Abstract
Adult stem cells play a critical role in the maintenance of tissue homeostasis and prevention of aging. While the regenerative potential of stem cells with low cellular turnover, such as adipose-derived stem cells (ASC), is increasingly recognized, the study of chronological aging in ASCs is technically difficult, and remains poorly understood. Here, we employ our model of chronological aging in primary human ASCs to examine genome-wide transcriptional networks. We demonstrate first that the transcriptome of aging ASCs is distinctly more stable than that of age-matched fibroblasts, and further, that age-dependent modifications in cell cycle progression and translation initiation specifically characterize aging ASCs in conjunction with increased nascent protein synthesis and a distinctly shortened G1 phase. Our results reveal novel chronological aging mechanisms in ASCs that are inherently different from differentiated cells, and that may reflect an organismal attempt to meet the increased demands of tissue and organ homeostasis during aging.
Keywords: human ASC, adipose-derived stem cell, aging, transcriptome, translation initiation, cell cycle
The Role of Cell Cycle Control in Early Chronological Human Stem Cell Aging
Aging modulates transcription and translation in primary human ASCs resulting in altered regulation of cell cycle with shortened G1 phase. These changes reflect an organismal attempt to meet the increased demands of tissue and organ homeostasis during aging. The critical role played by cell cycle control serves as an important target for the prevention and treatment of aging and metabolic diseases.
Introduction
Aging is a complex process characterized by a progressive decline in physiological integrity secondary to impaired cellular function. Further, aging is the primary risk factor for the development of most adult diseases, including cancer and degenerative disorders [1]. Adult stem cells play a critical role in the maintenance of organismal health and the prevention of age-associated diseases and their chronological deterioration results in compromised regenerative capacities [2]. Unfortunately, most research efforts have been devoted to studying aging in differentiated cells and the mechanisms specific to stem cell aging have not been well characterized [3].
During aging, adult stem cells, unlike differentiated cells, must maintain a delicate balance between self-renewal and differentiation, the former linked to tumorigenesis and the latter to a diminished regenerative capacity [1, 2, 4]. Because the regulation of cellular processes frequently differ between stem cells and differentiated cells [5], it is likely that the mechanisms governing cellular process in aging stem cells are also distinct. Therefore, it is important to study how aging specifically affects stem cells to identify mechanisms that are unique to these cells. Furthermore, it is important to distinguish between two classes of adult stem cells, defined by either a high or low cell turnover. Notably, most studies of stem cell aging examine the mechanisms of replicative aging in stem cells with high cell turnover as opposed to normal chronological aging. Yet, the therapeutic potential of stem cells with low cell turnover, such as adipose-derived stem cells (ASC), is increasingly recognized as potentially superior in part because of their low oncogenic potential.
Adipose-derived stem cells are adult mesenchymal stem cells that display similar differentiation properties to highly replicating bone marrow-derived stem cells (BMSCs) [6, 7]. Yet, ASCs offer multiple advantages over other stem cells for both investigational and therapeutic applications. First, ASCs can be easily isolated with high yield from subcutaneous adipose tissue and are readily expandable by in vitro culturing. Secondly, ASCs consistently maintain their concentration and proliferation rates with advancing age [8]. Finally, ASCs represent a more biologically relevant model with which to study the natural chronological mechanisms of aging, as compared to fast replicating stem cells that age largely by replicative exhaustion and senescence. Despite the benefits of studying aging in an ASC-based model, few studies have been conducted with primary human ASCs isolated from cohorts of healthy individuals. Multiple factors, including restricted access to human tissues and inter-sample heterogeneity, have previously rendered these studies challenging. As a consequence, little is known about natural chronological aging in primary human ASCs.
In this work, we studied chronological aging in primary human ASCs isolated from healthy patients via RNA sequencing (RNA-Seq) technology. Comparisons to age-matched dermal fibroblasts and replicative senescent IMR-90 cells revealed novel ASC-specific modifications during early chronological aging with more active transcriptional profiles of cell cycle and translation initiation pathways. Accordingly, aging ASCs demonstrated increased nascent protein synthesis and a shortened G1 phase that may reflect an organismal attempt to meet increased homeostatic demands during aging. Together, our results support a model in which ASC transcriptional integrity is largely maintained in aging human adipose tissue and reveal novel ASC-specific chronological aging mechanisms.
Materials and Methods
Tissue Procurement
Subcutaneous abdominal adipose tissues were excised from consented healthy female patients undergoing elective abdominoplasty (University of Pennsylvania IRB approval Protocol number 812150). The specimens were immediately transferred to the laboratory. The adipose tissues were dissected from skin and stored at −70°C in 50 ml conical tubes until ASC isolation. No cryopreservation or other agents were used in the freezing of the whole adipose tissue specimens. Skin specimens were processed for dermal fibroblasts without storage at −70°C.
ASC Isolation and Culturing
ASCs were isolated from 20 tissue samples of female individuals between 24 to 64 years, according to a standard collagenase protocol [8]. The isolated ASCs were cultured in Dulbecco’s Modified Eagle Medium/F12 (Gibco Life Technologies Co., Norwalk, CT) supplemented with 1% Penicillin/Streptomycin (Gibco Life Technologies Co.) and 10% FBS (Serum Source International, Charlotte, NC) at 37°C with 5% CO2. The culture media were changed every three days. All analyses were conducted with early passage ASCs (p<4).
Dermal Fibroblast Isolation and Culturing
Fibroblasts were isolated from 9 female donors ages 25 to 64, using the method described in [9]. The cells were cultured under 5% CO2 at 37°C, and culture media were changed every three days.
RNA-Sequencing
Approximately 500,000 ASCs and fibroblasts were isolated. Total RNA was extracted from the cells using TRIzol (Thermo Fisher Scientific, Grand Island, NY) and quality was examined by electrophoresis. Poly (A)+ RNA were isolated from the total RNA samples using the NEBNext Poly (A) mRNA Magnetic Isolation Module (New England Biolabs, Ipswich, MA). RNA-Seq libraries were generated for each sample using the NEBNext Ultra Directional RNA Library Prep Kit with NEBNext® Multiplex Oligos for Ilumina according to the manufacture’s protocol (New England Biolabs, Ipswich, MA). The libraries were pooled and sequenced on the NextSeq 500 desktop sequencer (Illumina Inc., San Diego, CA) (Please see supplemental methods for details).
