Abstract
Ataxia-telangiectasia (A-T) is an autosomal recessive chromosome breakage disorder caused by mutations in the ATM gene. Typically it presents in early childhood with progressive cerebellar dysfunction along with immunodeficiency and oculocutaneous telangiectasia. An increased risk of malignancy is also associated with the syndrome and, rarely, may be the presenting feature in small children. We describe a 17-year-old boy with slurred speech, mild motor delays and learning disability diagnosed with atypical A-T in the setting of T-cell acute lymphoblastic leukemia. Suspicion for A-T was raised after review of a peripheral blood karyotype demonstrating rearrangements involving chromosomes 7 and/or 14. The diagnosis was confirmed after molecular testing identified a novel homozygous missense variant in ATM (c.5585T>A; p.Leu1862His) that resulted in protein instability and abolished serine/threonine protein kinase activity. To our knowledge, this is the first report of concurrent A-T and lymphoid malignancy diagnoses in an older child or adult with only mild neurological disease. Our experience suggests that screening for the disorder should be considered in any individual with lymphoid malignancy and neurological findings, especially as radiation and certain chemotherapy protocols are contraindicated in A-T.
INTRODUCTION
Ataxia-telangiectasia (A-T) is an autosomal recessive disorder classically characterized by progressive cerebellar dysfunction, immunodeficiency, oculocutaneous telangiectasias, and increased risk of malignancy, particularly of lymphoid origin.1 Laboratory abnormalities include elevated alpha-fetoprotein, chromosomal instability, and radiosensitivity. Neurological symptoms (i.e., gait and truncal ataxia, oculomotor apraxia, and choreoathetosis) typically present between age 1 and 4. By age 10, most children are wheelchair bound.2 Telangiectasias are usually evident by age 6.2 Rarely, in young children, malignancy may be the presenting feature of the disorder.1, 3
In 1995, the gene responsible for A-T, ATM (ataxia telangiectasia mutated), was identified.4 Subsequently, it became evident that phenotypic spectrum of A-T included atypical and adult-onset forms of the disorder.5 However, to our knowledge, lymphoid malignancy has not been reported as the initial recognized finding of A-T in late childhood or adulthood. We report a case of A-T in a 17-year-old boy with only mild neurological findings, T-cell acute lymphoblastic leukemia (T-ALL), and a novel ATM mutation.
CLINICAL REPORT
The patient initially came to the attention of Pediatric Oncology after presenting to the Pediatric Emergency Department at age 17 years for a 2 week history of cough and an 8 day history of bilateral facial swelling. He had been prescribed a course of amoxicillin for a presumed dental infection with no response. At the time, the patient’s reported medical history was otherwise notable for some learning disability and slurred speech.
In the Emergency Department, the patient was tachycardic with a heart rate of 118 bpm. His vital signs were otherwise normal. The patient’s physical examination was notable for significant bilateral facial edema with submandibular lymphadenopathy and poor dentition. Decreased breath sounds in the lower two thirds of the right lung with coarse breath sounds at the apex were also appreciated. A chest radiograph demonstrated a large right-sided pleural effusion, a right mid lung airspace opacity, and a left-sided tracheal deviation. A subsequent CT scan of the neck and chest revealed a large right anterior mediastinal mass with complete occlusion of the superior vena cava and narrowing of the right mainsteam bronchus, a large right pleural effusion with compressive atelectasis of the right lung, and multiple regions of lymphadenopathy. Initial white blood cell count was 177.11 × 10(3)/µl. A bone aspiration and biopsy was performed to confirm the presumed diagnosis of T-ALL.
While reviewing the electronic medical record, Pediatric Oncology noted that the patient had previously been evaluated by the Clinical Genetics service at age 14 years for a history of learning disability, dysarthria, and mild motor delays. He rolled over at 7 months, sat without support at 12 months, and walked at 14 months. No gait difficulties were ever appreciated. The patient’s verbal milestones were initially on target but, at about age 3 years, some difficulties with speech were noted by his family. In addition, at the age of 9 years, the patient had failed a year in school, reportedly due to difficulties with reading. At the age of 17, he was in a special education high school class with 10 other students. The patient’s genetic evaluation at age 14 included a peripheral blood karyotype and a SNP microarray. Cytogenetic testing demonstrated rearrangements involving chromosomes 7, 14, or 7 and 14 in approximately 27% of the cells raising suspicion for a possible chromosome breakage disorder such as A-T or Nijmegen Breakage Syndrome (which is caused by autosomal recessive mutations in NBN). The microarray revealed several regions of homozygosity, including one overlying the ATM gene on chromosome 11. No apparent deletion or long contiguous stretch of homozygosity was appreciated over the NBN gene. After this initial evaluation by Clinical Genetics, the patient was lost to follow up until his presentation to the Pediatric Emergency Department.
