Abstract
A study of genome-wide gene expression in major depressive disorder (MDD) was undertaken in a large population-based sample to determine whether altered expression levels of genes and pathways could provide insights into biological mechanisms that are relevant to this disorder. Gene expression studies have the potential to detect changes that may be due to differences in common or rare genomic sequence variation, environmental factors or their interaction. We recruited a European-ancestry sample of 463 individuals with recurrent MDD and 459 controls, obtained self-report and semi-structured interview data about psychiatric and medical history and other environmental variables, sequenced RNA from whole blood and genotyped a genome-wide panel of common SNPs. We used analytical methods to identify MDD-related genes and pathways using all of these sources of information. In analyses of association between MDD and expression levels of 13,857 single autosomal genes, accounting for multiple technical, physiological and environmental covariates, a significant excess of low p-values was observed, but there was no significant single-gene association after genome-wide correction. Pathway-based analyses of expression data detected significant association of MDD with increased expression of genes in the interferon α/β signaling pathway. This finding could not be explained by potentially confounding diseases and medications (including antidepressants) or by computationally-estimated proportions of white blood cell types. Although cause-effect relationships cannot be determined from these data, the results support the hypothesis that altered immune signaling plays a role in the pathogenesis, manifestation, and/or the persistence and progression of MDD.
Keywords: Major Depressive Disorder, MDD, RNA-sequencing, transcriptomic, pathway expression signature, interferon-I signaling
Introduction
MDD is a complex phenotype, with moderate heritability estimates (31-42%) (1). Physiological correlates include alterations in neurohormonal secretion, structural brain anatomy and markers of immune function and inflammation. In contrast to the other two most-studied psychiatric disorders in adults (bipolar disorder and schizophrenia), or traits with comparable heritability such as Type 2 diabetes (26%) (2), it has been challenging to identify relevant genetic factors for MDD from genome-wide association studies (GWAS), with the largest GWAS to date (with over 9,000 MDD cases) detecting no significant associations with common single nucleotide polymorphisms (SNPs) (3). This might be explained by MDD’s phenotypic heterogeneity (4, 5) and complex genetic architecture (6), and its association with environmental factors that are likely to interact with genetics (7).
Whole-transcriptome studies offer another type of genome-wide search for disease-related mechanisms by measuring mRNA expression levels of each gene in a relevant tissue. While expression data do not directly disentangle cause-effect relationships, altered expression levels in disease can reflect the effect of common and/or rare genetic sequence variation, environmental factors, the effects of the disease processes itself, and interaction between genetic variation and environmental factors. We applied deep RNA sequencing (RNA-seq) to whole-blood RNA in a large sample from a population-based survey research panel, including cases on and off psychiatric medication (unlike most clinical samples). This ascertainment method, which made whole blood the only feasible tissue for expression profiling, had the advantage of providing information on subjects’ “ real-life” state (in contrast with cell lines or postmortem tissues), with the limitations of requiring statistical correction of many state-related, possibly confounding variables, and of having limited potential to identify some brain-specific mechanisms. On the other hand, there is increasing evidence implicating dysregulation of glucocorticoid and immune responses in MDD, and white blood cells may be a particularly a suitable tissue for studying the relevant immunological factors.
We present here the largest whole-transcriptome study of MDD to date and the first using RNA-seq. After collecting psychiatric, demographic, environmental and medical information, we studied 922 European-ancestry individuals (463 cases, 459 controls) with RNA-seq of whole blood RNA and with a GWAS assay. Three types of analyses of association to MDD were then carried out. (i) Analyses of individual gene expression levels produced a significant excess of low p-values, but no significant association of a single gene after correction for genome-wide testing. (ii) Analyses of expression levels across pre-defined pathways and gene sets detected significant genome-wide association with the interferon α/β signaling pathway. (iii) Joint evaluation of gene expression and SNP genotypes identified significant association to CINP, a gene involved in the cell cycle arrest, possibly induced by interferon (8, 9).
Methods
Subject recruitment and assessment
(See Supplementary Methods and Batte et al., (in revision, Genome Research) for additional methodological details.) Recruitment was carried out under IRB-approved protocols. Cases (recurrent or chronic MDD) and controls were recruited by Knowledge Networks, Inc. (KN) (Menlo Park, CA) from a nationally representative panel of ~60,000 individuals. KN invited 14,463 individuals (self-reported Caucasian ancestry, ages 21-60) for screening. Respondents were screened online with lifetime self-report versions of the depression and alcohol/substance dependence modules of the Composite International Diagnostic Interview-Short Form (CIDI-SF) (10) plus screening items for psychotic and bipolar disorders. After excluding those who reported current dependence or lifetime psychotic or bipolar disorder, individuals who tentatively met inclusion criteria (see below) were invited to participate. Those who consented to be contacted answered additional online questions about height, weight, childhood trauma and smoking. Among those who completed blood draw by a national phlebotomy company (after giving written informed consent), 650 prospective cases and 589 prospective controls were interviewed by telephone with a modified Structured Clinical Interview for DSM-IV (SCID) (full depression, bipolar, alcohol, substance and anxiety modules; psychosis screen; and a more detailed illness and medication history); PHQ-9 (current depression) (11) and GAD-7 (current anxiety) (12) scales; and a family history screen (MDD, bipolar disorder and suicide). Most telephone interviews were completed within 1-2 months of online screening and blood draw, with delays of several months in a small proportion of cases. (Sample sizes at each stage are provided in Supplementary Methods.)
Clinical data were reviewed by clinical site principal investigators (MMW, JBP) and the overall PI (DFL). After exclusions for substance dependence, non-affective psychosis and bipolar disorder, we identified 475 cases with MDD with two or more lifetime episodes or one episode lasting ≥ 2 years, and 474 controls who never experienced a two-week period with depressed mood plus 2 or more other MDD criteria. After exclusions for genotypically non-European ancestry, unusual medical comorbidities and RNA-seq and GWAS quality control (QC) analyses, 922 individuals were included in further analyses (463 cases and 459 controls).
