Abstract
Background
Patient-derived xenografts have recently become a powerful tool for cancer research and may be used to guide personalized therapy. Thus far, patient derived xenografts have been grown from tumor tissue obtained after surgical resection. However, many cancer patients never undergo surgery for a variety of reasons. We hypothesized that xenograft tumors could be grown from smaller volumes of patient tissue, such as those obtained during diagnostic biopsies.
Methods
Surgical specimens were obtained after resection of primary or metastatic lesions of the following cancers: pancreatic carcinoma, non-small-cell lung cancer, bladder (urothelial) carcinoma, and melanoma. At least 10 cases of each cancer were included in this study. To mimic clinical biopsies, small fragments of the surgical specimens were biopsied with a 22G needle and the needle contents were injected subcutaneously in immunocompromised mice. The tumor fragment from which the biopsy was taken was also implanted subcutaneously in the contralateral side of the same mouse as a control.
Results
Success rates of the traditional method of xenograft implantation ranged from 27.3–70%. Success rates of the fine needle aspirate technique ranged from 0%–36.4%. An attempt to engraft a percutaneous core needle liver biopsy of a metastatic pancreatic adenocarcinoma also was successful.
Conclusions
We have found that it is possible to engraft fine needle aspirates and core biopsies of solid tumors in order to generate patient-derived xenografts. This may open up xenografting to a wider cancer patient population than previously possible.
Graphical Abstract
We examined the feasibility of creating PDXs with fine needle aspirates of pancreatic, bladder, lung, and melanoma tumors. The significance of this report is the possibility to generate PDXs without patients undergoing invasive surgeries.
Introduction
Progress in cancer research has depended on the continual development of increasingly relevant preclinical models of human cancers, ranging from human and animal cell lines grown in vitro to genetically engineered animal models of cancer. However, there remains a high failure rate of new cancer treatments that are found to be successful in pre-clinical trials. This is attributed to the inability of many pre-clinical models to recreate the heterogeneity seen in human patient tumors1. Recently, patient-derived xenograft (PDX) models have gained popularity in cancer research due to their ability to better recapitulate the heterogeneity of human tumors2–5. Furthermore, PDX models have been shown to predict clinical responses to chemotherapeutic drugs more accurately than other platforms, which centralizes the role of PDX models in a new generation of personalized cancer therapy1,6,3.
One major limitation of PDX models is the need for surgical resection of a patient’s cancer tissue in order to generate xenografts. Many cancer patients do not receive any surgical resection of tumor tissue, and surgical rates vary greatly by the type of cancer. In an NHS study, surgical resections rates ranged from 80% of uterine and female breast cancer patients, to less than 10% of patients with cancer of the liver, bladder, prostate, lung, and pancreas7. Therefore, we hypothesized that new PDX tumors would be able to be established from small volumes of tumor tissue, such as that which would be obtained during fine needle aspirates or core needle biopsies obtained during diagnostic workup. Recently, Allaway et al demonstrated that it was feasible to engraft fine needle aspirate samples of pancreatic adenocarcinoma obtained during EUS-FNA procedures8. We also confirmed these findings in pancreatic adenocarcinoma, and explored the possibility of FNA xenografts in urothelial, lung, and melanoma specimens.
Materials and Methods
Patient-derived tissue and ethics statement
Surgically resected tumor specimens were obtained from patients with pancreatic carcinomas, non-small cell lung carcinomas, melanomas, and urothelial cell carcinomas of the bladder. All surgically resected tumors were collected after written patient consent and in accordance with the institutional review board-approved protocols of the University of Texas M.D. Anderson Cancer Center (protocols: Pancreatic Cancer LAB07-0854, Lung LAB10-0704, Bladder LAB08-0179, and Melanoma LAB07-0875). All surgically resected specimens were accepted as they became available, and no samples were excluded based on pre-treatment status. All animals were housed and maintained under guidelines established by the American Association of Laboratory and Animal Care, and animal experiments were performed in accordance with the NIH-Animal Care and Use Committee (ACUC) guidelines (protocol 09-07-10131) after The University of Texas M.D. Anderson Cancer Center IRB approval.