A focused history and examination performed during the patient’s second assessment by Clinical Genetics was notable for a lack of findings typically associated with A-T. Aside from slurred speech and some learning disability, the neurologic features of the syndrome (i.e., gait and truncal ataxia, oculomotor apraxia, choreoathetosis) were absent. In addition, a history of recurrent infections was denied. No cutaneous telangiectasias were appreciated. A dilated fundus examination was also performed and no ocular telangiectasias reported. However, a chromosome breakage disorder, most likely A-T, was strongly suspected based on the patient’s prior karyotype and microarray results. Rapid molecular testing was obtained to confirm the diagnosis as radiation and certain chemotherapy regimens are contraindicated with chromosome breakage disorders. Given the patient’s atypical presentation, expedited whole exome sequencing, rather than targeted gene testing, was pursued. An elevated alpha-fetoprotein (AFP) measurement (117.9 ng/mL, normal range 0.6 – 3.9 ng/mL), was also obtained.
MATERIALS AND METHODS
Sequencing and Analysis
A 3 week expedited whole exome sequencing and analysis protocol was performed utilizing DNA extracted from the patient and his mother as previously described.6, 7 Research findings were confirmed by Sanger sequencing in a CLIA certified laboratory.
EBV-Transformed Lymphoblast Cell Lines
Peripheral blood samples were obtained with informed consent and under the supervision of the local institutional review board committee. Mononuclear cells were collected by density centrifugation with Ficoll (GE Healthcare), and B lymphocytes were transformed with the Epstein-Barr virus (EBV) following standard protocols. Briefly, mononuclear cells from the peripheral blood were suspended in EBV containing B95-8 conditional media for 2 weeks with the addition of RPMI (+10%FBS) media as needed. The outgrowing EBV-immortalized B cells were verified by flow cytometry analyses and sequenced for patient derived ATM mutations.8 Two independent peripheral blood samples were collected after the successful induction therapy.
Western Blot
Western blotting was performed to assess ATM kinase expression and activity in irradiated EBV-transformed lymphoblast cell lines. Cells were treated with 10Gy IR then harvested in a lysis buffer containing 50mM Tris.HCl (pH 7.3), 137mM NaCl, 1mM NaF, 1mM NaVO4, 10% glycerol, 1% Triton X-100, 0.2% Sarkosyl, protease inhibitor cocktail (Sigma P8340), 1mM PMSF, and 6.25U/ul Benzonase (EMD Millipore 71205). Cells were incubated in this lysis buffer for 1hr on ice, then spun down at 4°C for 15 min at 13,600 ×g. Supernatants were transferred to a fresh tube, assessed for protein concentration using the Bio-Rad Protein Assay (Bio-Rad 500-0006), and analyzed by gel electrophoresis. Antibodies against phosphorylated Kap1 (Bethyl A300-767A), total Kap1 (Cell signaling 4124S), vinculin (Sigma V284), and total ATM (Sigma A1106) were used at 1:1000 dilution.
RESULTS
Whole exome sequencing identified a homozygous variant of unknown significance in the patient in the ATM gene (c.5585T >A; p.Leu1862His). This previously unreported missense variant was not present in over 3,092 internal control individuals from the Columbia University Institute of Genomic Medicine or in two external databases of approximately 67,200 control individuals [National Heart, Lung, and Blood Institute Exome Sequencing Project (March 2013 release) (esp.gs.washington.edu) and the Exome Aggregation Consortium (January 2015 release) (exac.broadinstitute.org)]. The missense variant in ATM was predicted to be deleterious (score −5.67) by Provean (provean.jcvi.org), damaging (score 0.000) by SIFT (sift.jcvi.org), and disease causing (probability 0.9988) by Mutation Taster (mutationtaster.org).