RNA-Seq Data Analysis
Data were aligned to the human genome build 19 with STAR v2.4.1d, with index built using transcriptome information from ENSEMBL. Data were normalized at the read level, prior to quantification, using a resampling strategy PORT v0.8 (https://github.com/itmat/Normalization) (please see supplemental methods for details). Differential expression analysis was performed between the young and old groups (in pairwise comparisons) by calculating Mann-Whitney p-values for each gene and then performing a Benjamini-Hochberg correction for multiple testing, to produce q-values for each gene. The distribution of q-values was then compared between ASC and Fibroblasts. An equal number of samples were used for each tissue. Similarly, the distribution of T-Statistics between young and old was compared to the distribution of T-Statistics comparing two random sets of samples (referred to as permuted data in Results).
We utilized Ingenuity Pathway Analysis (IPA) software (Qiagen, CA) and Database for Annotation, Visualization and Integrated Discovery (DAVID) [10, 11] for pathway analysis. Multiexperiment Viewer (MeV) was used for hierarchical clustering analysis [12].
All data were deposited in the standard public repository GEO (the Gene Expression Omnibus). Both raw data and quantified spreadsheets are available. The GEO accession number is GSE86244.
Nascent Protein Synthesis Analysis
ASCs and fibroblasts were inoculated to 70% confluence in 48-well plates, and allowed to grow for 24 hr. Click-IT Plus OPP Alexa Fluor 488 Protein Synthesis Assay Kit (ThermoFisher Scientific, Grand Island, NY) was used, according to the manufacture’s instruction, to detect the incorporation of O-propargyl-puromycin. Fluorescent signals from both Alexa Fluor 488 and DAPI were quantified using the Synergy H1 Multi-Mode Reader (BioTek, Winooski, VT). Signal intensities of Alexa Fluor 488 incorporated in protein chains were normalized to the intensities of DAPI signal of cell nuclei. Representative images of cells incorporated Alexa Fluor 488 and DAPI were acquired using a Nikon Diaphot inverted microscope equipped with digital camera and the NIS-Elements Microscope Imaging software (Nikon Instruments Inc., Melville, NY).
Western Blot Analysis
Whole cell extracts were prepared from 80% confluent ASCs and fibroblasts by on-plate cell lysis with 2x Laemmli sample buffer containing protease and phosphatase inhibitors (Roche Diagnostics Corporation, Indianapolis, IN). Western blot analysis was performed as described before [13]. Primary antibodies utilized: rabbit polyclonal anti-pho-eIF2-α, anti-eIF2-α, anti-Tubulin (Cell Signaling Technology, Danvers, MA), and mouse monoclonal anti-RPL29 (Abacam, Cambridge, MA). Secondary antibodies utilized: HRP conjugated anti-rabbit IgG and HRP conjugated anti-mouse (Cell Signaling Technology). The blots were developed using SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific). Protein bands were visualized using Amersham Imager 600 (GE Healthcare Bio-Sciences). Intensity of each band was analyzed using Image J [14].
Indirect Immunofluorescence
Indirect immunofluorescence procedure was utilized as described in [15]. Briefly, cells were fixed, permeabilized and blocked before incubation with primary antibodies. To detect H3K9 and Nup 93, rabbit anti-Histone H3 (tri methyl K9) antibody (Abcam, Cambridge, MA) and rabbit anit-Nup93 antibody (Santa Cruz Biotechnology, Dallas, Texas) were used. Alexa 488 goat anti-rabbit (Thermo Fisher Scientific) were used as the secondary antibody and ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific) was used to label cell nuclei. Nikon flouresence microscope equipped with digital camera and the NIS-Elements Microscope Imaging software (Nikon Instruments Inc., Melville, NY) were used to acquire cell images.
Cell Cycle Analysis
ASCs and fibroblasts isolated from the young and old donors were analyzed for DNA content after ethanol fixation and stained with propidium iodide using a modified method described by Crissman and Steinkamp [16] (please see supplemental methods for details). The BD FACSCanto II system was used to collect fluorescence data. Cells were gated on forward scatter versus side scatter and fluorescence data were obtained from 20,000 events per sample. Histogram plots of fluorescence (DNA) and percentages of cells in different phases of the cell cycle were calculated using the ModFit LT (Verity Software House, Topsham, ME).
Results
Global Gene Expression Profiles of Aging ASCs Are Significantly More Stable than Those of Aging Fibroblasts and IMR-90 Cells
Twenty ASC samples and nine dermal fibroblast samples were obtained from females aged 24 to 64 (Table 1A). Genome-wide expression profiles of all samples were examined by RNA-Seq analyses. IMR-90 cells from early (p19) and late (p50) passages were included as a control for replicative aging and senescence. IMR-90 cells were derived from a human fetal lung fibroblast cell line and have been routinely used to study replicative senescence [17, 18] as these cells enter permanent cell cycle arrest after a limited number of cell divisions [19]. As shown in Table 1B, ASCs demonstrate the highest number of overall detected transcripts, IMR90 cells the lowest, and fibroblasts an intermediate number. However, if transcripts with low read counts (5 and less) are excluded from the analysis, the order is reversed. Only 42% of ASC transcripts demonstrate a read count >5, fibroblasts 46%, and IMR90 cells 61%. Therefore, ASCs exhibit an overall higher percentage of genes expressed at low levels as compared to differentiated cells.
Table 1A.
Sample Features.
| ASCs | Fibroblasts | ||
|---|---|---|---|
| Age (Young Group) | Age (Old Group) | Age (Young Group) | Age (Old Group) |
| 24 | 50 | 25 | 53 |
| 27 | 50 | 25 | 58 |
| 30 | 52 | 27 | 59 |
| 33 | 53 | 32 | 64 |
| 33 | 53 | 34 | |
| 34 | 55 | ||
| 35 | 61 | ||
| 37 | 63 | ||
| 39 | 64 | ||
| 39 | 64 | ||
Table 1B.