Establishment of patient-derived cancer xenografts from tumor fragments and biopsy samples
The establishment of direct heterotopic xenograft tumors in NOD/SCID mice has been described in detail previously5. After gross determination of viable tumor by the attending pathologist, specimens were resected and kept in ice cold cell culture media composed of RPMI 1640 supplemented with 10% FBS (Hyclone Laboratories, Waltham, MA) and 2% penicillin/streptomycin (Sigma-Aldrich). The time from surgical resection to placement of samples in cold media (warm ischemia time) was under 30 minutes, and the time in cold media was under one hour. All NOD/SCID mice were purchased from the National Cancer Institute (Bethesda, Maryland). 4–6 week old NOD/SCID mice were anesthetized with inhaled isoflurane. Patient tumors in cold media were minced into fragments 1–2mm3 in volume. In order to simulate a clinical biopsy, each tumor fragment was biopsied by passing a 22 gauge needle three times through the tissue, while gently aspirating with a 1 cc syringe. Subsequently, liquid matrigel (BD Biosciences) was aspirated with the syringe to make a total volume of approximately 100 microliters. The syringe contents were then injected subcutaneously in the flank of the NOD/SCID mouse. The same tumor fragment that was biopsied was then implanted subcutaneously in the contralateral flank of the same mouse as a control, along with approximately 100 microliters of cell culture media and liquid matrigel (Figure 1). Two to five NOD/SCID mice were implanted with patient tumor fragments for each surgical case. The mice were monitored for tumor growth by palpation of implantation sites on a weekly basis. The time to tumor formation (TTF) was recorded when a subcutaneous nodule was palpated on the mouse’s flank. Tumors were harvested when they reached a maximum diameter of 1.5 cm.
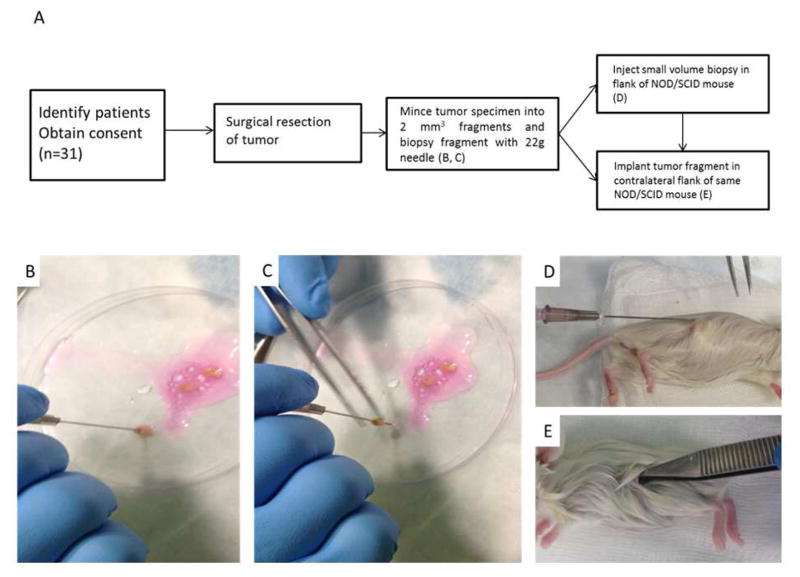
Figure 1.
Experimental design and fine needle aspirate xenograft procedure. (A) Experimental flowsheet. After obtaining the tumor tissue, mince into ~2 mm3 fragments in cell culture media/matrigel. (B,C) Position a 1cc syringe with 22-gauge needle over the fragment and advance the fragment up the needle while gently aspirating. Aspirate more cell culture media/matrigel for a total of ~100 μl. (D) Inject syringe contents subcutaneously on flank of mouse. (E) Implant same tumor fragment in contralateral flank of same NOD/SCID mouse.
Establishment of patient-derived xenograft from diagnostic clinical biopsy
A patient with metastatic pancreatic adenocarcinoma underwent percutaneous ultrasound guided biopsy of a liver lesion by interventional radiology. The biopsy was obtained using an 18 gauge Tru-Cut Biopsy Needle (Carefusion, San Diego, CA). The biopsy fragment was immediately transferred to ice cold cell culture media. The fragment, which originally measured approximately 1.0 mm x 0.2 mm, was divided into three fragments and implanted subcutaneously into the flanks of three NOD/SCID mice. The mice were monitored for tumor growth by palpation of implantation sites on a weekly basis. Tumors were harvested when they reached a maximum diameter of 1.5 cm.
Results
Engraftment rates
PDX engraftment rates ranged from 27.3–70%, in the traditional method of xenograft implantation and 0–36.4% with the biopsy method. Pancreatic and melanoma cases were the most likely to engraft, while bladder and NSCLC tumors were the least likely to engraft with the biopsy method (Table 1). The details and clinical characteristics of all attempted PDX cases are listed in Table 2. Second generation PDXs were engrafted from two biopsy-derived melanoma tumors. In these mice, every fragment that was implanted successfully established a tumor which histologically resembled the previous generation (data not shown).