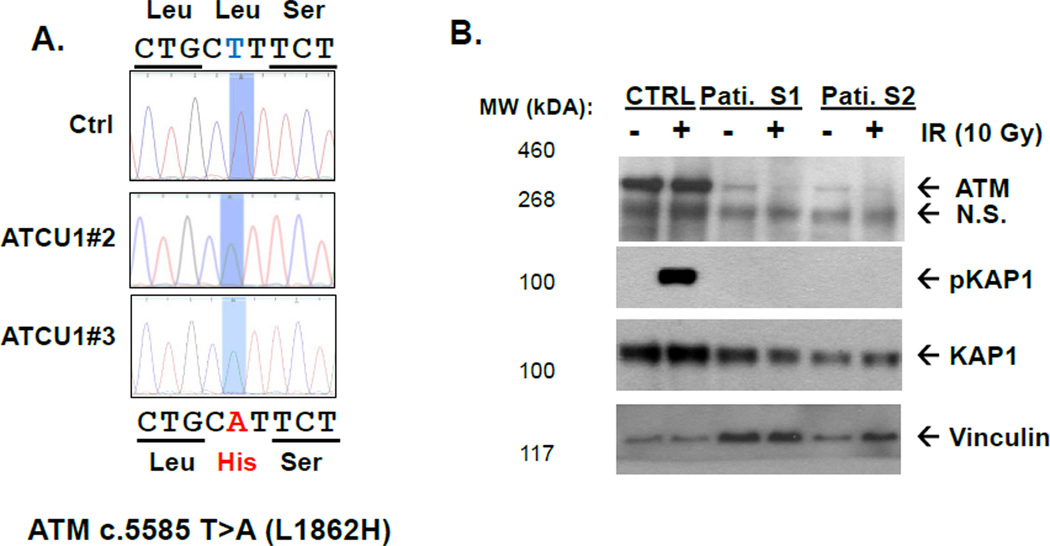
The patient cells showed severe decreased expression of the ATM protein at the expected size (<5% of wild type protein level) and abolished ATM kinase activity as measured by irradiation induced phosphorylation of Kap1, a relatively specific substrate of ATM (Figure 1B).9 This same phenotype was replicated in two independently derived EBV-transformed lymphoblast cell lines from the same patient (ATCU1#1 and #2) (Figure 1A).
Figure 1. ATM protein and kinase activity of ATCU1 derived lymphobolastcytes.
A). Genomic sequencing of patient derived lymphoblastic cell lines verified the homozygous ATM c.5585 T>5 (p.Leu1862His) mutation. The control (Ctrl) is an EBV transformed lymphoblast cell line from a health donor. A color version of this figure is available online.
B). Western blotting analyses for total ATM (Sigma MAT3), phosphorylated – KAP1 (Bethyl), total KAP1 and Vinculin as loading control.
DISCUSSION
Classically ataxia-telangiectasia (A-T) is characterized by cerebellar dysfunction, immunodeficiency, telangiectasias, and an increased risk for malignancy, especially leukemia and lymphoma. Although lymphoid malignancy may be the initial presenting feature of A-T, this has primarily been described in small children.1, 3 We report a 17-year-old boy diagnosed with A-T in the setting of T-cell acute lymphoblastic leukemia (T-ALL). The patient’s presentation was atypical and A-T was only suspected after a prior karyotype demonstrating chromosomal instability was reviewed. A cautious clinical diagnosis was made primarily based on the patient’s laboratory findings (cytogenetic testing, elevated AFP) and his newly diagnosed malignancy.