RNA-Sequencing and genotype data
Whole blood was collected in PaxGene tubes (Qiagen, Valencia, CA) for RNA and in acid-citrate-dextrose tubes for DNA. PaxGene tubes contain a proprietary cell lytic agent for immediate release of RNA from cells; mRNA levels remain stable for years with appropriate storage (13). Extracted DNA was genotyped with the Illumina Omni1-Quad microarray (Centrillion Biosciences, Mountain View, CA). Post-QC genotypes were available for 720,591 autosomal SNPs. RNA was extracted; most globin RNA was removed (GLOBINclear Kits, Life Technologies, Grand Island, NY) (Supplementary Figure S1-4); and Illumina TrueSeq kits (La Jolla, CA) were used for RNA purification (polyA selection), chemical fragmentation, single-stranded cDNA conversion, DNA library preparation and oligonucleotide barcoding. Sequencing was carried out with an Illumina HiSeq 2000 (50 or 51bp single-ended reads, 3 multiplexed libraries/lane) yielding 70 million reads (average) per individual. Reads were mapped to the NCBI v37 H. sapiens reference genome using Tophat (14). Complete genotype and RNA-seq data are available form https://nimhgenetics.org (see data access note below).
Gene expression was quantified with HTSeq using the “intersection_strict” criteria (15). Only uniquely aligned reads were used to quantify gene expression levels. Reads were assigned to 21,578 of the 22,339 annotated protein-coding genes (NCBI v37). Analyses included 13,857 autosomal genes with ≥100 individuals with ≥10 reads (in total from anywhere across the transcript). Effects of technical covariates (e.g., per-individual 5’ bias, GC bias, sequencing depth, and percent globin reads) and biological covariates (e.g., estimates of blood cell type proportions and time of day of blood draw) (see Table S1 for complete list) were removed by ridge regression of logarithm-transformed read counts. Cell-type proportions were inferred using a method based on non-negative least squares (NNLS) (16), making use of external microarray data on cell-type specific expression signatures (17) (Supplementary Methods).
Association of gene expression levels with MDD
A likelihood ratio test (LRT) was used to determine the significance of association between the expression levels of each gene and MDD. Using LRT, the strength of the association is determined by comparing the likelihood of the null (“background”) model that includes only a set of confounding factors (see below) with the likelihood of the full model (confounding factors plus expression value) (Supplementary Methods). Final p-values were obtained using permutation analyses (8,000 initial permutations or 1,000,000 permutations for 3 genes with p=0 in the initial 8,000) (Figure S5). FDR was used for multiple hypothesis correction (18).
In total we corrected for 39 “background” covariates (Table 1, S2). These consist of 24 environmental, physiological, and medical covariates, including BMI, gender, smoking and age; manually-curated indicators of intake of medications and substances (reported by ≥30 individuals), including cholesterol-lowering and antihypertensive medication classes, thyroid medication, cannabis and alcohol use, many of which were more common in cases and had a non-negligible effect on gene expression (Table S2, Figure S10). Other covariates include five genotypic principal components (PCs) reflecting population structure (19) (Supplementary Methods, Figure S4); and ten hidden confounding factors obtained as PCs of the residual expression data (Figure S6). We decided to include expression PCs because previous studies have shown that removal of expression PCs results in improved power for detecting eQTLs (20) and identifying true-positive co-expression relationships between genes (21). We also evaluated whether there was an excess of low p-values (deviation from the expected uniform distribution), by estimating the proportion of true positives using the π1 statistic (22).
Table 1. Covariate values for cases and controls.
| Cases | Controls | p-value | |
|---|---|---|---|
| Covariate name | Mean (SD) | (case vs. control) | |
| Age at interview | 44.77 (10.69) | 44.64 (11.0) | ns |
| BMI | 30.25 ( 7.74) | 27.87 (6.45) | 2.63E-07 |
| Number of cigarettes per day | 2.49 ( 6.76) | 1.12 (4.55) | 8.78E-06 |
| Number of blood pressure meds | 0.31 (0.66) | 0.22 (0.53) | 0.06 |
| Current alcohol use | 0.61 (0.78) | 0.69 (0.76) | 0.04 |
| N (%) | |||
| N (cases or controls) | 463 | 459 | |
| Female gender | 360 (0.778) | 288 (0.627) | 4.08E-07 |
| Ate before blood draw | 323 (0.698) | 339 (0.739) | ns |
| Exercised before blood draw | 74 (0.160) | 92 (0.200) | 0.064 |
| Smoked before blood draw | 67 (0.145) | 34 (0.074) | 4.01E-04 |
| ACE Inhibitor | 36 (0.078) | 30 (0.065) | ns |
| Anticholinergic meds | 23 (0.050) | 12 (0.026) | 0.044 |
| Antihistaminic meds | 60 (0.130) | 39 (0.085) | 0.018 |
| Beta blocker | 44 (0.095) | 14 (0.031) | 3.45E-05 |
| Cholesterol lowering meds | 64 (0.138) | 50 (0.109) | ns |
| Decongestant | 41 (0.089) | 10 (0.022) | 4.94E-06 |
| Decongestant or stimulant | 52 (0.112) | 10 (0.022) | 1.27E-08 |
| Diuretic | 29 (0.063) | 25 (0.054) | ns |
| Non-steroidal anti-inflammatory | 46 (0.099) | 19 (0.041) | 4.05E-04 |
| Oral hypoglycemic | 30 (0.065) | 16 (0.035) | 0.026 |
| Oral contraceptive | 45 (0.097) | 39 (0.085) | ns |
| Protein pump inhibitor | 52 (0.112) | 32 (0.070) | 0.016 |
| Thyroid medication | 54 (0.117) | 34 (0.074) | 0.018 |
| Opiate medication or use | 46 (0.099) | 9 (0.020) | 1.18E-07 |
| Cannabis use (past 2 weeks) | 23 (0.050) | 9 (0.020) | 9.61E-03 |
The variables shown in this table were used as background in the likelihood ratio tests of association of MDD with expression levels of genes or pathways (Figure S6); ten expression principal components and five genotypic principal components were also included. P-values were computed by Fisher’s exact tests (binary variables) or Spearman’s rank correlation tests (non-binary variables). Note that some of the medication categories are partially overlapping. Table S2 shows levels of association of each of these variables (and of genotypic principal components) with expression principal components.