Table 1.
Success rates of patient-derived xenograft engraftment using the traditional xenograft method and the experimental fine needle aspirate method (TTF=time to tumor formation, FNA=fine needle aspirate).
| Pancreas | Melanoma | Bladder | NSCLC | |
|---|---|---|---|---|
|
| ||||
| Traditional xenograft growth (% success) | 7/10 (70%) | 5/11 (45.5%) | 3/11 (27.3%) | 8/17 (47.1%) |
| TTF in days (range) | 41 (29–53) | 39 (26–54) | 74 (26–125) | 58 (26–175) |
|
| ||||
| Biopsy xenograft growth (% success) | 3/9 (33.3%) | 4/11 (36.4%) | 0/11 (0%) | 1/17 (5.8%) |
| TTF in days (range) | 70 (39–147) | 57 (46–85) | N/A | 60 (60) |
Table 2.
Xenograft Patient Information
| Patient | Sex | Age (years) | Xenograft source | Prior therapy | Engraftment rate with traditional method (mice) | TTF with traditional method (days) | Engraftment rate with biopsy method (mice) | TTF with biopsy method (days) | |
|---|---|---|---|---|---|---|---|---|---|
| Pancreas | MDA-PATX109 | M | 56 | Primary | Yes | 3/5 | 48 | 0/5 | N/A |
| MDA-PATX112 | F | 72 | Primary | No | 3/5 | 29 | 1/5 | 147 | |
| MDA-PATX113 | M | 59 | Met | Yes | 5/5 | 29 | 5/5 | 48 | |
| MDA-PATX115 | M | 51 | Primary | No | 1/5 | 33 | 0/5 | N/A | |
| MDA-PNETX22 | F | 64 | Primary | Yes | 0/5 | N/A | 0/5 | N/A | |
| MDA-PATX116 | M | 70 | Primary | Yes | 0/5 | N/A | 0/5 | N/A | |
| MDA-PATX117 | M | 79 | Primary | Yes | 4/5 | 43 | 0/5 | N/A | |
| MDA-PATX118 | F | 71 | Primary | No | 4/5 | 25 | 3/5 | 39 | |
| MDA-PATX121 | F | 64 | Met | Yes | 3/3 | 53 | N/A | N/A | |
| MDA-PATX128 | M | 67 | Primary | Yes | 0/5 | N/A | 0/5 | N/A | |
| Melanoma | MDA-MTX1 | F | 65 | Met | Yes | 0/5 | N/A | 0/5 | N/A |
| MDA-MTX2 | M | 67 | Met | Yes | 0/5 | N/A | 0/5 | N/A | |
| MDA-MTX3 | M | 75 | Met | Yes | 2/5 | 48 | 2/5 | 85 | |
| MDA-MTX4 | F | 57 | Met | Yes | 0/5 | N/A | 0/5 | N/A | |
| MDA-MTX5 | F | 72 | Met | Yes | 2/5 | 26 | 2/5 | 46 | |
| MDA-MTX6 | M | 30 | Met | Yes | 4/5 | 38 | 2/5 | 48 | |
| MDA-MTX7 | M | 69 | Met | No | 2/5 | 27 | 3/5 | 50 | |
| MDA-MTX8 | M | 68 | Met | No | 0/5 | N/A | 0/5 | N/A | |
| MDA-MTX9-1 | M | 66 | Met | No | 4/5 | 54 | 0/5 | N/A | |
| MDA-MTX9-2 | M | 66 | Met | No | 0/5 | N/A | 0/5 | N/A | |
| NSCLC | MDA-TC168 | M | 52 | Primary | No | 1/2 | 40 | 0/2 | N/A |
| MDA-TC170 | M | 78 | Primary | No | 1/3 | 41 | 0/3 | N/A | |
| MDA-TC171 | M | 77 | Primary | No | 1/2 | 175 | 0/2 | N/A | |
| MDA-TC172 | M | 44 | Primary | No | 1/3 | 41 | 2/3 | 60 | |
| MDA-TC173 | F | 67 | Primary | No | 0/2 | N/A | 0/2 | N/A | |
| MDA-TC174 | F | 63 | Primary | No | 0/3 | N/A | 0/3 | N/A | |
| MDA-TC175 | F | 74 | Primary | No | 0/3 | N/A | 0/3 | N/A | |
| MDA-TC177 | M | 59 | Primary | Yes | 0/2 | N/A | 0/2 | N/A | |
| MDA-TC181 | F | 60 | Primary | No | 0/3 | N/A | 0/3 | N/A | |
| MDA-TC184 | F | 59 | Primary | Yes | 0/2 | N/A | 0/2 | N/A | |
| MDA-TC185 | M | 69 | Primary | No | 1/3 | 42 | 0/3 | N/A | |
| MDA-TC186 | M | 73 | Primary | No | 0/2 | N/A | 0/2 | N/A | |
| MDA-TC187 | M | 49 | Primary | Yes | 1/4 | 69 | 0/4 | N/A | |
| MDA-TC188 | F | 67 | Primary | Yes | 2/3 | 29 | 0/3 | N/A | |
| MDA-TC190 | F | 59 | Primary | No | 1/3 | 26 | 0/3 | N/A | |
| MDA-TC193 | M | 71 | Primary | No | 0/2 | N/A | 0/2 | N/A | |
| Bladder | MDA-UTX1 | M | 72 | Primary | No | 1/5 | 125 | 0/5 | N/A |
| MDA-UTX2 | M | 77 | Primary | No | 2/5 | 26 | 0/5 | N/A | |