ATM encodes a serine/threonine protein kinase involved in the cellular response to DNA double strand breaks. Targets for ATM catalytic activity include proteins involved in cell-cycle checkpoint arrest (i.e., Chk1 and Chk2), DNA repair (i.e., BRCA1 and RAD51), apoptosis (i.e., p53), and chromatin relaxation (i.e., KAP1).10 Both heterozygous and homozygous hereditary mutations in ATM result in an increased risk for cancer due to impaired DNA damage repair and genomic instability. Leukemia and lymphoma are the most common cancers reported in individuals with A-T, however breast, thyroid, pancreatic, and other neoplasms have also been observed. Systemic genetic analyses of T-ALL in A-T patients are not yet available. Genomic analyses of ATM deficient murine immature T cell lymphomas reveal frequent activating mutations in Notch1 (~50%), deletion of PTEN (~25%), and mutation in FBW7 and RUNX1, which share significant similarity with sporadic T-ALL from human patients.11, 12
The classic and severe forms of A-T are attributed to homozygous or compound heterozygous truncating mutations that result in no detectable levels of the ATM. Milder, late onset, or atypical forms of the syndrome appear to be secondary to missense or leaky splice site mutations that allow for some residual activity of the kinase.13 Over 470 mutations in ATM have been reported in individuals with A-T.14 While there are no identifiable hotspots for truncating mutations and most mutations are private, cancer associated missense mutations (mostly somatic) of ATM have been found to cluster around the C-terminal kinase domain.15 Molecular testing in our patient identified a novel homozygous missense mutation in the ATM gene (c.5585T>A; p.Leu1862His) in the N terminal domain of the protein. Western blot analyses of the patient’s lymphoblastic cell lines revealed severely reduced protein level and kinase activity. A western blot performed on EBV-transformed lymphoblast derived cells demonstrated reduced ATM protein stability with <5% of wild type levels of protein detected. In addition, upon irradiation, ATM mediated phosphorylation of Kap1 was absence. These laboratory findings confirmed the clinical diagnosis of A-T. The N-terminal region of ATM contains many alpha-helical HEAT repeats (helix-turn-helix motifs). Crystal structural of the highly related DNA-dependent protein kinase (DNA-PK) suggests that the N-terminal HEAT domains facilitate the bending and are essential for the C-shape of the protein.16 In fact the majority A-T associated missense mutation of ATM severely reduce ATM protein stability. Expression of kinase inactivated ATM protein has been found to be incompatible for embryonic development using mouse models.15
Importantly, A-T was initially suspected in our patient because a karyotype had been previously performed on peripheral blood lymphocytes. The chromosome 7 and 14 rearrangements typical of chromosomal breakage disorders (e.g. A-T) are only appreciated when a T cell mitogen, such as phytohemagglutinin, is used to stimulate T cell division. When a karyotype is performed on total bone marrow, mitogen stimulation is not required, as the myeloid progenitors in bone marrow, but not necessarily the T cells, are actively dividing. Therefore routine cytogenetic analysis of bone marrow may miss the characteristic chromosomal rearrangements of A-T and a high index of suspicion is necessary to make the diagnosis of atypical cases with malignancy. Our experience suggests that evaluation for A-T should be considered in any individual with a lymphoid malignancy and neurologic dysfunction or other features of A-T. This recommendation includes older children and adults and those with mild or atypical findings. It is especially important to identify individuals with the A-T prior to the initiation of treatment for cancer is vital as radiation and certain chemotherapy regimens may be fatal and/or contraindicated.
Acknowledgments
We would like to thank Slavé Petrovski and David B. Goldstein for their analysis of the whole exome sequencing data and their comments on the manuscript.
We would like to also acknowledge the following individuals or groups for the contributions of control samples: E. Cirulli; D. Goldstein; D. Daskalakis; R Buckley; S. Hirose; M. Hauser; D. Levy; J.Hoover-Fong, N. L. Sobreira and D. Valle; A. Poduri; T. Young and K. Whisenhunt; Z. Farfel, D. Lancet, and E. Pras; G. Cavalleri; N. Delanty; G. Nestadt; J. Samuels, Y. Wang; V. Shashi; M. Carrington; M. Harms, T. Miller, A. Pestronk, R. Bedlack, R. Brown, N. Shneider, S. Gibson, J. Ravits, A. Gitler, J. Glass, F. Baas, S. Appel and E. Simpson, G. Rouleau; The Murdock Study Community Registry and Biorepository; the Carol Woods and Crosdaile Retirement Communities; ALS Sequencing Consortium; National Institute of Allergy and Infectious Diseases Center for HIV/AIDS Vaccine Immunology (CHAVI) (U19-AI067854), National Institute of Allergy and Infectious Diseases Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (UM1-AI100645); and the Epi4K Consortium and Epilepsy Phenome/Genome Project.
