Stippled areas represent proteins affected in ΔAREA, which are regulated on an equal level (+/- 20%) or stronger in ΔAREB. For these proteins, the regulations might be an indirect effect due to the reduced levels of AreB in the ΔAREA mutant.
(TIF)
Acknowledgments
We thank Jana Marie Boysen for technical assistance during HPLC analysis. We are particularly grateful to Brian Williamson for critical reading of this manuscript.
Data Availability
All transcriptome files are available from the Gene Express Omnibus (GEO) repository database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80480). All proteome files are available from the ProteomeXchange Consortium database (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier <PXD004664>.
Funding Statement
This project was funded by the Deutsche Forschungsgemeinschaft (www.dfg.de), Grant Number TU101/12-2, BT. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sun SK, Snyder WC. The bakanae disease of the rice plant Fusarium: diseases, biology and taxonomy The Pennsylvania State University Press, University Park; 1981; 104–113. [Google Scholar]
- 2.Kurosawa E. Experimental studies on the nature of the substance secreted by the bakanae fungus. Nat Hist Soc Formosa. 1926;16: 213–227. [Google Scholar]
- 3.Yabuta T. Biochemistry of the “bakanae” fungus of rice. Agric Hortic. 1935;10: 17–22. [Google Scholar]
- 4.Tudzynski B, Hölter K. Gibberellin biosynthetic pathway in Gibberella fujikuroi: evidence for a gene cluster. Fungal Genet Biol. 1998;25: 157–170. 10.1006/fgbi.1998.1095 [DOI] [PubMed] [Google Scholar]
- 5.Wiemann P, Willmann A, Straeten M, Kleigrewe K, Beyer M, Humpf H-U, et al. Biosynthesis of the red pigment bikaverin in Fusarium fujikuroi: genes, their function and regulation. Mol Microbiol. 2009;72: 931–946. 10.1111/j.1365-2958.2009.06695.x [DOI] [PubMed] [Google Scholar]
- 6.Studt L, Wiemann P, Kleigrewe K, Humpf H-U, Tudzynski B. Biosynthesis of fusarubins accounts for pigmentation of Fusarium fujikuroi perithecia. Appl Environ Microbiol. 2012;78: 4468–4480. 10.1128/AEM.00823-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desjardins AE, Plattner RD, Nelson PE. Production of fumonisin B and moniliformin by Gibberella fujikuroi from rice from various geographic areas. Appl Environ Microbiol. 1997;63: 1838–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiemann P, Sieber CMK, von Bargen KW, Studt L, Niehaus E-M, Espino JJ, et al. Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013;9: e1003475 10.1371/journal.ppat.1003475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niehaus E-M, Kleigrewe K, Wiemann P, Studt L, Sieber CMK, Connolly LR, et al. Genetic manipulation of the Fusarium fujikuroi fusarin gene cluster yields insight into the complex regulation and fusarin biosynthetic pathway. Chem Biol. 2013;20: 1055–1066. 10.1016/j.chembiol.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 10.Niehaus E-M, Janevska S, von Bargen KW, Sieber CMK, Harrer H, Humpf H-U, et al. Apicidin F: characterization and genetic manipulation of a new secondary metabolite gene cluster in the rice pathogen Fusarium fujikuroi. PLoS ONE. 2014;9: e103336 10.1371/journal.pone.0103336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niehaus E-M, von Bargen KW, Espino JJ, Pfannmüller A, Humpf H-U, Tudzynski B. Characterization of the fusaric acid gene cluster in Fusarium fujikuroi. Appl Microbiol Biotechnol. 2014;98: 1749–1762. 10.1007/s00253-013-5453-1 [DOI] [PubMed] [Google Scholar]
- 12.Nazari F, Sulyok M, Kobarfard F, Yazdanpanah H, Krska R. Evaluation of emerging Fusarium mycotoxins beauvericin, enniatins, fusaproliferin and moniliformin in domestic rice in Iran. Iran J Pharm Res. 2015;14: 505–512. [PMC free article] [PubMed] [Google Scholar]