| MDA-UTX3 | M | 55 | Primary | No | 0/5 | N/A | 0/5 | N/A | |
| MDA-UTX4 | M | 59 | Primary | No | 0/5 | N/A | 0/5 | N/A | |
| MDA-UTX5 | M | 61 | Primary | No | 0/5 | N/A | 0/5 | N/A | |
| MDA-UTX6 | F | 67 | Primary | No | 0/5 | N/A | 0/5 | N/A | |
| MDA-UTX7 | F | 74 | Primary | No | 0/5 | N/A | 0/5 | N/A | |
| MDA-UTX8 | M | 65 | Primary | No | 0/5 | N/A | 0/5 | N/A | |
| MDA-UTX9 | M | 67 | Primary | Yes | 1/5 | 70 | 0/5 | N/A | |
| MDA-UTX10 | M | 63 | Primary | No | 0/5 | N/A | 0/5 | N/A |
Time to tumor formation
Xenografts derived from pancreatic adenocarcinoma specimens grew in an average time of 41 days for the traditional implantation method and 70 days for the biopsy implantation method. In melanoma specimens, traditional xenografts grew in 39 days versus 57 days for the biopsy method. In bladder cases, traditional xenografts grew in 74 days, while no biopsy xenografts grew. In NSCLC, the traditional xenografts grew in 58 days versus 60 days for the biopsy xenograft (however, only one biopsy xenograft successfully grew (Table 1)). In all tumor types, there was a lag in tumor formation of FNA-derived xenografts. However, once tumors formed, the growth rates were similar between the two tumors (data not shown).
Pre-treatment of patient tumors has differing effects on tumor engraftment in each cancer
Our group previously showed in pancreatic adenocarcinoma xenografts that pre-treated patient tumors with chemotherapy and/or radiation have lower engraftment rates than treatment-naïve tumors9. A similar trend was observed in the pancreas specimen group of this study. Of the 10 PDA specimens, three were untreated, all three (100%) successfully engrafted via the standard approach, and 2/3 (67%) grew FNA xenografts. Seven PDA specimens were pre-treated, four of which grew xenografts via the traditional method (57%), and only one grew a FNA xenograft (14%). However, the pretreatment of melanoma did not seem to affect engraftment rates. Out of the 10 melanoma specimens, four were treatment naïve, two (50%) grew traditional xenografts and one (25%) grew FNA xenografts. In the six pre-treated melanoma samples, three (50%) traditional melanoma xenografts engrafted and three (50%) FNA xenografts engrafted. In NSCLC, pretreatment also did not seem to affect traditional xenograft establishment. Out of 16 NSCLC specimens, twelve were untreated. Six (50%) traditional xenografts engrafted and one (17%) also grew a FNA xenograft. In the remaining four pre-treated specimens, two (50%) successfully engrafted, while none grew FNA xenografts (0%). In bladder carcinoma, 9/10 specimens were treatment naïve, and 2 of these 9 (22%) grew traditional xenografts, but no FNA xenografts. There was only one pre-treated specimen, and it grew a traditional xenograft (100%), but not an FNA xenograft (0%). The details of each case and engraftment rates can be found in Table 2.
Histology of biopsy xenografts resembles that of traditional xenografts
The histology of xenografts derived from fine needle aspirates accurately resembled the histology of human tumors and xenografts implanted with the traditional method in every case (Figure 2), despite the biopsy xenografts starting with far fewer implanted cells. Both the traditionally implanted xenograft and the fine needle aspirate xenograft resembled the histology of the original patient tumor.