The collection of control samples and data was funded in part by: New York-Presbyterian Hospital; the Columbia University College of Physicians and Surgeons; the Columbia University Medical Center; Biogen, Inc.; B57 SAIC-Fredrick Inc M11-074; The Ellison Medical Foundation New Scholar award AG-NS-0441-08; National Institute of Mental Health (K01MH098126, R01MH097993, R01MH097971, RC2MH089915); National Institute of Allergy and Infectious Diseases (1R56AI098588-01A1); National Human Genome Research Institute (U01HG007672); National Institute of Neurological Disorders and Stroke (U01NS077303, U01NS053998, U01NS077274, U01NS077276, U01NS077303, U01NS077364, U01NS077367, and U01NS077275); National Institute of Allergy and Infectious Diseases Center (U19-AI067854, UM1-AI100645); and the Bill and Melinda Gates Foundation.
The laboratory characterization was in part supported by NIH 5R01CA158073 to SZ. SZ is the recipient of the Leukemia Lymphoma Society Scholar Award and JLC is supported by NIH F31CA183504.
REFERENCES
- 1.Schoenaker MH, Suarez F, Szczepanski T, Mahlaoui N, Loeffen JL. Treatment of acute leukemia in children with ataxia telangiectasia (A-T) European journal of medical genetics. 2016 doi: 10.1016/j.ejmg.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Gatti R. In: GeneReviews(R) Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, et al., editors. Seattle (WA): 1993. [Google Scholar]
- 3.Bielorai B, Fisher T, Waldman D, Lerenthal Y, Nissenkorn A, Tohami T, et al. Acute lymphoblastic leukemia in early childhood as the presenting sign of ataxia-telangiectasia variant. Pediatric hematology and oncology. 2013;30:574–582. doi: 10.3109/08880018.2013.777949. [DOI] [PubMed] [Google Scholar]
- 4.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 5.Teive HA, Moro A, Moscovich M, Arruda WO, Munhoz RP, Raskin S, et al. Ataxia-telangiectasia - A historical review and a proposal for a new designation: ATM syndrome. Journal of the neurological sciences. 2015;355:3–6. doi: 10.1016/j.jns.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Need AC, Shashi V, Hitomi Y, Schoch K, Shianna KV, McDonald MT, et al. Clinical application of exome sequencing in undiagnosed genetic conditions. Journal of medical genetics. 2012;49:353–361. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu X, Petrovski S, Xie P, Ruzzo EK, Lu YF, McSweeney KM, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genetics in medicine : official journal of the American College of Medical Genetics. 2015;17:774–781. doi: 10.1038/gim.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ventura M, Gibaud A, Le Pendu J, Hillaire D, Gerard G, Vitrac D, et al. Use of a simple method for the Epstein-Barr virus transformation of lymphocytes from members of large families of Reunion Island. Human heredity. 1988;38:36–43. doi: 10.1159/000153752. [DOI] [PubMed] [Google Scholar]
- 9.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nature cell biology. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 10.Choi M, Kipps T, Kurzrock R. ATM Mutations in Cancer: Therapeutic Implications. Molecular cancer therapeutics. 2016;15:1781–1791. doi: 10.1158/1535-7163.MCT-15-0945. [DOI] [PubMed] [Google Scholar]
- 11.Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nature reviews. Cancer. 2016;16:494–507. doi: 10.1038/nrc.2016.63. [DOI] [PubMed] [Google Scholar]
- 12.Zha S, Bassing CH, Sanda T, Brush JW, Patel H, Goff PH, et al. ATM-deficient thymic lymphoma is associated with aberrant tcrd rearrangement and gene amplification. The Journal of experimental medicine. 2010;207:1369–1380. doi: 10.1084/jem.20100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor AM, Byrd PJ. Molecular pathology of ataxia telangiectasia. Journal of clinical pathology. 2005;58:1009–1015. doi: 10.1136/jcp.2005.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Human mutation. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto K, Wang J, Sprinzen L, Xu J, Haddock CJ, Li C, et al. Kinase-dead ATM protein is highly oncogenic and can be preferentially targeted by Topo-isomerase I inhibitors. eLife. 2016;5 doi: 10.7554/eLife.14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]