Transcripts Expression in ASCs, Fibroblasts and INR-90 Cells.
| Expressed | More than 5 Counts | > 2 Fold Change in old | > 2 Fold up | > 2 fold down | |
|---|---|---|---|---|---|
| ASC | 34125 | 14408 (42%) | 374 (1%) | 279 | 95 |
| Fibroblast | 33652 | 15463 (46%) | 2037 (6%) | 1180 | 857 |
| IMR-90 | 25970 | 16072 (62%) | 2438 (9%) | 660 | 1778 |
%: percentage of the “expressed”.
When the expression profiles of old and young cohorts are compared within each cell type, ASCs demonstrate the lowest number of genes with >2-fold difference between old and young, a total of 1.1%. In contrast, fibroblasts demonstrate > 6%, and IMR90 > 9% of genes with >2-fold difference between old and young cohorts (Table 1B). Deeper analyses reveal that ASC transcriptome profiles differ significantly from those of fibroblasts and IMR90 cells in age-related regulatory trends and are overall more stable with advancing age. For example, in aging ASCs, only 279 genes are up-regulated, and only 95 are down-regulated. In aging fibroblasts, 1180 genes are up-regulated and 857 are down-regulated (Table 1B). In senescent IMR90 cells, the number of down-regulated genes (1778) is significantly greater than that of up-regulated genes (660), a profile opposite to the one observed in ASCs. Thus, age-dependent gene expression patterns in ASCs are distinct from that of both aging fibroblasts, and senescent IMR90 cells.
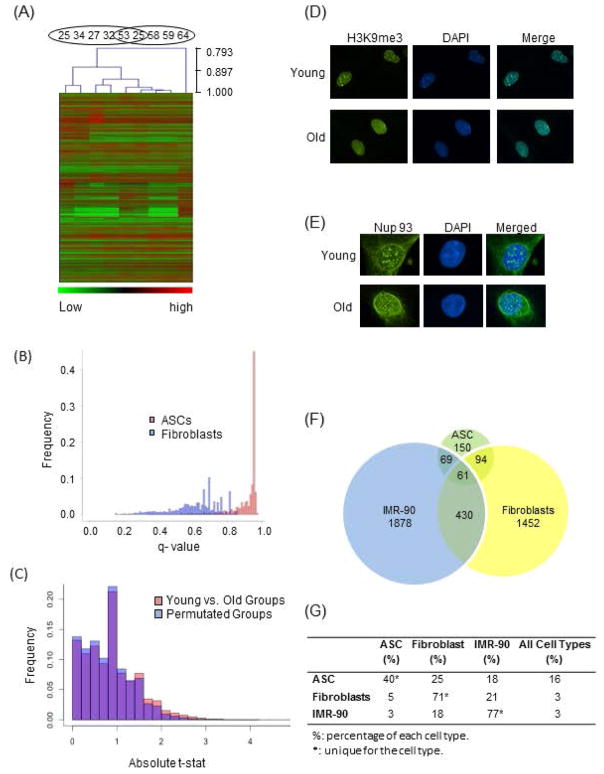
To explore these observations further, age-specific hierarchical clustering analyses were performed on ASCs and fibroblasts after low count transcripts were excluded. As shown in Fig 1A, fibroblasts demonstrate age-dependent transcriptional clustering with minimal mixing, whereas ASCs do not. To further validate this observation, we performed a global expression t-test analysis on ASC and fibroblast cohorts. Each gene’s q-value was obtained by comparing the transcript level in the young group to that in the old group and the q-value frequencies were plotted for both (Fig 1B). A larger number of genes in fibroblasts demonstrate altered expression levels at lower q-values than in ASCs. These results denote a higher statistical confidence of altered age-related expression in differentiated cells and together confirm that the transcriptome of aging ASCs is more stable than those of aging differentiated cells.
Fig 1. Comparison of Aging Impact on Transcriptome of ASCs and Terminally Differentiated Cells and Assessment of Nuclear Envelop and Chromatin Structure of ASCs.
(A) Hierachical clustering (Pearson correlation) was performed on fibroblast samples from 9 individuals, 5 young and 4 old. The age of the individuals are shown on top. Only transcripts with more than 5 counts were used. (B) Distributions of Benjamini and Hochberg q-values of Fibroblasts (blue) and ASCs (pink). The q-values were computed by comparing average transcript levels between the young and old groups of both fibroblast and ASC samples. (C) For each gene, a t-statistic was calculated to determine the significance in altered expression levels between the young and old ASC groups. The same test was performed on two groups of randomly assigned samples regardless of ages. The difference in t-statistics of the two tests was plotted. The difference is shown in pink when the value of grouped by age is larger than that of grouped by permutation, and in blue when grouped by permutation is larger than grouped by age. (D) Nuclear envelope structure is revealed by indirect immunofluorescence using anti-Nup 93 antibody, Nup93 panel – antibody labeling of Nup93 protein, DAPI panel – DAPI labeling of nuclear DNA, merged panel – overlay of Nup93 with DAPI. (E) Senescence-associated heterochromatin foci (SAHFs) were analyzed with immunofluorescence using anti-H3K9me3 antibody, H3K9me3 panel - antibody labeling of H3K9me3, DAPI panel – DAPI staining of nuclear DNA, merged panel – overlay of H3K9me3 with DAPI. (F) Genes with greater than 2 fold change in expression levels during early aging are compared among ASCs, fibroblasts and IMR-90. The numbers indicate amount of genes that are cell type specific or shared between 2 or 3 cell types. (G) For each cell type, the percentages of genes that are cell specific or shared by different cell types are shown. Percentages of cell type specific genes for each cell type are denoted with *.
To further substantiate whether our analysis was sufficiently sensitive enough to detect changes in gene expression profiles during early aging, we compared a subset of differentially expressed genes in the fibroblasts with known aging-associated genes in fibroblasts of mammals, identified in a meta-analysis of expression profiles [20]. Of the 73 previously reported genes, 9 were identified in our data with a q-value < 0.4, and all exhibited the same reported expression trend. At q < 0.5, 16 of 73 were identified and 13 of 16 demonstrated the same reported expression trend (Supplemental Table 1). These observations held true despite the fact that our fibroblast data sets examined only early chronological aging and early age-specific transcriptional profiles, rather than late- or replicative-aging profiles.