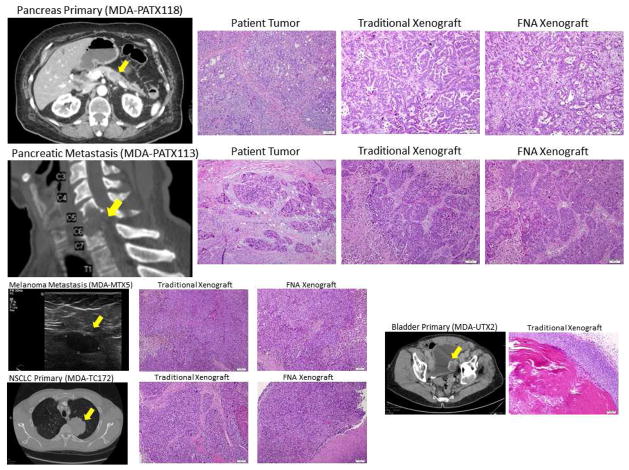
Figure 2.
Examples of tumor sources and histology of resultant xenografts. The histology of fine needle aspirate xenografts resembles that of traditionally implanted xenografts and patient tumors. Representative haematoxylin and eosin micrographs are shown. Bars are 100 microns.
Biopsy xenografts can be grown from pancreatic ductal adenocarcinoma metastases
After it was shown that it was possible to grow xenograft tumors from biopsy samples of primary tumors, we then turned our attention to sampling metastatic sites cancer patients. In one rare case, MDA-PATX113, we obtained surgically resected tumor tissue from a cervical spine metastasis from a patient with metastatic pancreatic adenocarcinoma. This sample was implanted in the same way as the previously processed primary tumors. In this case, every tumor sample that was implanted successfully grew, on both the traditional side and the FNA side of each mouse (Figure 2).
We then sought to grow a pancreatic adenocarcinoma xenograft grown from an outpatient biopsy. This patient, MDA-PATX121, underwent percutaneous ultrasound guided core needle biopsy of a liver lesion of metastatic pancreatic adenocarcinoma. The original sample was a cylindrical specimen measuring approximately 5mm in length and 1mm in diameter. The tumor core was divided into 3 pieces and implanted into three NOD/SCID mice, all of which grew palpable xenograft tumors by 57 days (Figure 3). One important difference in this case was that the core biopsy fragments were larger and therefore contained more cells than the fine needle aspirates implanted in the other cases, however these core needle biopsy fragments were still far smaller than the ~3mm3 tumor fragments that have been used in previously described methods4,5. In short, the two pancreatic adenocarcinoma cases established from metastatic sites both engrafted successfully, and moreover, all mice that were implanted successfully grew tumors at every implantation site, whether by traditional fragment, fine needle aspirate, or core needle biopsy fragment. This suggests that metastatic sites may more readily establish xenografts, but more cases are needed to confirm this finding.
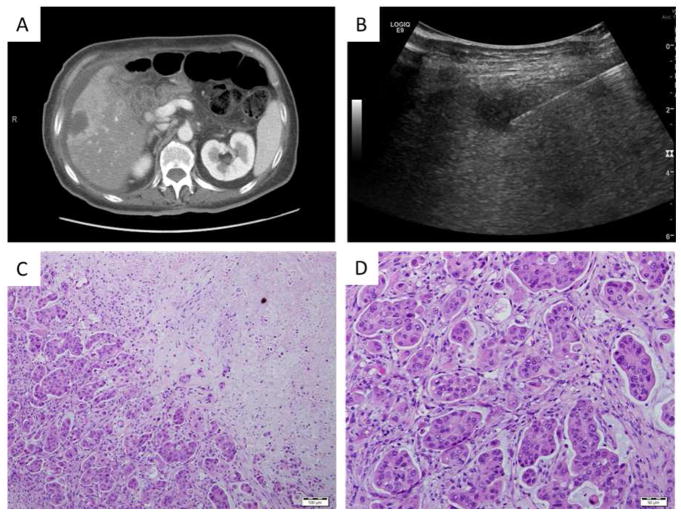
Figure 3.
MDA-PATX121, a xenograft grown from a percutaneous biopsy of a pancreatic adenocarcinoma metastatic to the liver. (A) CT imaging of liver metastasis of pancreatic adenocarcinoma. (B) Ultrasound image of biopsy procedure. (C,D) Representative photomicrographs of haematoxylin and eosin staining of xenograft. Bars are (C) 100 microns and (D) 50 microns.