- 13.Rösler SM, Sieber CMK, Humpf H-U, Tudzynski B. Interplay between pathway-specific and global regulation of the fumonisin gene cluster in the rice pathogen Fusarium fujikuroi. Appl Microbiol Biotechnol. 2016;100: 5869–82. 10.1007/s00253-016-7426-7 [DOI] [PubMed] [Google Scholar]
- 14.Studt L, Janevska S, Niehaus E-M, Burkhardt I, Arndt B, Sieber CMK, et al. Two separate key enzymes and two pathway-specific transcription factors are involved in fusaric acid biosynthesis in Fusarium fujikuroi. Environ Microbiol. 2016;18: 936–956. 10.1111/1462-2920.13150 [DOI] [PubMed] [Google Scholar]
- 15.Niehaus E-M, Studt L, von Bargen KW, Kummer W, Humpf H-U, Reuter G, et al. Sound of silence: the beauvericin cluster in Fusarium fujikuroi is controlled by cluster-specific and global regulators mediated by H3K27 modification. Environ Microbiol. 2016;18: 4282–4302. 10.1111/1462-2920.13576 [DOI] [PubMed] [Google Scholar]
- 16.Haas H, Marzluf GA. NRE, the major nitrogen regulatory protein of Penicillium chrysogenum, binds specifically to elements in the intergenic promoter regions of nitrate assimilation and penicillin biosynthetic gene clusters. Curr Genet. 1995;28: 177–183. [DOI] [PubMed] [Google Scholar]
- 17.Min K, Shin Y, Son H, Lee J, Kim J-C, Choi GJ, et al. Functional analyses of the nitrogen regulatory gene areA in Gibberella zeae. FEMS Microbiol Lett. 2012;334: 66–73. 10.1111/j.1574-6968.2012.02620.x [DOI] [PubMed] [Google Scholar]
- 18.Giese H, Sondergaard TE, Sørensen JL. The AreA transcription factor in Fusarium graminearum regulates the use of some nonpreferred nitrogen sources and secondary metabolite production. Fungal Biol. 2013;117: 814–821. 10.1016/j.funbio.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 19.Li J, Pan Y, Liu G. Disruption of the nitrogen regulatory gene AcareA in Acremonium chrysogenum leads to reduction of cephalosporin production and repression of nitrogen metabolism. Fungal Genetics and Biology. 2013;61: 69–79. 10.1016/j.fgb.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 20.Mihlan M, Homann V, Liu T-WD, Tudzynski B. AREA directly mediates nitrogen regulation of gibberellin biosynthesis in Gibberella fujikuroi, but its activity is not affected by NMR. Mol Microbiol. 2003;47: 975–991. [DOI] [PubMed] [Google Scholar]
- 21.Kudla B, Caddick MX, Langdon T, Martinez-Rossi NM, Bennett CF, Sibley S, et al. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990;9: 1355–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu YH, Marzluf GA. Site-directed mutagenesis of the “zinc finger” DNA-binding domain of the nitrogen-regulatory protein NIT2 of Neurospora. Mol Microbiol. 1990;4: 1847–1852. [DOI] [PubMed] [Google Scholar]
- 23.Ravagnani A, Gorfinkiel L, Langdon T, Diallinas G, Adjadj E, Demais S, et al. Subtle hydrophobic interactions between the seventh residue of the zinc finger loop and the first base of an HGATAR sequence determine promoter-specific recognition by the Aspergillus nidulans GATA factor AreA. EMBO J. 1997;16: 3974–3986. 10.1093/emboj/16.13.3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caddick MX, Arst HN. Deletion of the 389 N-terminal residues of the transcriptional activator AREA does not result in nitrogen metabolite derepression in Aspergillus nidulans. J Bacteriol. 1998;180: 5762–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tudzynski B. Nitrogen regulation of fungal secondary metabolism in fungi. Front Microbiol. 2014;5: 656 10.3389/fmicb.2014.00656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narendja F, Goller SP, Wolschek M, Strauss J. Nitrate and the GATA factor AreA are necessary for in vivo binding of NirA, the pathway-specific transcriptional activator of Aspergillus nidulans. Mol Microbiol. 2002;44: 573–583. [DOI] [PubMed] [Google Scholar]