A final validation of the ASC transcriptome data set was conducted via a permutation t-test analysis of differential gene expression of the old and young ASC groups. When the age-based groups (young versus old) are compared with age-independent permuted (randomly shuffled) groups, more changes in gene-expression from the permuted groups demonstrate higher statistical confidence levels than age-based groups (Fig 1C), confirming once again minimal age-related transcriptome modifications in ASCs. As predicted by their stable transcription profiles during early aging, our ASCs also maintain stable nuclear envelope structures (Fig 1D), lacking senescence-associated heterochromatin foci (SAHFs) and other signs of aging such as nuclear envelope deformities (Fig 1E), and are able to proliferate and differentiate in a robust manner.
Cell Cycle and Mitosis-Related Genes Are More Actively Transcribed in Aging ASCs and Correlate with a Distinctly Shortened G1 Phase
Despite the global transcriptional stability of aging ASCs, we identified a small subset of genes demonstrating age-related expression alterations in ASCs. When the subset of genes displaying more than 2 fold changes between young and old in all three cell types are analyzed, ASCs demonstrate the least number of genes with >2-fold change and thus the highest percentage of shared gene expression among the three classes of cells (Figs 1F and 1G). Only 40% of the genes with >2-fold change are unique to ASCs, as compared to 71% and 77% in fibroblasts and IMR-90 cells, respectively. This may reflect the global “stemness” expression profiles of ASCs.
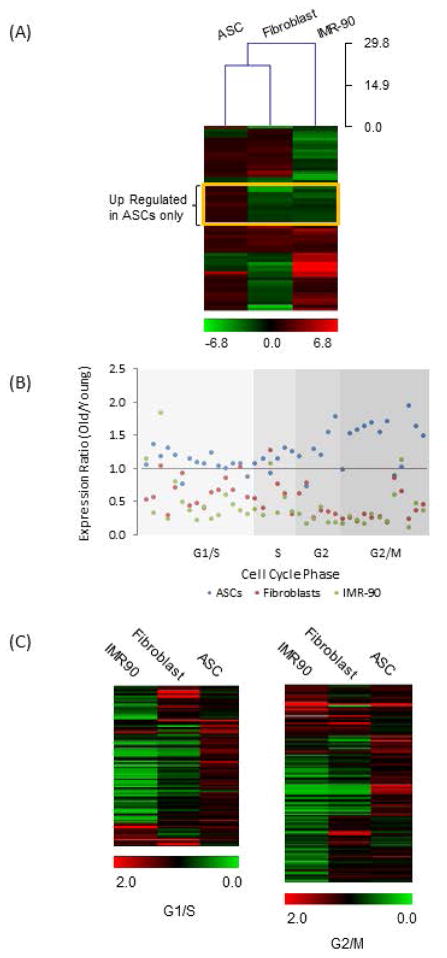
For the 61 differentially-expressed genes common to all three cell types (Fig 1F), a logarithm ratio of expression for each gene in old vs. young groups was calculated for each cell type, and compared using hierarchical clustering analysis (Fig 2A). We find that in aging ASCs, 80% (49) of these genes are up-regulated, in contrast to only 43% (26) in fibroblasts and 48% (29) in IMR-90 cells, representing yet again a distinct regulatory pattern in aging ASCs. When the 61 genes were analyzed for gene ontology (GO) term enrichment using DAVID [10, 11], 52 of them were present in the database and 42 matched with GOTERM bioprocesses. Prominent among the matched processes are cell cycle, cell growth and cell adhesion pathways (Supplemental Table 2). Significantly, we identified a subgroup of 13 genes, framed by the yellow rectangle in Fig 2A, that were specifically up-regulated in aging ASCs and down-regulated in aging differentiated cells (Fig 2A). Within this group, 10 genes (77%) are involved in cell cycle and mitosis suggesting a general up-regulation of these pathways in aging ASCs.
Fig 2. Comparison of Age-Dependent Expression Regulation of Genes Involved in Cell Cycle Progression in Different Cell Types.
(A) The ratios of transcript levels between average old and young groups were computed in log2 scale and compared using Hierachical clustering with Euclidean correlation. The result is shown in the heat map. The scale bar at the bottom displays the range of ratio in color. Unique trends to ASC are framed in yellow. (B) Expression ratio of old versus young was calculated for genes reported to periodically express in human cell cycle. The genes were grouped according to phase of cell cycle. Ratios of expression in old verses young ASCs are in blue, fibroblasts in red and IMR90 cells in green. (C) Expression ratio of old versus young was calculated for genes classified by MSigDB as involved in G1/S and G2/M. Heat maps were generated to display the expression ratio. Scale bars denote the range of ratios in color. Cell types are indicated at the top of the heat maps.
To investigate the role of these pathways further, we examined the transcript levels of genes that are periodically expressed during the cell cycle in synchronized Hela cells [21]. Thirty-nine out of fifty-two of these genes were detected in all three cell types. Expression ratios of old versus young were analyzed according to cell cycle phases (Fig 2B). A clear up-regulatory trend was evident for 85% of these genes in aging ASCs (blue), but only 8% in aging fibroblasts (red) and 10% in senescent IMR90 cells (green), consistent with more active cell cycle profiles in aging ASCs (Supplemental Table 3). For additional validation of these observations, we expanded our analysis to genes that are globally involved in the cell cycle pathway. Genes classified as involved in G1/S and G2/M were obtained from The Molecular Signatures Database (MSigDB) [22, 23]. For genes involved in G1/S, 109 of 112 listed were detected in ASCs and fibroblasts, and 106 were detected in IMR90 cells (Supplemental Table 4). For genes involved in G2/M, 136 out of 139 listed were detected in ASCs and fibroblasts, and 135 in IMR90 cells (Supplemental Table 4). Expression ratios of these genes in old versus young cohorts are displayed as heat maps (Fig 2C). As predicted, aging ASCs demonstrate age related up-regulation of a majority of these genes, specifically 72% of genes involved in G1/S and 62% of those involved in G2/M. In contrast, only 34% of genes involved in G1/S and 46% in G2/M are up-regulated in aging fibroblasts. Senescent IMR90 cells demonstrate the lowest number of up-regulated genes, 16% of those involved in G1/S and 29% involved in G2/M (Table 2). These results are consistent with an increase in cell cycle activities in aging ASCs.