Association of expression of gene pathways
Association of gene pathways was analyzed using two methods, based on all 1,325 canonical pathways from MSigDB (23) (c2.cp.v3.1) that contained 5-100 expressed genes in our data. Gene Set Enrichment Analysis (GSEA) (24) assessed the significance of each pathway using 5,000 permutations of MDD status for each pathway, where the log(LRT p-values) was used as the score for each gene. Hypergeometric tests were used to evaluate over-representation of pathways among subsets of the top N genes (ranked by association p-value) compared to all expressed genes. To ensure the robustness of this result, we repeated the analysis with varying N (N={30, 60, 100, 150, 300, 500}). FDR was used for multiple hypothesis correction (18), accounting for the number of assessed pathways.
Association of MDD with genetic variation and relationship with gene expression
Association between MDD and each genotyped SNP was evaluated using a standard logistic regression test, adjusted for five genotypic PCs (Figure S4). We also looked up genotypic association p-values in the independent Psychiatric Genomics Consortium (PGC) MDD GWAS (9240 cases and 9519 controls) (3). In a joint analysis of gene expression and genotypes, we identified genes whose genetic effect on MDD may be mediated through altering gene expression. Here, we used as a test statistic the least significant of two p-values for association of MDD with (i) expression of that gene; and (ii) genotype of its strongest expression quantitative trait locus (eQTL) SNP (Supplementary Methods). By evaluating the maximum (the least significant) of these two p-values as a single test statistic, this analysis identifies relationships where both expression and genotype support the association between the gene and MDD. Significance was evaluated by computing this statistic for 1,000,000 permutations of MDD status for each gene. To derive stable estimates of p-values, a Weibull extreme value distribution was fit to the permutation test-statistics and used as the null distribution to estimate the probability of observing the statistic seen in the real data.
Clinical variables
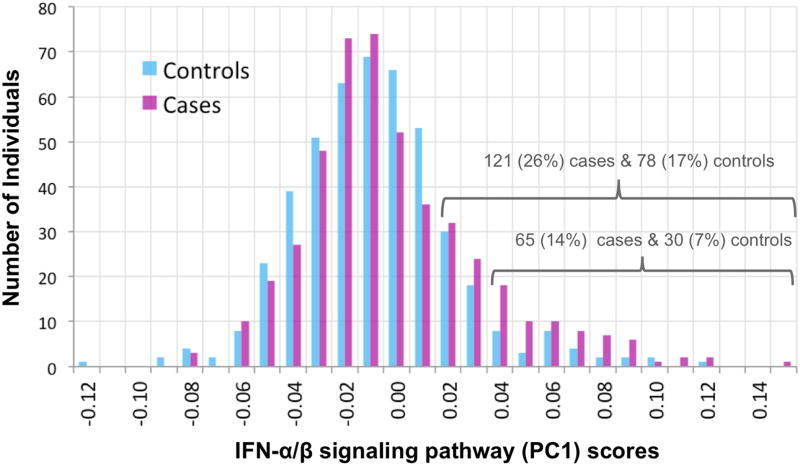
Factor scores (Principal Components Analysis) were computed separately from clinical variables and childhood trauma questionnaire responses, and association of these scores with interferon α/β signaling pathway (PC1) scores (Figure 1 legend) was analyzed by ANOVA, corrected for age and sex (Supplementary Methods).
Figure 1. IFN-α/β signaling pathway PC1 scores.
Shown are the distributions of values of a score that summarizes expression levels of genes in the interferon -α/β signaling pathway for cases (magenta) and controls (cyan). Each bar indicates the number of individuals with a score between the X-axis value and the next higher value. Scores were computed as PC1 from Principal Components Analysis of normalized read counts for the 20 genes (shown in Table 3) in the pathway with p<0.05 for association with MDD individually (among the 49 genes, of 64 in the pathway, that passed the inclusion criterion of ≥10 total reads in ≥100 individuals). GSEA identified a significant association (0.05 FDR) between interferon α/β pathway gene expression and MDD (note that GSEA uses expression data for all genes, and not the summary PC1 score which is shown in this figure). There is an excess of cases with higher scores, as shown by the numbers over the brackets. Raw read counts were initially corrected for technical and biological covariates (Table S1: specimen-specific sequencing variables, RNA quality, white cell type proportion estimates, time of blood draw). Analysis of case-control difference included additional covariates (see Methods) including medication and substance classes seen in ≥30 subjects (Tables 2 and S2). Case scores were not predicted by clinical variables or childhood trauma scores (Supplementary Results). Case-control differences and enrichment of top gene subsets for this pathway were not explained by excluding 92 individuals with rarer medical diagnoses or medications, estimating white cell type proportions by a second method, controlling for intake of three anti-depressant classes, or controlling for substance abuse/dependence or steroid medications.
Results
Association of single genes with MDD status
Analysis of single genes did not identify a genome-wide significant association (p < 3.6e-6, correcting for 13,857 genes). An excess of small p-values was observed (Figure S7). We quantified this excess by estimating the proportion of true positive tests (π1=0.13) (22), which indicated that expression levels of many genes are modestly associated with MDD. Additionally, we performed a power analysis to estimate the theoretical effect size with likelihood ratio tests given the current cohort size (25). We estimated that we have 80% power to detect genome-wide significant p-values (<3.6e-6) for individual genes with an expected estimated odds ratio of >=1.6 (log fold change of ~0.5). This analysis suggests that, should there be a true association from blood expression profiles, the odds ratio of is likely <1.6. At a less stringent threshold (0.25 FDR), there were 29 associated genes (Table 2), with biological functions including innate immune processes, vesicle trafficking, cell cycle regulation and splicing. The top two genes were MINOS1 (organization of mitochondrial inner membrane, p < 5e-6) and COPG (part of the Coatomer complex involved in vesicle trafficking between ER and Golgi, p < 8e-6.