- 27.Muro-Pastor MI, Strauss J, Ramón A, Scazzocchio C. A paradoxical mutant GATA factor. Eukaryotic Cell. 2004;3: 393–405. 10.1128/EC.3.2.393-405.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger H, Basheer A, Böck S, Reyes-Dominguez Y, Dalik T, Altmann F, et al. Dissecting individual steps of nitrogen transcription factor cooperation in the Aspergillus nidulans nitrate cluster. Mol Microbiol. 2008;69: 1385–1398. 10.1111/j.1365-2958.2008.06359.x [DOI] [PubMed] [Google Scholar]
- 29.Muro-Pastor MI, Gonzalez R, Strauss J, Narendja F, Scazzocchio C. The GATA factor AreA is essential for chromatin remodelling in a eukaryotic bidirectional promoter. EMBO J. 1999;18: 1584–1597. 10.1093/emboj/18.6.1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger H, Pachlinger R, Morozov I, Goller S, Narendja F, Caddick M, et al. The GATA factor AreA regulates localization and in vivo binding site occupancy of the nitrate activator NirA. Mol Microbiol. 2006;59: 433–446. 10.1111/j.1365-2958.2005.04957.x [DOI] [PubMed] [Google Scholar]
- 31.Michielse CB, Pfannmüller A, Macios M, Rengers P, Dzikowska A, Tudzynski B. The interplay between the GATA transcription factors AreA, the global nitrogen regulator and AreB in Fusarium fujikuroi. Mol Microbiol. 2014;91: 472–493. 10.1111/mmi.12472 [DOI] [PubMed] [Google Scholar]
- 32.Haas H, Angermayr K, Zadra I, Stöffler G. Overexpression of nreB, a new GATA factor-encoding gene of Penicillium chrysogenum, leads to repression of the nitrate assimilatory gene cluster. J Biol Chem. 1997;272: 22576–22582. [DOI] [PubMed] [Google Scholar]
- 33.Conlon H, Zadra I, Haas H, Arst HN, Jones MG, Caddick MX. The Aspergillus nidulans GATA transcription factor gene areB encodes at least three proteins and features three classes of mutation. Mol Microbiol. 2001;40: 361–375. [DOI] [PubMed] [Google Scholar]
- 34.Wong KH, Hynes MJ, Davis MA. Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi. Eukaryot Cell. 2008;7: 917–925. 10.1128/EC.00076-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong KH, Hynes MJ, Todd RB, Davis MA. Deletion and overexpression of the Aspergillus nidulans GATA factor AreB reveals unexpected pleiotropy. Microbiology (Reading, Engl). 2009;155: 3868–3880. [DOI] [PubMed] [Google Scholar]
- 36.Dzikowska A, Kacprzak M, Tomecki R, Koper M, Scazzocchio C, Weglenski P. Specific induction and carbon/nitrogen repression of arginine catabolism gene of Aspergillus nidulans—functional in vivo analysis of the otaA promoter. Fungal Genet Biol. 2003;38: 175–186. [DOI] [PubMed] [Google Scholar]
- 37.Macios M, Caddick MX, Weglenski P, Scazzocchio C, Dzikowska A. The GATA factors AREA and AREB together with the co-repressor NMRA, negatively regulate arginine catabolism in Aspergillus nidulans in response to nitrogen and carbon source. Fungal Genet Biol. 2012;49: 189–198. 10.1016/j.fgb.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 38.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13: 3999–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis MA, Small AJ, Kourambas S, Hynes MJ. The tamA gene of Aspergillus nidulans contains a putative zinc cluster motif which is not required for gene function. J Bacteriol. 1996;178: 3406–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Small AJ, Todd RB, Zanker MC, Delimitrou S, Hynes MJ, Davis MA. Functional analysis of TamA, a coactivator of nitrogen-regulated gene expression in Aspergillus nidulans. Mol Genet Genomics. 2001;265: 636–646. [DOI] [PubMed] [Google Scholar]
- 41.Kukuczka B, Magneschi L, Petroutsos D, Steinbeck J, Bald T, Powikrowska M, et al. Proton Gradient Regulation5-like1-mediated cyclic electron flow is crucial for acclimation to anoxia and complementary to nonphotochemical quenching in stress adaptation. Plant Physiol. 2014;165: 1604–1617. 10.1104/pp.114.240648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bibbins M, Crepin VF, Cummings NJ, Mizote T, Baker K, Mellits KH, et al. A regulator gene for acetate utilisation from Neurospora crassa. Mol Genet Genomics. 2002;267: 498–505. 10.1007/s00438-002-0682-5 [DOI] [PubMed] [Google Scholar]