Discussion
The solid tumors from which xenografts were attempted in this paper are inherently heterogeneous, with cells in the same tumor having varying phenotypes and genetic characteristics10–18. The success rate of establishing a xenograft depends on presence of enough cells that are able to initiate new tumor formation19–21. Therefore, engraftment rates were highly variable, as many groups have reported previously5,22,23. A review by Hidalgo et al. reported engraftment rates ranging from 3%–90% on a variety of different methodologies and tumor types22. Even within each case, the growth of the traditional xenograft did not predict the growth of the biopsy xenograft, and vice versa. There was a diverse tumor growth success rate among all mice implanted with the same tumor. For example, each mouse in one implantation group had the possibility of growing only the traditional xenograft, only the biopsy xenograft, both, or neither (data not shown). In this regard, a limitation of our study is that the FNA method may not adequately capture tumor heterogeneity since the FNAs were performed on the single 2mm3 fragment which was implanted on the contralateral flank. In future practice, pooling FNAs from different quadrants of the tumor is more likely to represent the heterogeneity of the entire tumor, which may lead to more predictable engraftment. In terms of cell numbers, we previously demonstrated that a PDX can be established with as few as 100 purified ALDH+ cells and retain the stroma architecture of the human tumor and traditional xenograft19. Lee et al. developed a method quantifying cell numbers from 22G FNA passages of mouse xenografts and detected cell counts of >1000 cells per aspiration27. Therefore one FNA passage through a tumor should be enough to establish a xenograft, and multiple passes may increase success rates.
It is notable that xenografts established with the FNA method resulted in the same overall tumor and stroma architecture as the patient tumor. Suetsugu et al., using pancreatic adenocarcinoma PDX tumors previously established by us, showed that tumors implanted in red fluorescent protein (RFP)-expressing nude mice acquired RFP positive stroma, including cancer associated fibroblasts and tumor-associated macrophages. The same phenomenon happened in further passage into green fluorescent protein (GFP)-expressing nude mice, and again into cyan fluorescent protein (CFP)-expressing nude mice. Each passage retained fluorescent stroma, blood vessels, and stromal cells from each generation of color, indicating that the stroma of PDX tumors is murine in nature24. Taken together, this supports the idea that the stromal architecture of a patient-derived xenograft is largely determined by the phenotype of the cancer cells in how they recruit murine stroma, and transplanting large amounts of patient stroma may not be necessary for xenograft formation.
There are several other possibilities for variable engraftment rates of xenografts. One potential issue was that we implanted both samples in the same mouse. Groups have reported endocrine effects of tumors impacting the growth of distant tumors on the same animal25,26. However, in those studies, tumors were generated from different cell lines with different phenotypes, while both of our tumors were derived from a relatively similar area of a single patient’s tumor. While there could be significant intra-tumoral heterogeneity, we felt that the tumors were composed similarly enough as to not impact the growth of each other. The only way to assure lack of interference between multiple tumors on the same animal is to only implant one sample per mouse, however this requires considerable extra resources. One more reason for variable engraftment rates of patient-derived xenografts is pre-treatment of the patient’s tumor with chemotherapy and/or radiation. We have shown previously that pre-treatment of a pancreatic adenocarcinoma tumor with neoadjuvant chemotherapy can decrease the engraftment rate by 40%9. Although we did not have large enough sample sizes for statistical analysis, this trend was re-captured in our study. Pre-treatment did not seem to affect engraftment rates of the other tumors, but larger sample sizes are necessary to truly determine the effect of pre-treatment on FNA xenografts.
Patient derived tumor xenografts derived from pancreatic adenocarcinoma, melanoma and NSCLC were more likely to form compared to urothelial carcinomas, which had lower growth rates. These differing success rates likely reflect the relative growth rates of these cancers. NSCLC and bladder cancer cases that were attempted at engraftment were collected from less advanced disease than those from pancreas and melanoma. In the case of NSCLC, this resulted in a small volume of tumor tissue being available for xenografting. Therefore, only 2–3 mice were implanted with tumor tissue, rather than 5 mice as in the pancreas, melanoma, and bladder sets. This may have caused the success rate to be lower than what would have been possible if tumor fragments were implanted in 5 mice for each case. The high success rate of metastatic lesion engraftment suggests that more advanced disease will be more likely to form a xenograft; however, more cases are needed to confirm this observation. Therefore, a weakness of this study is the lack of cases attempted with more advanced/metastatic bladder and NSCLC cancers.