Table 2.
Percentage of Genes Up-regulated in Indicated Subsets of Genes.
Genes previously reported to periodically express in human cell cycle.
Genes involved in G1/S and G2/M classified by MSigDB.
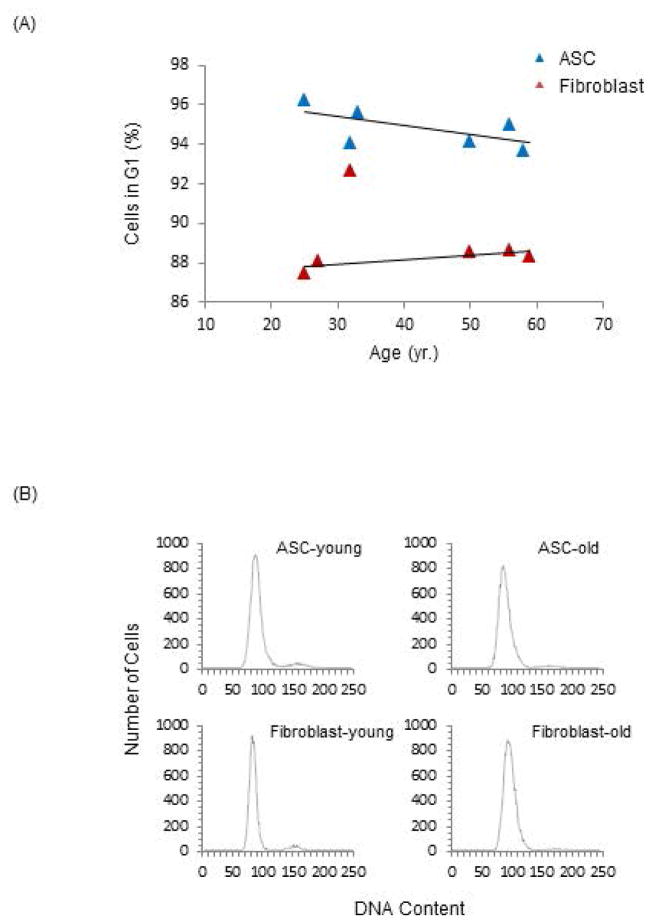
The expression profile of cell cycle genes in aging ASCs led us to examine cell cycle phase distributions in old and young ASCs and fibroblasts (see Method). As demonstrated in Fig 3, a smaller percentage of aging ASCs are in G1 phase compared with young ASCs, a trend that is reversed in fibroblasts. These results suggest a shortening of the G1 phase in aging ASCs, indicating a faster progression to subsequent cell cycle phases and cell proliferation. These subtle but important age-related cell cycle modifications are consistent with our prior data supporting robust viability and proliferative capacities specific to aging ASCs that are not present in aging differentiated cells [8].
Fig 3. Analyses of Cell Cycle in the ASCs and Fibroblasts.
(A) Cell cycles were analyzed by flow cytometry using propidium iodide (PI) labeled cells. The percentage of cells in G1 phase in the ASC and fibroblast populations were plotted against age. The cell types are indicated in the legend. (B) Representative histograms of PI labeled cell populations of young and old ASCs as well as fibroblasts were generated with the Y-axis representing cell numbers and the X-axis representing DNA content.
Translation Initiation Pathways Are Specifically Positively Regulated by Differential Transcription in Aging ASCs
To identify differentially regulated pathways with high statistical confidence, we conducted a t-test on the RNA-Seq expression data of each gene between old and young cohorts, as well as between the permuted ASC groups, as previously described. Differentially expressed genes between the young and old ASCs were identified, and 1013 of them with false discovery rates (FDR) less than 75% were selected for pathway analysis using Ingenuity Pathway Analysis software (Qiagen, CA). The eukaryotic initiation factor-2 (eIF2) signaling pathway was identified as the one with the highest statistical significance (Supplemental Table 5), with 55 out of 104 differentially expressed genes mapping to this pathway in ASCs, p = 10−20. The 55 genes distinctly fall into two classes, down-regulated ribosomal genes and up-regulated translation initiation factor genes in the old versus young ASC groups. Supplemental Tables 6A and 6B list the top 28 genes based on fold changes. Significantly, the expression pattern of the genes encoding ribosomal protein in aging ASCs is clearly reversed from that in aging fibroblasts, while the pattern for translation initiation genes is mixed between the two aging cell types. These data suggest that translation initiation and protein synthesis are differentially and specifically regulated in an age-dependent manner in ASCs, and importantly, contrast from aging fibroblasts.