- 43.Kurat CF, Lambert J-P, Petschnigg J, Friesen H, Pawson T, Rosebrock A, et al. Cell cycle-regulated oscillator coordinates core histone gene transcription through histone acetylation. Proc Natl Acad Sci USA. 2014;111: 14124–14129. 10.1073/pnas.1414024111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kouzminova E, Selker EU. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 2001;20: 4309–4323. 10.1093/emboj/20.15.4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucl Acids Res. 2004;32: 5539–5545. 10.1093/nar/gkh894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13: 227–232. 10.1038/nrg3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Roch KG, Johnson JR, Florens L, Zhou Y, Santrosyan A, Grainger M, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004;14: 2308–2318. 10.1101/gr.2523904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Legube G, Trouche D. Regulating histone acetyltransferases and deacetylases. EMBO Rep. 2003;4: 944–947. 10.1038/sj.embor.embor941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bömke C, Tudzynski B. Diversity, regulation, and evolution of the gibberellin biosynthetic pathway in fungi compared to plants and bacteria. Phytochemistry. 2009;70: 1876–1893. 10.1016/j.phytochem.2009.05.020 [DOI] [PubMed] [Google Scholar]
- 50.Proctor RH, Desjardins AE, Plattner RD, Hohn TM. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genetics and Biology. 1999;27: 100–112. 10.1006/fgbi.1999.1141 [DOI] [PubMed] [Google Scholar]
- 51.Seo J-A, Proctor RH, Plattner RD. Characterization of four clustered and coregulated genes associated with fumonisin biosynthesis in Fusarium verticillioides. Fungal Genetics and Biology. 2001;34: 155–165. 10.1006/fgbi.2001.1299 [DOI] [PubMed] [Google Scholar]
- 52.Arst HN, Cove DJ. Nitrogen metabolite repression in Aspergillus nidulans. Mol Gen Genet. 1973;126: 111–141. [DOI] [PubMed] [Google Scholar]
- 53.Marzluf GA. Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev. 1997;61: 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ment D, Alkan N, Luria N, Bi F-C, Reuveni E, Fluhr R, et al. A Role of AREB in the Regulation of PACC-Dependent Acid-Expressed-Genes and Pathogenicity of Colletotrichum gloeosporioides. Mol Plant Microbe Interact. 2015;28: 154–166. 10.1094/MPMI-09-14-0252-R [DOI] [PubMed] [Google Scholar]
- 55.Marroquin-Guzman M, Wilson RA. GATA-Dependent Glutaminolysis Drives Appressorium Formation in Magnaporthe oryzae by Suppressing TOR Inhibition of cAMP/PKA Signaling. PLoS Pathog. 2015;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krappmann S, Braus GH. Nitrogen metabolism of Aspergillus and its role in pathogenicity. Med Mycol. 2005;43 Suppl 1: S31–40. [DOI] [PubMed] [Google Scholar]
- 57.Magasanik B, Kaiser CA. Nitrogen regulation in Saccharomyces cerevisiae. Gene. 2002;290: 1–18. [DOI] [PubMed] [Google Scholar]
- 58.Georis I, Feller A, Tate JJ, Cooper TG, Dubois E. Nitrogen catabolite repression-sensitive transcription as a readout of TOR pathway regulation: the genetic background, reporter gene and GATA factor assayed determine the outcomes. Genetics. 2009;181: 861–874. 10.1534/genetics.108.099051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Letting DL, Rakowski C, Weiss MJ, Blobel GA. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol Cell Biol. 2003;23: 1334–1340. 10.1128/MCB.23.4.1334-1340.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.López-Berges MS, Schäfer K, Hera C, Di Pietro A. Combinatorial function of velvet and AreA in transcriptional regulation of nitrate utilization and secondary metabolism. Fungal Genet Biol. 2014;62: 78–84. 10.1016/j.fgb.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 61.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AWJ. The genetics of Aspergillus nidulans. Adv Genet. 1953;5: 141–238. [DOI] [PubMed] [Google Scholar]