In both cases where tissue was obtained from metastases of pancreatic adenocarcinoma, MDA-PATX113 and MDA-PATX121, every fine needle aspirate or core biopsy fragment that was implanted grew into a large xenograft. This adds weight to the relevance of this approach, since metastatic lesions are overall less likely to be surgically resected, leaving little chance for xenografts to ever be generated from them. Allaway et al. also showed that engraftment of FNA specimens from metastatic pancreatic adenocarcinoma sites was also more successful than FNA specimens of the primary site8. In the future clinicians and researchers could consider obtaining biopsy tissue from metastatic tumor sites since it potentially has a high chance of successfully establishing a xenograft. However, we did not examine whether this this was an effect of cell number or tumor biology of cell number versus tumor biology of primary tumors versus metastases, which is a limitation of this study and warrants further exploration.
In this study, we have shown that it is possible to generate xenografts from small volumes of tumor samples that can be obtained during an outpatient fine needle aspiration or core needle biopsy. This will enable tumor xenografts to be grown from patients that cannot be surgical candidates. The successful growth of a PDX from surgically resected tumor predicts clinical recurrence in pancreatic adenocarcinoma, and these patients are unlikely to be operated on additional times28. This method potentially opens the door to xenograft research on these recurrences in order to compare them to the xenograft grown from the primary tumor. Also, with abundant xenograft tissue, laboratories could also generate cell lines from each clinical scenario, as we have shown previously29. These PDX tumors and cell lines can be beneficial for future pre-clinical studies, and also guide personalized therapy for current patients by testing different chemotherapeutics on mice bearing tumors generated from the patient’s own tumor tissue4. The future direction for this project will be attempting xenografts of more biopsy samples from different cancers, especially metastases, obtained during outpatient diagnostic procedures rather than surgical resections.
Acknowledgments
This work was supported by the University of Texas M.D. Anderson Moonshot Gap Initiative as well as National Institutes of Health (NIH) grant T32CA009599 (D.R.). We would like to thank Mayrim Rios-Perez, M.D., Alexander Ondari, M.D., and Flavio Baio, M.D., for critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnson JI, Decker S, Zaharevitz D, Rubinstein LV, Venditti JM, Schepartz S, et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer. 2001;84:1424–1431. doi: 10.1054/bjoc.2001.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9:338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubio-Viqueira B, Hidalgo M. Direct in vivo xenograft tumor model for predicting chemotherapeutic drug response in cancer patients. Clin Pharmacol Ther. 2009;85:217–221. doi: 10.1038/clpt.2008.200. [DOI] [PubMed] [Google Scholar]
- 4.Rubio-Viqueira B, Jimeno A, Cusatis G, Zhang X, Iacobuzio-Donahue C, Karikari C, et al. An In vivo Platform for Translational Drug Development in Pancreatic Cancer. Clin Cancer Res. 2006;12:4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 5.Kim MP, Evans DB, Wang H, Abbruzzese JL, Fleming JB, Gallick GE. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat Protoc. 2009;4:1670–1680. doi: 10.1038/nprot.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hidalgo M, Bruckheimer E, Rajeshkumar NV, Garrido-Laguna I, Oliveira ED, Rubio-Viqueira B, et al. A Pilot Clinical Study of Treatment Guided by Personalized Tumorgrafts in Patients with Advanced Cancer. Mol Cancer Ther. 2011;10:1311–1316. doi: 10.1158/1535-7163.MCT-11-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. [accessed 13 Aug 2014];NHS treated cancer patients receiving major surgical resections. http://www.ncin.org.uk/publications/data_briefings/major_resection.