Aging ASCs Exhibit Increased Nascent Protein Synthesis in Contrast to Aging Fibroblasts
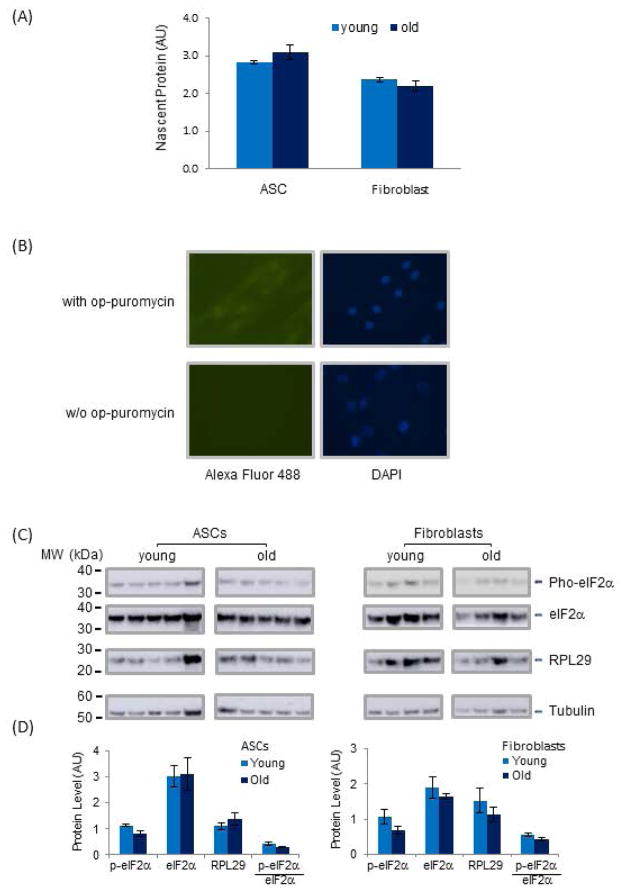
To further investigate the contrasting trend of down-regulated ribosomal genes and up-regulated translation initiation factor genes in aging ASCs, we investigated nascent protein synthesis in ASCs and fibroblasts from young and old cohorts via op-puromycin incorporation study (Figs 4A and 4B). As predicted by the increased transcription of translation initiation factors in aging ASCs, we observed a strong trend towards increased (10%) nascent protein synthesis (p < 0.07, n = 5) in old ASCs (Fig 4A), a trend not observed in aging fibroblasts. Further, despite lower transcript levels of Ribosomal Protein L29 (RPL29) in aging ASCs, the protein level of RPL29 is increased in this cohort, consistent with increased nascent protein synthesis (Figs 4C and 4D). The increase in nascent protein synthesis is further consistent with decreased phosphorylation of eIF2-α at Ser-51, as detected by Western blot in aging ASCs (Figs 4C and 4D). Phosphorylation of eIF2-α at Ser-51 represents one of the inhibitory mechanisms of translation initiation and its diminution indicates increased nascent protein synthesis. Interestingly, although eIF2-α phosphorylation was also found to be lower in aging fibroblasts (Figs 4C and 4D), this did not result in an increase in nascent protein synthesis, suggesting that the coordinated regulation of translation initiation is cell type-specific and likely involves different regulatory mechanisms in aging ASCs and differentiated cells.
Fig 4. General Nascent Protein Synthesis and Translational Regulation and Post-translational Modification of Proteins Involved in Translation Initiation and Ribosome Functions in ASCs and Fibroblasts.
(A) Nascent protein synthesis was measured by op-puromycin incorporation in nascent protein chains in ASCs and fibroblasts. The detected levels of nascent protein were normalized to the intensity of DAPI staining of cell nuclei in the same cell populations. P value for ASC samples was <0.07 n=5. (B) Representative fluorescence images of cells plated in wells with or without op-puromycin. Cell nuclei were detected by DAPI labeled DNA and nascent protein chains were detected by Alexa Fluor 488 fluorescence attached to op-puromycin. (C) Levels of eIF2-α, phosphorylated eIF2-α, RPL29 and Tubulin in ASCs and fibroblasts were examined by Western blots. For each cell type, samples were grouped according to age. Levels of tubulin served as internal controls for total protein content in the samples. (D) Levels of each protein shown in (C) were quantified by densitometry and shown in arbitrary unit. Protein quantities were normalized to tubulin to correct for different total protein content in the samples.
Discussion
In this study, we investigated primary human ASCs from patients of different ages, without extensive in vitro manipulation or expansion, as a novel model for elucidating natural chronological stem cell aging pathways. Targeted replicative exhaustion and senescence have been used traditionally as in vitro models of cell aging. Senescent cells, however, are present in organisms of all ages, and there is no proven causal relationship between cellular senescence and normal human aging. Similarly, immortalized and/or replicatively exhausted cell lines that have been used to model aging because of easy accessibility and fast expansion do not closely resemble their original in vivo environment. We argue here that these models do not faithfully recapitulate gradual chronological aging and fail short of elucidating features of aging that can readily be clinically translated. Toward that end, our data confirm that late passage IMR-90 cells, a model of replicative senescence, demonstrate transcriptional profiles distinctively different from those of aging primary ASCs. Unlike other adult human stem cells, primary human ASC tissue concentrations and proliferation rates remain constant with advancing age [8], and thus age-related functional changes in their regenerative potential may be attributed to direct molecular modifications rather than to changes in cell numbers or proliferation, making these cells a more robust and biologically relevant model with which to study natural aging [2, 24–27].
By examining the genome wide transcription profiles of primary human ASCs from young and old cohorts as well as age-matched terminally differentiated fibroblasts and IMR-90 cells of high and low passage, we describe here features unique to chronological human ASC aging. Overall, our results demonstrate that ASCs maintain the most stable and robust transcription profiles among the three cell types during aging. Aging ASCs further express a larger number of genes at low level (count of 5 or less in our RNAseq data). Consistent with this, Ramalho-Santos et al reported that stem cells express a significantly higher number of genes with unknown functions compared to differentiated cell types in their study of “Stemness” of stem cells by transcriptional profiling [28].
While we validated here that the stable expression profiles of aging ASCs represent their natural functional stability during early chronological aging, inherent human heterogeneity may have limited our detection of extremely small shifts in age-related transcriptional modification. Age-related clinical and gene expression changes have been observed to begin in humans at the age of 40 and thus we chose to designate the young cohort as below 40 years of age and the old cohort as above age 50 [29, 30]. The fact that age-related expression alterations were clearly detected in the age-matched fibroblasts, however, argues that our approach was indeed adequate for detecting age-related changes in ASC gene expression.
As further validation of our chronological aging model, although the transcriptome of older ASCs is distinctively different and more stable than those of older fibroblasts and IMR-90 cells, we successfully identify a subset of genes with age-related differential expression common to all three cell types. The expression patterns of these genes, which are largely involved in cell cycle and mitosis, differ strikingly between ASCs and differentiated cells and demonstrate robust transcriptional up-regulation in aging ASCs. Specifically, 85% of the genes expressed periodically during cell cycle progression are upregulated in old ASCs compared to only 8% in fibroblasts and 10% in senescent IMR90 cells. Furthermore, 72% of genes expressed in G1/S and 62% of those expressed in G2/M [22, 23] are upregulated in old ASCs but considerably downregulated in the differentiated cells. The increased expression of cell cycle genes in old ASCs correlates with a faster G1 phase progression in these cells. In contrast, differentiated cells display silencing of these pathways and a slower G1 phase progression. These observations emphasize the distinct mechanisms regulating aging in primary stem cells versus differentiated cells that likely reflect the difference in chronological aging in ASCs and replicative senescence in differentiated cells. Significantly, these features were not masked by human sample heterogeneity, underscoring the pivotal role played by this pathway in early chronological ASC aging and the importance of this primary human stem cell model of chronological aging.