- 62.Wiemann P, Albermann S, Niehaus E-M, Studt L, von Bargen KW, Brock NL, et al. The Sfp-type 4’-phosphopantetheinyl transferase Ppt1 of Fusarium fujikuroi controls development, secondary metabolism and pathogenicity. PLoS ONE. 2012;7: e37519 10.1371/journal.pone.0037519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Geissmann TA, Verbiscar AJ, Phinney BO, Cragg G. Studies on the biosynthesis of gibberellins from (−)-kaurenoic acid in cultures of Gibberella Fujikuroi. Phytochemistry. 1966;5: 933–947. [Google Scholar]
- 64.Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11: 53–55. 10.1002/yea.320110107 [DOI] [PubMed] [Google Scholar]
- 65.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA. 2006;103: 10352–10357. 10.1073/pnas.0601456103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schumacher J. Tools for Botrytis cinerea: New expression vectors make the gray mold fungus more accessible to cell biology approaches. Fungal Genet Biol. 2012;49: 483–497. 10.1016/j.fgb.2012.03.005 [DOI] [PubMed] [Google Scholar]
- 67.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn New York: Cold Spring Harbor Laboratory: Cold Spring Harbor; 1989. [Google Scholar]
- 68.Pöggeler S, Masloff S, Hoff B, Mayrhofer S, Kück U. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr Genet. 2003;43: 54–61. 10.1007/s00294-003-0370-y [DOI] [PubMed] [Google Scholar]
- 69.Staben C, Jensen B, Singer M, Pollock J, Schechtman M, Kinsey J. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet Newsl. 1989;36: 79–81. [Google Scholar]
- 70.Tudzynski B, Mende K, Weltring KM, Kinghorn JR, Unkles SE. The Gibberella fujikuroi niaD gene encoding nitrate reductase: isolation, sequence, homologous transformation and electrophoretic karyotype location. Microbiology (Reading, Engl). 1996;142 (Pt 3): 533–539. [DOI] [PubMed] [Google Scholar]
- 71.Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81: 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fisher RA. On the Interpretation of x2 from Contingency Tables, and the Calculation of P. Journal of the Royal Statistical Society. 1922;85: 87–94. [Google Scholar]
- 73.Yates F. The Analysis of Multiple Classifications with Unequal Numbers in the Different Classes. Journal of the American Statistical Association. 1934;29: 51–66. [Google Scholar]
- 74.Gacek-Matthews A, Noble LM, Gruber C, Berger H, Sulyok M, Marcos AT, et al. KdmA, a histone H3 demethylase with bipartite function, differentially regulates primary and secondary metabolism in Aspergillus nidulans. Molecular Microbiology. 2015;96: 839–860. 10.1111/mmi.12977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Todd RB, Fraser JA, Wong KH, Davis MA, Hynes MJ. Nuclear accumulation of the GATA factor AreA in response to complete nitrogen starvation by regulation of nuclear export. Eukaryotic Cell. 2005;4: 1646–1653. 10.1128/EC.4.10.1646-1653.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wiśniewski JR, Zielinska DF, Mann M. Comparison of ultrafiltration units for proteomic and N-glycoproteomic analysis by the filter-aided sample preparation method. Anal Biochem. 2011;410: 307–309. 10.1016/j.ab.2010.12.004 [DOI] [PubMed] [Google Scholar]
- 77.Bergner SV, Scholz M, Trompelt K, Barth J, Gäbelein P, Steinbeck J, et al. STATE TRANSITION7-dependent phosphorylation is modulated by changing environmental conditions, and its absence triggers remodeling of photosynthetic protein complexes. Plant Physiol. 2015;168: 615–634. 10.1104/pp.15.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kalli A, Hess S. Effect of mass spectrometric parameters on peptide and protein identification rates for shotgun proteomic experiments on an LTQ-orbitrap mass analyzer. Proteomics. 2012;12: 21–31. 10.1002/pmic.201100464 [DOI] [PubMed] [Google Scholar]