Table 2. Top MDD-associated genes at 0.25 FDR.
| Gene Name | p-value | Direction | Function |
|---|---|---|---|
| MINOS1 | 5.00E-06 | − | Mitochondrial component |
| COPG | 8.00E-06 | + | Vesicle trafficking (Golgi-to-ER) |
| SYF2 | 2.93E-05 | − | Splicing factor |
| EHMT2 | 1.26E-04 | + | Histone methylation |
| UBAP2L | 1.26E-04 | + | Transcription activation/acetylation |
| MAFK | 1.26E-04 | − | Transcription activation/repressor |
| MX1 | 1.26E-04 | + | Immune function |
| C1ORF101 | 1.26E-04 | + | (not available) |
| SNRNP35 | 1.26E-04 | − | Splicing factor |
| FAM120AOS | 1.47E-04 | − | (not available) |
| NOP2 | 1.48E-04 | + | Cell cycle regulation |
| SRSF5 | 1.64E-04 | − | Splicing factor |
| TNFRSF10B | 2.52E-04 | + | Apoptosis, immune function |
| PIPOX | 2.52E-04 | + | (not available) |
| OAS1 | 2.52E-04 | + | Immune function |
| RABEPK | 2.52E-04 | + | Vesicle trafficking |
| SEMA3C | 2.52E-04 | + | Axon guidance |
| CINP | 2.52E-04 | − | Cell cycle regulation |
| H6PD | 3.78E-03 | − | ER, converting cortisone to cortisol |
| CCNI | 3.78E-03 | − | Cell cycle regulation |
| CROCC | 3.78E-03 | − | Cell cycle |
| IFIT3 | 3.78E-03 | + | Immune function |
| MRFAP1 | 3.78E-03 | − | Cell cycle regulation |
| STIP1 | 5.04E-04 | + | Axon guidance |
| MORC3 | 5.04E-04 | + | RNA binding |
| SDK1 | 5.04E-04 | − | Axon guidance |
| RGPD1 | 5.04E-04 | + | RNA binding |
| NXT1 | 5.04E-04 | + | RNA binding and transport |
| PACSIN1 | 5.04E-04 | + | Immune function, vesicle trafficking |
The table shows the top 29 genes (0.25 FDR) associated with MDD status. “Direction indicates whether the association was with increased (+) or decreased (-) expression.
Pathway enrichment analysis
In analyses of the enrichment of pathways among sets of top genes, all tested subsets of 60-500 genes were significantly enriched (0.05 FDR) for one pathway: interferon (IFN) α/β signaling (REACTOME) (Figures 1, S8, Tables 3, 4 and S7). GSEA also identified only this pathway at 0.05 FDR by permutation tests (un-corrected p = 2.5e-5, Bonferroni-correct p < 0.05). Of the 64 genes in this pathway annotated in MSigDB (23), 49 were adequately expressed here, of which 20 had a nominal p < 0.05 (Table 4). Overexpression was observed for 34 of the 49 genes including 19 of the 20 nominally significant genes (Tables 4 and S7). No clinical variable was observed to predict IFN pathway (PC1) scores in cases (see Figure 1 legend). (See Supplementary Data1 for the list of top 10 pathways).
Table 3. Enrichment p-values for association of IFN α/β signaling pathway and MDD.
| Pathway | N=30 | N=60 | N=100 | N=150 | N=200 | N=300 | N=500 |
|---|---|---|---|---|---|---|---|
| Interferon α/β signaling pathway | 0.25 FDR | 1.0E-06 | 7.0E-07 | 3.0E-11 | 2.0E-11 | 3.0E-13 | 3.0E-16 |
Using the hypergeometric test, over-representation of pathways among the top N associated genes was assessed for significance. The table shows results for the significantly over-represented pathway at 0.05 FDR (see Supplementary Table S7 for complete list of genes and single gene association p-values).
Table 4. IFN α/β signaling pathway genes with the strongest associations with MDD.
| Gene Name | p-value | Rank among all genes | Direction |
|---|---|---|---|
| MX1 | 1.26E-04 | 7 | + |
| OAS1 | 2.52E-04 | 15 | + |
| IFIT3 | 3.78E-04 | 22 | + |
| PTPN6 | 1.26E-03 | 43 | + |
| ADAR | 1.26E-03 | 45 | + |
| IRF7 | 1.64E-03 | 53 | + |
| IFIT1 | 3.15E-03 | 100 | + |
| USP18 | 3.78E-03 | 121 | + |
| ISG15 | 3.78E-03 | 122 | + |
| OAS2 | 4.16E-03 | 127 | + |
| IRF8 | 6.93E-03 | 185 | + |
| IFIT2 | 7.30E-03 | 201 | + |
| OAS3 | 7.56E-03 | 208 | + |
| MX2 | 8.44E-03 | 226 | + |
| IRF9 | 1.07E-02 | 274 | + |
| IFI6 | 1.13E-02 | 285 | + |
| OASL | 1.44E-02 | 352 | + |
| XAF1 | 1.49E-02 | 362 | + |
| IFI35 | 1.65E-02 | 404 | + |
| IFNAR2 | 4.99E-02 | 1030 | − |
The table shows the top twenty genes (those associated with nominal p-value < 0.05) in the interferon α/β signaling pathway, along with p-values, direction of effect, and association rank among all expressed genes.
Post-hoc re-assessment of potential confounders
Possible sources of spurious association with the interferon α/β signaling pathway were explored in post hoc analyses:
Unusual medication factors. We excluded 91 additional subjects with medications or illnesses with any potential impact on the immune system that were too rare for individual adjustment (Table S4, e.g., insulin, histamine-2 or leukotriene antagonists, antibiotics, immune suppressants; autoimmune diagnoses). Enrichment of interferon α/β signaling pathway among subsets of 60-500 genes remained significant (0.05 FDR) (Table S3).
Cell type proportions. Initial normalization of the data removed the effect of inferred cell type proportions (Supplementary Methods), and as expected, we did not observe any correlation between these estimates and the residual expression levels of interferon pathway genes. (In raw data, IFN signaling is most strongly correlated with the proportion of activated dendritic cells, but that proportion is not predicted by MDD status.) As an additional check, we used an alternative computational method (ridge regression instead of NNLS) to estimate cell-type proportions. The enrichment of interferon α/β signaling pathway among the top 100, 300, and 500 genes remained significant (0.05 FDR), after accounting for re-estimated cell-type proportions in the LRT (Supplementary Results).
Anti-depressants. In cases, we computed LRT p-values for association of all gene expression levels with three separate antidepressant classes taken by ≥30 individuals: serotonin reuptake inhibitors, serotonin-norepinephrine reuptake inhibitors, bupropion (Table S3, S5). Of the 20 nominally-significant interferon signaling genes (Table 4), none was among the top 300 genes associated with any of these classes, and no enrichment for this pathway (p>.5) was observed in sets of top 30-500 associated genes (whereas 17 of the top IFN signaling genes were among the top 300 in the primary analysis of association with MDD).