Deeper analyses of differentially expressed genes between old and young ASCs identified significant modifications of the translation initiation and protein synthesis pathways, with old ASCs demonstrating global downregulation of ribosomal genes and upregulation of translation initiation factor genes. The surprising opposing transcription patterns of these closely related cellular processes in aging ASCs posed an intriguing question that was further investigated at the protein level. These analyses revealed increased nascent protein synthesis, higher levels of the RPL29 protein, and reduced inhibitory phosphorylation of eIF2-α in old ASCs, consistent with increased translation activity in these cells. The subtle contrasting levels of ribosomal mRNA and protein could be due to the temporal maintenance of the steady-state, heterogeneity of samples, or concurrent transcriptional and posttranscriptional regulatory mechanisms. Previous studies have demonstrated that the correlation between mRNA and protein levels is highly variable, and 40% variance is not uncommon [31].
Taken together, our results indicate that in aging ASCs, increased nascent protein synthesis is the result of coordinated regulation of translation initiation and ribosome protein content. These synergistic modifications suggest that multiple regulatory events at the transcriptional, translational and post-translational levels act together to enable aging ASCs to maintain their “stemness” and tissue homeostasis. Further, these modifications are in agreement with a shortening of the G1 phase in these cells, whereby an increase in protein synthesis would facilitate a more rapid transition to the next phase of the cell cycle. In contradistinction, we did not observe similarly coordinated cellular processes in aging fibroblasts, indicating that these age-dependent regulatory events are cell type-specific. Kowalczyk et al similarly observed a lower frequency of old hematopoietic stem cells (HSCs) in the G1 phase when compared with young HSCs [32]. Although ASCs differ from HSCs in many aspects, it is conceivable that both of these adult stem cells employ a similar protective mechanism during chronological aging, further underscoring the critical role played by cell cycle control during primary human stem cell aging.
In addition to the eIF2 pathway, several others were found to be modulated during ASC aging, specifically, eIF4 and p70S6K, mTOR (mechanistic target of rapamycin), and CCR3 signaling, as well as breast cancer regulation by stathmin-1 (Supplemental Table 5). Among these, mTOR and breast cancer regulation by stathmin-1 are particularly germane. The mTOR pathway includes ribosomal proteins and translation factors that are involved in the regulation of protein synthesis, many of which are shared with the eIF2 pathway [33–35]. It is possible that both pathways contribute to the altered regulation of protein synthesis and G1 duration during ASC aging. Stmn1 (Stathmin-1) is a major regulator of microtubule dynamics that is necessary for the regulation of the mitotic spindle and thus could similarly contribute to the shifts in cell cycle regulation in aging ASCs [36, 37].
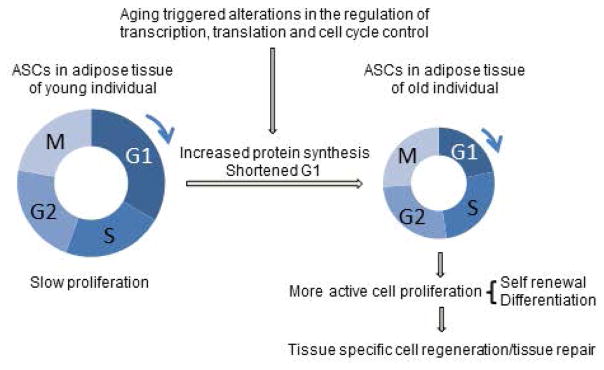
In summary, our results support a model in which cell cycle regulation plays a central role in the chronological aging of primary human adipose derived stem cells (Fig 5). Aging ASCs are characterized by a globally stable transcriptome with distinct and specific alterations in the expression of genes involved in cell cycle progression and protein synthesis that correlate with a shortened G1 phase and an increase in nascent protein synthesis. Recent genetic studies have implicated the stem niche as having a critical contribution to cell cycle regulation in stem cells [38]. Consequently, age-associated changes in the ASC niche microenvironment may lead to alterations in ASC cell cycle control that in turn could influence cell fate decisions, self-renewal or differentiation capacity, or changes in lineage differentiation potentials. An increase in protein synthesis during early chronological aging in ASCs, may support ASC homeostasis to satisfy an age-related increased demand for adipocyte and adipose tissue regeneration. Adipocyte regeneration is a dynamic process [39, 40]. The critical pioneering studies examined adipocyte and lipid turnover rate during human life span and elegantly demonstrated that the definitive number of adipocytes is established during childhood and adolescence. In adulthood, the population of adipocytes remains stable with approximately 10% being renewed annually via ASC differentiation. This rate is controlled by a dynamic equilibrium between adipocyte regeneration and cell death suggesting strict regulation of adipocyte cell number in adulthood. The effect of aging on adipocyte and adipose tissue homeostasis remains poorly understood, however, it is widely accepted that age-related adipose dysfunction strongly contributes to a myriad of metabolic and other disorders. Because ASCs are primarily responsible for adipocyte generation in adults, the characterization of transcriptome and cell cycle alterations during ASC aging will enhance our mechanistic understanding of adipose tissue homeostasis and metabolism, in addition to basic mechanisms of stem cell aging, and thereby contribute to the prevention and treatment of aging and metabolic diseases.
Fig 5. Role of Cell Cycle Control in Early Chronological Aging.
Cell cycle progressions in young and old ASCs are depicted by corresponding circles with arrows pointing to the direction of progression. Relevant cellular processes are connected by straight arrows.
Summary
Our unique access to continuous sources of human adipose tissues from healthy patients with a wide age span has enabled us to develop a model of chronological aging in primary human ASCs. We employed this model to examine genome-wide transcription networks and identified age-dependent and ASC-specific alterations. We demonstrate first that the transcriptome of aging ASCs is distinctly more stable than that of age-matched fibroblasts, and further, that age-dependent modifications in cell cycle progression and translation initiation specifically characterize aging ASCs in conjunction with increased nascent protein synthesis and a distinctly shortened G1 phase. Our data reveals a critical role for cell cycle control in early human stem cell aging and offers insights into the development of novel therapeutic avenues for the prevention and treatment of aging-related diseases.