- 79.Martens L, Chambers M, Sturm M, Kessner D, Levander F, Shofstahl J, et al. mzML--a community standard for mass spectrometry data. Mol Cell Proteomics. 2011;10: R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deutsch E. mzML: a single, unifying data format for mass spectrometer output. Proteomics. 2008;8: 2776–2777. 10.1002/pmic.200890049 [DOI] [PubMed] [Google Scholar]
- 81.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24: 2534–2536. 10.1093/bioinformatics/btn323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bald T, Barth J, Niehues A, Specht M, Hippler M, Fufezan C. pymzML--Python module for high-throughput bioinformatics on mass spectrometry data. Bioinformatics. 2012;28: 1052–1053. 10.1093/bioinformatics/bts066 [DOI] [PubMed] [Google Scholar]
- 83.Kremer LPM, Leufken J, Oyunchimeg P, Schulze S, Fufezan C. Ursgal, universal python module combining common bottom-up proteomics tools for large-scale analysis. J Proteome Res. 2016;15: 788–794. 10.1021/acs.jproteome.5b00860 [DOI] [PubMed] [Google Scholar]
- 84.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, et al. Open mass spectrometry search algorithm. J Proteome Res. 2004;3: 958–964. 10.1021/pr0499491 [DOI] [PubMed] [Google Scholar]
- 85.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20: 1466–1467. 10.1093/bioinformatics/bth092 [DOI] [PubMed] [Google Scholar]
- 86.Kim S, Mischerikow N, Bandeira N, Navarro JD, Wich L, Mohammed S, et al. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol Cell Proteomics. 2010;9: 2840–2852. 10.1074/mcp.M110.003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barth J, Bergner SV, Jaeger D, Niehues A, Schulze S, Scholz M, et al. The interplay of light and oxygen in the reactive oxygen stress response of Chlamydomonas reinhardtii dissected by quantitative mass spectrometry. Mol Cell Proteomics. 2014;13: 969–989. 10.1074/mcp.M113.032771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4: 923–925. 10.1038/nmeth1113 [DOI] [PubMed] [Google Scholar]
- 89.Käll L, Storey JD, MacCoss MJ, Noble WS. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res. 2008;7: 29–34. 10.1021/pr700600n [DOI] [PubMed] [Google Scholar]
- 90.Höhner R, Barth J, Magneschi L, Jaeger D, Niehues A, Bald T, et al. The metabolic status drives acclimation of iron deficiency responses in Chlamydomonas reinhardtii as revealed by proteomics based hierarchical clustering and reverse genetics. Mol Cell Proteomics. 2013;12: 2774–2790. 10.1074/mcp.M113.029991 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
The F. fujikuroi Wt and the ΔAREA and ΔAREB deletion mutants were cultivated for 3 days in ICI liquid cultures with 6 mM (Nitrogen limitation) or 60 mM (nitrogen sufficiency) glutamine as sole nitrogen source. Data is based on proteome analysis. Shown are differentially up-regulated and down-regulated proteins at nitrogen limitation (6) and nitrogen sufficiency (60) in ΔAREA and ΔAREB compared to Wt.
(TIF)
The F. fujikuroi Wt and the ΔAREA and ΔAREB deletion mutants were cultivated for 3 days in ICI liquid cultures with 6 mM (-N) or 60 mM (+N) gln as sole nitrogen source. Data is based on proteome analysis. Differentially regulated proteins in the mutants compared to Wt under respective nitrogen conditions were functionally classified according to their most prominent FunCat category. Stippled areas represent proteins affected in ΔAREA, which are regulated on an equal level (+/- 20%) or stronger in ΔAREB. For these proteins, the regulations might be an indirect effect due to the reduced levels of AreB in the ΔAREA mutant.
(TIF)
Data Availability Statement
All transcriptome files are available from the Gene Express Omnibus (GEO) repository database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80480). All proteome files are available from the ProteomeXchange Consortium database (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier <PXD004664>.