Drugs of abuse and steroids. Enrichment analyses were repeated (Table S3) with additional covariates to account for substance dependence (derived from covariates, Supplementary Methods) and for Fagerstrom nicotine dependence score; and for classes of steroid medications (primarily inhaled steroids for asthma or allergies). Interferon α/β signaling genes were enriched (0.05 FDR) in the subsets of top 100-500 genes, after accounting for these covariates in the LRT.
Association of genetic variation with MDD status
One common SNP showed genome-wide significant genotypic association with MDD (rs11232553, chr11:80,941,531; p<3e-8; 1.8MB downstream of MIR708 and 660 KB upstream of MIR4300), but no association for this SNP was observed in the much larger PGC MDD dataset (Supplementary Results). The joint analysis that combines expression data with genotype (Methods) identified one genome-wide significant gene, CINP --- 0.05 FDR; uncorrected p=2.5e-4 for expression level alone, 3.2e-04 for genotypic association of its top eQTL [rs2896439], 1e-6 for the combined test. A modest association of rs2896439 in the PGC cohort (p=0.035) suggests caution in interpreting our result. Three other genes achieved FDR< 0.1: SCAI (cell migration and regulation of cell cycle), SDK1 (axon guidance and implicated in HIV-associated nephropathy) and RABEPK (endosome to Golgi trafficking). We did not detect evidence of a relationship between genetic variation affecting interferon pathway genes and MDD in either analysis (joint analysis or genotype-only) of this cohort, or in a targeted analysis of the PGC dataset (Supplementary Results).
Discussion
Type I Interferon signaling and MDD
Significant association was observed between MDD and expression of interferon α/β signaling pathway genes in a large population-based sample, using whole-blood RNA reflecting the physiological state at the time of blood draw. Primary analyses removed the effects of a large set of technical, biological, medical and hidden covariates, and post hoc analyses did not identify measured factors that explained the finding.
IFN-α and IFN-β are type I interferons (IFN-I), the main cytokines of the innate immune system that respond primarily to viral infection and to malignant cells. They activate genes that interfere with viral replication, activate other immune responses to infection and inhibit cell growth (26). A “weak” (chronic) signaling mode, that is observed in the absence of known pathogens, may increase the efficiency of response to stimuli, but may also play a role in autoimmune and neuroinflammatory disorders (27). The binding of IFN-I to interferon receptors (IFNAR) can activate two main transcriptional complexes: IFN-α-activated-factor (AAF), which mainly mediates activation of other cytokines, and IFN-stimulated gene factor 3 (ISGF3), which mainly mediates antiviral activity. Here, we observed the upregulation of ISGF3 induced genes in MDD cases (Table 4), including IRF9, a component of the ISGF3 complex (28).
Increased IFN-I signaling in MDD is consistent with previous data implicating dysregulation of cytokines in depression. The most direct observation is that patients receiving IFN-I therapies (IFN-α for hepatitis C or IFN-β for multiple sclerosis) often develop clinically significant depression (29-32). Previous studies have associated changes in secreted cytokines and inflammatory markers, including IFN-I, (33, 34)with a reduction in tryptophan, the precursor of serotonin, in both cerebral spinal fluid (CSF) (34) and plasma (35).
More broadly, increased IFN-I signaling is relevant to a set of inter-related findings regarding the role of immune system in the pathophysiology of MDD (reviewed by Zunszain et al. (36)): (i) Glucocorticoid dysregulation: Hypersecretion of cortisol (stimulated by corticotropin releasing factor, CRF) is observed in MDD, possibly caused or mediated by glucocorticoid receptor (GR) resistance (36). (ii) Immune/inflammatory dysregulation: Levels of circulating inflammatory cytokines are increased, particularly tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6) (37-39). Also, plasma levels of a non-specific inflammation marker, C-reactive protein, predicted depression in a Danish sample of over 73,000 individuals (40). (iii) Hippocampal volume is reduced with longer durations of MDD(41), due either to cell loss (implicating inflammation-related apoptosis (36, 42) ) or to reduced cellular volume (36).
Existing studies suggest multiple hypotheses concerning the causal relationships among these findings. An initiating factor could be stress-induced glucocorticoid secretion with resulting neuroinflammation, cytokine production and apoptosis (43). Alternatively, dysregulated cytokine production could directly cause depression (perhaps mediated by tryptophan depletion) and also increase GR resistance (44). Indeed it has been proposed that sequence variants which predispose to depression in modern society may have been useful ancient adaptations to pathogens (45). It is reasonable to assume that there are multiple causal inter-relationships, reflecting a high degree of phenotypic and etiologic heterogeneity in MDD.
The finding of increased expression levels of IFN-I signaling pathway in this study is consistent with all of the above hypotheses. It could represent a key causal or mediating factor, and/or the end-result of a complex process. However, a third possibility is that increased expression of IFN-I signaling is caused by a confounding factor that is downstream of, or simply correlated with MDD. We did not find genetic variants impacting IFN pathway genes (either protein-coding variants or known eQTLs) to be significantly associated with MDD in this cohort. Note there are multiple IFN-I gene subtypes in a cluster on chromosome 9, and all were insufficiently expressed for analysis in this study. They have highly homologous sequences, so that many sequencing reads in this region were excluded here because we counted only uniquely-mapped reads.
We also should not overlook the possibility that, as the primary function of the IFN-I signaling system is response to viral infection, a subset of MDD patients could have unrecognized chronic and/or active infections, whose effects could be influenced by many genetic sequence variants and by other environmental factors. This hypothesis cannot be directly addressed by the present data, but suggests an interesting future research direction.
Comparison with previous studies
MDD transcriptome studies of postmortem brain tissue have suggested alterations in glutamatergic and GABA-ergic pathways (46)and in synaptic genes relevant to the anatomical finding of reduced dendritic spines (47). Interpretation is complicated by confounding factors (postmortem changes and drug exposures) and small samples (typically fewer than 30 cases).