Supplementary Material
Acknowledgments
The authors acknowledge research support from NIH (grant K08 AG042496 to IP).
We thank Raia Dierova and Robert Gersch for their assistance with ASC isolation and Qiwen Hu for submitting raw data to Gene Expression Omnibus (GEO) database.
Footnotes
Author Contributions:
Xiaoyin Shan: manuscript writing, experimental design, collection and assembly of data, data analysis and interpretation,
Cleresa Roberts: collection of data
Eun Ji Kim: Data analysis
Arianna Brenner: Data analysis
Gregory Grant: Data analysis and manuscript editing
Ivona Percec: Conception and design, provision of study material and patients, data analysis and interpretation, manuscript writing and final approval of manuscript
Disclosure of Potential Conflicts of Interest
None declared.
References
- 1.Signer RA, Morrison SJ. Mechanisms that regulate stem cell aging and life span. Cell Stem Cell. 2013;12:152–165. doi: 10.1016/j.stem.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oh J, Lee YD, Wagers AJ. Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nat Med. 2014;20:870–880. doi: 10.1038/nm.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tavernarakis N. Ageing and the regulation of protein synthesis: a balancing act? Trends Cell Biol. 2008;18:228–235. doi: 10.1016/j.tcb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Buszczak M, Signer RA, Morrison SJ. Cellular differences in protein synthesis regulate tissue homeostasis. Cell. 2014;159:242–251. doi: 10.1016/j.cell.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 6.Lindroos B, Suuronen R, Miettinen S. The potential of adipose stem cells in regenerative medicine. Stem Cell Rev. 2011;7:269–291. doi: 10.1007/s12015-010-9193-7. [DOI] [PubMed] [Google Scholar]
- 7.Zuk PA, Zhu M, Ashjian P, et al. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devitt SM, Carter CM, Dierov R, et al. Successful isolation of viable adipose-derived stem cells from human adipose tissue subject to long-term cryopreservation: positive implications for adult stem cell-based therapeutics in patients of advanced age. Stem Cells Int. 2015;2015:146421. doi: 10.1155/2015/146421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huschtscha LI, Napier CE, Noble JR, et al. Enhanced isolation of fibroblasts from human skin explants. Biotechniques. 2012;53:239–244. doi: 10.2144/0000113939. [DOI] [PubMed] [Google Scholar]
- 10.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 11.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 13.Shan X, Quaile MP, Monk JK, et al. Differential expression of PDE5 in failing and nonfailing human myocardium. Circ Heart Fail. 2012;5:79–86. doi: 10.1161/CIRCHEARTFAILURE.111.961706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shan X, Wang H, Margulies KB. Apoptosis signal-regulating kinase 1 attenuates atrial natriuretic peptide secretion. Biochemistry. 2008;47:10041–10048. doi: 10.1021/bi800972z. [DOI] [PubMed] [Google Scholar]
- 16.Crissman HA, Steinkamp JA. Rapid, one step staining procedures for analysis of cellular DNA and protein by single and dual laser flow cytometry. Cytometry. 1982;3:84–90. doi: 10.1002/cyto.990030204. [DOI] [PubMed] [Google Scholar]
- 17.Sherwood SW, Rush D, Ellsworth JL, et al. Defining cellular senescence in IMR-90 cells: a flow cytometric analysis. Proc Natl Acad Sci U S A. 1988;85:9086–9090. doi: 10.1073/pnas.85.23.9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Q, Fischer A, Reagan JD, et al. Oxidative DNA damage and senescence of human diploid fibroblast cells. Proc Natl Acad Sci U S A. 1995;92:4337–4341. doi: 10.1073/pnas.92.10.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 20.de Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitfield ML, Sherlock G, Saldanha AJ, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liberzon A. A description of the Molecular Signatures Database (MSigDB) Web site. Methods Mol Biol. 2014;1150:153–160. doi: 10.1007/978-1-4939-0512-6_9. [DOI] [PubMed] [Google Scholar]
- 23.Liberzon A, Subramanian A, Pinchback R, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyle M, Wong C, Rocha M, et al. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell. 2007;1:470–478. doi: 10.1016/j.stem.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Cerletti M, Jang YC, Finley LW, et al. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell. 2012;10:515–519. doi: 10.1016/j.stem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molofsky AV, Slutsky SG, Joseph NM, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sousa-Victor P, Gutarra S, Garcia-Prat L, et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506:316–321. doi: 10.1038/nature13013. [DOI] [PubMed] [Google Scholar]
- 28.Ramalho-Santos M, Yoon S, Matsuzaki Y, et al. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 29.Hsu VM, Stransky CA, Bucky LP, et al. Fat grafting’s past, present, and future: why adipose tissue is emerging as a critical link to the advancement of regenerative medicine. Aesthet Surg J. 2012;32:892–899. doi: 10.1177/1090820X12455658. [DOI] [PubMed] [Google Scholar]
- 30.Lu T, Pan Y, Kao SY, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 31.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kowalczyk MS, Tirosh I, Heckl D, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Proud CG. The mTOR pathway in the control of protein synthesis. Physiology (Bethesda) 2006;21:362–369. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 34.Thoreen CC, Chantranupong L, Keys HR, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cassimeris L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol. 2002;14:18–24. doi: 10.1016/s0955-0674(01)00289-7. [DOI] [PubMed] [Google Scholar]
- 37.Rubin CI, Atweh GF. The role of stathmin in the regulation of the cell cycle. J Cell Biochem. 2004;93:242–250. doi: 10.1002/jcb.20187. [DOI] [PubMed] [Google Scholar]
- 38.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 39.Spalding KL, Arner E, Westermark PO, et al. Dynamics of fat cell turnover in humans. Nature. 2008;453:783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 40.Arner P, Bernard S, Salehpour M, et al. Dynamics of human adipose lipid turnover in health and metabolic disease. Nature. 2011;478:110–113. doi: 10.1038/nature10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.