Previous blood cell gene expression studies—typically targeting 1-15 genes in samples of 25-100 MDD cases—have reported expression increases for cytokine and glucocorticoid-related genes and reductions for GR and neuroplasticity-related genes such as BDNF (48). One study reported increased blood expression levels of IFN-α1, IFN-α2 and IFN-β1 in 22 MDD cases and 11 controls (31). There are two reports from a larger study that included microarray whole-transcriptome and GWAS data for 215 MDD cases and their relatives (San Antonio Family Heart Study): RNF123 was the most significant gene in a bivariate linkage analysis of MDD and gene expression values (p>0.5 here) (49); and DISC1 SNPs showed genotypic association to MDD (50) (p=0.025 for DISC1 here but with increased expression, while previous studies predict reduced expression of DISC1). Whole-transcriptome association results have not been reported to date for that study. Interestingly, a recent study of subjective social isolation reported dysregulation of pro- and anti-inflammatory genes, including down-regulation of interferon I signaling pathway in lonely individuals (51, 52); we note that the impact of other unmeasured factors that modulate the interferon response (for example, age differences in the cohorts) may underlie the apparent discrepancy in the direction of effect of interferon I signaling.
Meta-analysis of studies of serum cytokine levels in MDD suggests significant increases in TNF-α and IL-6 (37); here, overexpression of TNFRSF10B was the thirteenth strongest individual gene finding (Table 2), while overexpression of IL-6 was not observed, although the strongest IL-6 findings have been in CSF(34). There have been studies of IFN-λ (without significant findings overall (37)), but not type I interferons. We were unable to measure serum biomarkers in the present study because the study was designed to collect a large national sample, and this design did not permit obtaining and then immediately centrifuging and preserving serum.
CINP and interferon
Joint analysis of association of MDD to expression levels and to genotypes detected an association with CINP (cyclin-dependent kinase 2 interacting protein). The antiviral component of interferon signaling results in cell-cycle arrest and inhibition of cell growth by inhibiting cyclin-dependent kinases (9), and specifically CDK2(53). CINP interacts with CDK2 as a component of the DNA damage integrity checkpoint, and its suppression has been linked to cell cycle arrest (8). As noted above, however, support for this association is very modest in the larger PGC MDD cohort.
Strengths and limitations
Strengths of this study design include deep RNA-seq data; a large population-based sample with a representative female:male ratio (2:1), including medicated and unmedicated cases; a relevant tissue type for immunological mechanisms; and correction for multiple measured and unmeasured covariates. Limitations include use of non-brain tissue; the use of mixed white blood cell types (as opposed to cell sorted data) and not having cell count measurements; and case-control differences in many physiological covariates (Table 1, S2). The detection of a single associated pathway, despite an excess of modestly significant p-values, also suggests that (as in GWAS) larger samples are needed. Our reliance on log-linear models to account for covariates could have left residual correlation due to non-linear effects. Also, we only adjusted for covariates relevant to at least 30 subjects in the primary analysis. Under-reporting of medical information by cases could have produced spurious results given our reliance on self-report (although none of those variables were associated with IFN-I signaling). IFN-I signaling could have been influenced by common viral infections unrelated to MDD, although these are seasonal and case status was not related to order of blood collection. We used two statistical estimates of white cell type proportions based on expression signatures from microarray data, but these estimates might not have been sufficiently accurate, and they were based on microarray measurements whereas our data is based on RNA-seq.
Finally, results were partially dependent on our approach to removing hidden confounding factors by accounting for expression PCs, which has been shown to produce more accurate estimations of eQTLs (20, 54) and of functional gene interactions (21). To avoid over-fitting, we removed the top 10 PCs based only on explained variance (Figure S6), ignoring MDD status.
In a post hoc assessment, we do observe weaker (non-significant) associations between the interferon pathway and MDD status without removing the effect of top expression PCs. While this indicates some sensitivity of our analysis to modeling choices, we have observed that the top expression PCs are correlated with confounding factors including age (Supplementary Figure S9, S10), and thus it is likely that the chosen procedure yields more meaningful results.
Conclusion
We identified a significant association between MDD and increased expression of genes involved in interferon α/β signaling. This supports hypotheses of dysregulated cytokine activity in MDD, but gene expression data cannot resolve critical cause-effect relationships. Increased IFN-I signaling could be a direct cause of depression, resulting from some combination of genetic sequence variants, psychological stress, or normal or abnormal responses to unknown viral infection or other physiological stressors. Increased signaling could also be down-stream of depression, or simply based on correlation of IFN-I signaling with unknown confounding variables that are correlated with depression. There are also plausible hypotheses (with several possible directionalities of effects) involving an interaction between immune dysregulation and increased resistance to and release of glucocorticoids. There is a need for studies that simultaneously measure clinical variables, genotypes, gene expression, viral sequences or antibodies, and proteins related to immune function and glucocorticoid dynamics.
Supplementary Material
Acknowledgments
This work was supported by NIH/NIMH Grants 5RC2MH089916 (DFL, PI; MMW, JBP, DK, AEU, Co-Investigators) and 3R01MH090941 (DK, Co-Investigator).
Footnotes
Data Access Data from this study are available to qualified scientists from the NIMH Center for Collaborative Genomic Studies on Mental Disorders, including raw and aligned RNAseq data, raw and normalized read counts per gene, and covariate data. Scientists who are interested in requesting data should consult https://www.nimhgenetics.org/access_data_biomaterial.php, which provides specific instructions about how to request data, including the name and email address of the program official who is responsible for handling requests. When inquiring about data access, please reference this study as the Depression Genes and Networks study (D. Levinson, PI).
Conflict of interest CM is an employee of Illumina, Inc. CDH was employed by Illumina, Inc. for a portion of the study and by Personalis for a portion. RM is employed by Centrillion Biosciences, Inc.
Supplementary Information is included (to be linked to the online version of the paper from Molecular Psychiatry’s website)
References
- 1.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. The American journal of psychiatry. 2000;157(10):1552–62. doi: 10.1176/appi.ajp.157.10.1552. Epub 2000/09/29. [DOI] [PubMed] [Google Scholar]
- 2.Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance--a population-based twin study. Diabetologia. 1999;42(2):139–45. doi: 10.1007/s001250051131. Epub 1999/03/04. [DOI] [PubMed] [Google Scholar]
- 3.Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Molecular psychiatry. 2013;18(4):497–511. doi: 10.1038/mp.2012.21. Epub 2012/04/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rush AJ. The varied clinical presentations of major depressive disorder. The Journal of clinical psychiatry. 2007;68(Suppl 8):4–10. Epub 2007/07/31. [PubMed] [Google Scholar]
- 5.Sullivan PF, de Geus EJ, Willemsen G, James MR, Smit JH, Zandbelt T, et al. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Molecular psychiatry. 2009;14(4):359–75. doi: 10.1038/mp.2008.125. Epub 2008/12/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.A mega-analysis of genome-wide association studies for major depressive disorder. Molecular psychiatry. 2012 doi: 10.1038/mp.2012.21. Epub 2012/04/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klengel T, Binder EB. Gene-environment interactions in major depressive disorder. Canadian journal of psychiatry Revue canadienne de psychiatrie. 2013;58(2):76–83. doi: 10.1177/070674371305800203. Epub 2013/02/28. [DOI] [PubMed] [Google Scholar]
- 8.Grishina I, Lattes B. A novel Cdk2 interactor is phosphorylated by Cdc7 and associates with components of the replication complexes. Cell Cycle. 2005;4(8):1120–6. Epub 2005/08/06. [PubMed] [Google Scholar]
- 9.Sangfelt O, Erickson S, Castro J, Heiden T, Gustafsson A, Einhorn S, et al. Molecular mechanisms underlying interferon-alpha-induced G0/G1 arrest: CKI-mediated regulation of G1 Cdk-complexes and activation of pocket proteins. Oncogene. 1999;18(18):2798–810. doi: 10.1038/sj.onc.1202609. Epub 1999/06/11. [DOI] [PubMed] [Google Scholar]
- 10.Kessler RC, Andrews G, Mroczek D, Ustun B, Wittchen H. The World Health Organization Composite International Diagnostic Interview short-form (CIDI-SF) International Journal of Methods in Psychiatric Research. 1998;7(4):171–85. [Google Scholar]
- 11.Kroenke K, Spitzer RL, Williams JB. The PHQ-9: validity of a brief depression severity measure. Journal of general internal medicine. 2001;16(9):606–13. doi: 10.1046/j.1525-1497.2001.016009606.x. Epub 2001/09/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spitzer RL, Kroenke K, Williams JB, Lowe B. A brief measure for assessing generalized anxiety disorder: the GAD-7. Archives of internal medicine. 2006;166(10):1092–7. doi: 10.1001/archinte.166.10.1092. Epub 2006/05/24. [DOI] [PubMed] [Google Scholar]
- 13.Debey-Pascher S, Hofmann A, Kreusch F, Schuler G, Schuler-Thurner B, Schultze JL, et al. RNA-stabilized whole blood samples but not peripheral blood mononuclear cells can be stored for prolonged time periods prior to transcriptome analysis. The Journal of molecular diagnostics : JMD. 2011;13(4):452–60. doi: 10.1016/j.jmoldx.2011.03.006. Epub 2011/06/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. Epub 2009/03/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anders S. HTSeq: Analysing high-throughput sequencing data with Python. 2010 doi: 10.1093/bioinformatics/btac166. http://www-huberemblde/users/anders/HTSeq/doc/overviewhtml. [DOI] [PMC free article] [PubMed]
- 16.Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z, Clark HF. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PloS one. 2009;4(7):e6098. doi: 10.1371/journal.pone.0006098. Epub 2009/07/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes and immunity. 2005;6(4):319–31. doi: 10.1038/sj.gene.6364173. Epub 2005/03/25. [DOI] [PubMed] [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57(1):289–300. [Google Scholar]
- 19.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38(8):904–9. doi: 10.1038/ng1847. Epub 2006/07/25. [DOI] [PubMed] [Google Scholar]
- 20.Stegle O, Parts L, Durbin R, Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS computational biology. 2010;6(5):e1000770. doi: 10.1371/journal.pcbi.1000770. Epub 2010/05/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furlotte NA, Kang HM, Ye C, Eskin E. Mixed-model coexpression: calculating gene coexpression while accounting for expression heterogeneity. Bioinformatics. 2011;27(13):i288–94. doi: 10.1093/bioinformatics/btr221. Epub 2011/06/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(16):9440–5. doi: 10.1073/pnas.1530509100. Epub 2003/07/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40. doi: 10.1093/bioinformatics/btr260. Epub 2011/05/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. Epub 2005/10/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shieh G. On power and sample size calculations for likelihood ratio tests in generalized linear models. Biometrics. 2000;56(4):1192–6. doi: 10.1111/j.0006-341x.2000.01192.x. Epub 2000/12/29. [DOI] [PubMed] [Google Scholar]
- 26.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nature reviews Immunology. 2005;5(5):375–86. doi: 10.1038/nri1604. Epub 2005/05/03. [DOI] [PubMed] [Google Scholar]
- 27.Taniguchi T, Takaoka A. A weak signal for strong responses: interferon-alpha/beta revisited. Nature reviews Molecular cell biology. 2001;2(5):378–86. doi: 10.1038/35073080. Epub 2001/05/02. [DOI] [PubMed] [Google Scholar]
- 28.Wang BX, Fish EN. The yin and yang of viruses and interferons. Trends in immunology. 2012;33(4):190–7. doi: 10.1016/j.it.2012.01.004. Epub 2012/02/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Udina M, Castellvi P, Moreno-Espana J, Navines R, Valdes M, Forns X, et al. Interferon-induced depression in chronic hepatitis C: a systematic review and meta-analysis. The Journal of clinical psychiatry. 2012;73(8):1128–38. doi: 10.4088/JCP.12r07694. Epub 2012/09/13. [DOI] [PubMed] [Google Scholar]
- 30.Schaefer M, Engelbrecht MA, Gut O, Fiebich BL, Bauer J, Schmidt F, et al. Interferon alpha (IFNalpha) and psychiatric syndromes: a review. Progress in neuro-psychopharmacology & biological psychiatry. 2002;26(4):731–46. doi: 10.1016/s0278-5846(01)00324-4. Epub 2002/08/22. [DOI] [PubMed] [Google Scholar]
- 31.Schlaak JF, Trippler M, Hoyo-Becerra C, Erim Y, Kis B, Wang B, et al. Selective hyper-responsiveness of the interferon system in major depressive disorders and depression induced by interferon therapy. PloS one. 2012;7(6):e38668. doi: 10.1371/journal.pone.0038668. Epub 2012/06/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felger JC, Cole SW, Pace TW, Hu F, Woolwine BJ, Doho GH, et al. Molecular signatures of peripheral blood mononuclear cells during chronic interferon-alpha treatment: relationship with depression and fatigue. Psychological medicine. 2012;42(8):1591–603. doi: 10.1017/S0033291711002868. Epub 2011/12/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raison CL, Borisov AS, Woolwine BJ, Massung B, Vogt G, Miller AH. Interferon-alpha effects on diurnal hypothalamic-pituitary-adrenal axis activity: relationship with proinflammatory cytokines and behavior. Molecular psychiatry. 2010;15(5):535–47. doi: 10.1038/mp.2008.58. Epub 2008/06/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raison CL, Borisov AS, Majer M, Drake DF, Pagnoni G, Woolwine BJ, et al. Activation of central nervous system inflammatory pathways by interferon-alpha: relationship to monoamines and depression. Biol Psychiatry. 2009;65(4):296–303. doi: 10.1016/j.biopsych.2008.08.010. Epub 2008/09/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes MM, Carballedo A, McLoughlin DM, Amico F, Harkin A, Frodl T, et al. Tryptophan depletion in depressed patients occurs independent of kynurenine pathway activation. Brain, behavior, and immunity. 2012;26(6):979–87. doi: 10.1016/j.bbi.2012.05.010. Epub 2012/06/12. [DOI] [PubMed] [Google Scholar]
- 36.Zunszain PA, Anacker C, Cattaneo A, Carvalho LA, Pariante CM. Glucocorticoids, cytokines and brain abnormalities in depression. Progress in neuro-psychopharmacology & biological psychiatry. 2011;35(3):722–9. doi: 10.1016/j.pnpbp.2010.04.011. Epub 2010/04/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dowlati Y, Herrmann N, Swardfager W, Liu H, Sham L, Reim EK, et al. A meta-analysis of cytokines in major depression. Biol Psychiatry. 2010;67(5):446–57. doi: 10.1016/j.biopsych.2009.09.033. Epub 2009/12/18. [DOI] [PubMed] [Google Scholar]
- 38.Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nature reviews Neuroscience. 2008;9(1):46–56. doi: 10.1038/nrn2297. Epub 2007/12/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller AH, Maletic V, Raison CL. Inflammation and its discontents: the role of cytokines in the pathophysiology of major depression. Biological psychiatry. 2009;65(9):732–41. doi: 10.1016/j.biopsych.2008.11.029. Epub 2009/01/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wium-Andersen MK, Orsted DD, Nielsen SF, Nordestgaard BG. Elevated C-reactive protein levels, psychological distress, and depression in 73, 131 individuals. JAMA Psychiatry. 2013;70(2):176–84. doi: 10.1001/2013.jamapsychiatry.102. Epub 2012/12/26. [DOI] [PubMed] [Google Scholar]
- 41.McKinnon MC, Yucel K, Nazarov A, MacQueen GM. A meta-analysis examining clinical predictors of hippocampal volume in patients with major depressive disorder. Journal of psychiatry & neuroscience : JPN. 2009;34(1):41–54. Epub 2009/01/07. [PMC free article] [PubMed] [Google Scholar]
- 42.Nadeau S, Rivest S. Glucocorticoids play a fundamental role in protecting the brain during innate immune response. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23(13):5536–44. doi: 10.1523/JNEUROSCI.23-13-05536.2003. Epub 2003/07/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heim C, Newport DJ, Mletzko T, Miller AH, Nemeroff CB. The link between childhood trauma and depression: insights from HPA axis studies in humans. Psychoneuroendocrinology. 2008;33(6):693–710. doi: 10.1016/j.psyneuen.2008.03.008. Epub 2008/07/08. [DOI] [PubMed] [Google Scholar]
- 44.Leonard B, Maes M. Mechanistic explanations how cell-mediated immune activation, inflammation and oxidative and nitrosative stress pathways and their sequels and concomitants play a role in the pathophysiology of unipolar depression. Neuroscience and biobehavioral reviews. 2012;36(2):764–85. doi: 10.1016/j.neubiorev.2011.12.005. Epub 2011/12/27. [DOI] [PubMed] [Google Scholar]
- 45.Raison CL, Miller AH. The evolutionary significance of depression in Pathogen Host Defense (PATHOS-D) Molecular psychiatry. 2013;18(1):15–37. doi: 10.1038/mp.2012.2. Epub 2012/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, et al. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PloS one. 2009;4(8):e6585. doi: 10.1371/journal.pone.0006585. Epub 2009/08/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang HJ, Voleti B, Hajszan T, Rajkowska G, Stockmeier CA, Licznerski P, et al. Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nature medicine. 2012;18(9):1413–7. doi: 10.1038/nm.2886. Epub 2012/08/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hepgul N, Cattaneo A, Zunszain PA, Pariante CM. Depression pathogenesis and treatment: what can we learn from blood mRNA expression? BMC medicine. 2013;11:28. doi: 10.1186/1741-7015-11-28. Epub 2013/02/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glahn DC, Curran JE, Winkler AM, Carless MA, Kent JW, Jr, Charlesworth JC, et al. High dimensional endophenotype ranking in the search for major depression risk genes. Biological psychiatry. 2012;71(1):6–14. doi: 10.1016/j.biopsych.2011.08.022. Epub 2011/10/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carless MA, Glahn DC, Johnson MP, Curran JE, Bozaoglu K, Dyer TD, et al. Impact of DISC1 variation on neuroanatomical and neurocognitive phenotypes. Molecular psychiatry. 2011;16(11):1096–104. 63. doi: 10.1038/mp.2011.37. Epub 2011/04/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(7):3080–5. doi: 10.1073/pnas.1014218108. Epub 2011/02/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cole SW, Hawkley LC, Arevalo JM, Sung CY, Rose RM, Cacioppo JT. Social regulation of gene expression in human leukocytes. Genome biology. 2007;8(9):R189. doi: 10.1186/gb-2007-8-9-r189. Epub 2007/09/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie RL, Gupta S, Miele A, Shiffman D, Stein JL, Stein GS, et al. The tumor suppressor interferon regulatory factor 1 interferes with SP1 activation to repress the human CDK2 promoter. The Journal of biological chemistry. 2003;278(29):26589–96. doi: 10.1074/jbc.M301491200. Epub 2003/05/07. [DOI] [PubMed] [Google Scholar]
- 54.Nica AC, Parts L, Glass D, Nisbet J, Barrett A, Sekowska M, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS genetics. 2011;7(2):e1002003. doi: 10.1371/journal.pgen.1002003. Epub 2011/02/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.