- 8.Allaway RJ, Fischer DA, de Abreu FB, Gardner TB, Gordon SR, Barth RJ, et al. Genomic characterization of patient-derived xenograft models established from fine needle aspirate biopsies of a primary pancreatic ductal adenocarcinoma and from patient-matched metastatic sites. Oncotarget. 2016;7:17087–17102. doi: 10.18632/oncotarget.7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim MP, Truty MJ, Choi W, Kang Y, Chopin-Lally X, Gallick GE, et al. Molecular Profiling of Direct Xenograft Tumors Established from Human Pancreatic Adenocarcinoma After Neoadjuvant Therapy. Ann Surg Oncol. 2012;19:S395–S403. doi: 10.1245/s10434-011-1839-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talmadge JE, Fidler IJ. Enhanced Metastatic Potential of Tumor Cells Harvested From Spontaneous Metastases of Heterogeneous Murine Tumors. J Natl Cancer Inst. 1982;69:975–980. [PubMed] [Google Scholar]
- 12.Dembinski JL, Krauss S. Characterization and functional analysis of a slow cycling stem cell-like subpopulation in pancreas adenocarcinoma. Clin Exp Metastasis. 2009;26:611–623. doi: 10.1007/s10585-009-9260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorunova L, Höglund M, Andrén-Sandberg Å, Dawiskiba S, Jin Y, Mitelman F, et al. Cytogenetic analysis of pancreatic carcinomas: Intratumor heterogeneity and nonrandom pattern of chromosome aberrations. Genes Chromosomes Cancer. 1998;23:81–99. doi: 10.1002/(sici)1098-2264(199810)23:2<81::aid-gcc1>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 14.Hahn SA, Seymour AB, Hoque ATMS, Schutte M, da Costa LT, Redston MS, et al. Allelotype of Pancreatic Adenocarcinoma Using Xenograft Enrichment. Cancer Res. 1995;55:4670–4675. [PubMed] [Google Scholar]
- 15.Yamano M, Fujii H, Takagaki T, Kadowaki N, Watanabe H, Shirai T. Genetic Progression and Divergence in Pancreatic Carcinoma. Am J Pathol. 2000;156:2123–2133. doi: 10.1016/S0002-9440(10)65083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan KS, Espinosa I, Chao M, Wong D, Ailles L, Diehn M, et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc Natl Acad Sci. 2009;106:14016–14021. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Natali P, Bigotti A, Cavaliere R, Liao S-K, Taniguchi M, Matsui M, et al. Heterogeneous Expression of Melanoma-associated Antigens and HLA Antigens by Primary and Multiple Metastatic Lesions Removed from Patients with Melanoma. Cancer Res. 1985;45:2883–2889. [PubMed] [Google Scholar]
- 18.Cornil I, Man S, Fernandez B, Kerbel RS. Enhanced Tumorigenicity, Melanogenesis, and Metastases of a Human Malignant Melanoma After Subdermal Implantation in Nude Mice. J Natl Cancer Inst. 1989;81:938–944. doi: 10.1093/jnci/81.12.938. [DOI] [PubMed] [Google Scholar]
- 19.Kim MP, Fleming JB, Wang H, Abbruzzese JL, Choi W, Kopetz S, et al. ALDH Activity Selectively Defines an Enhanced Tumor-Initiating Cell Population Relative to CD133 Expression in Human Pancreatic Adenocarcinoma. PLoS ONE. 2011;6:e20636. doi: 10.1371/journal.pone.0020636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patrawala L, Calhoun-Davis T, Schneider-Broussard R, Tang DG. Hierarchical Organization of Prostate Cancer Cells in Xenograft Tumors: The CD44+α2β1+ Cell Population Is Enriched in Tumor-Initiating Cells. Cancer Res. 2007;67:6796–6805. doi: 10.1158/0008-5472.CAN-07-0490. [DOI] [PubMed] [Google Scholar]
- 21.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 22.Hidalgo M, Amant F, Biankin AV, Budinská E, Byrne AT, Caldas C, et al. Patient-Derived Xenograft Models: An Emerging Platform for Translational Cancer Research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morton CL, Houghton PJ. Establishment of human tumor xenografts in immunodeficient mice. Nat Protoc. 2007;2:247–250. doi: 10.1038/nprot.2007.25. [DOI] [PubMed] [Google Scholar]
- 24.Suetsugu A, Katz M, Fleming J, Truty M, Thomas R, Moriwaki H, et al. Multi-color palette of fluorescent proteins for imaging the tumor microenvironment of orthotopic tumorgraft mouse models of clinical pancreatic cancer specimens. J Cell Biochem. 2012;113:2290–2295. doi: 10.1002/jcb.24099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McAllister SS, Gifford AM, Greiner AL, Kelleher SP, Saelzler MP, Ince TA, et al. Systemic Endocrine Instigation of Indolent Tumor Growth Requires Osteopontin. Cell. 2008;133:994–1005. doi: 10.1016/j.cell.2008.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller BE, Miller FR, Leith J, Heppner GH. Growth Interaction in Vivo between Tumor Subpopulations Derived from a Single Mouse Mammary Tumor. Cancer Res. 1980;40:3977–3981. [PubMed] [Google Scholar]
- 27.Lee H, Yoon T-J, Figueiredo J-L, Swirski FK, Weissleder R. Rapid detection and profiling of cancer cells in fine-needle aspirates. Proc Natl Acad Sci. 2009;106:12459–12464. doi: 10.1073/pnas.0902365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas RM, Truty MJ, Kim M, Kang Y, Zhang R, Chatterjee D, et al. The Canary in the Coal Mine: The Growth of Patient-Derived Tumorgrafts in Mice Predicts Clinical Recurrence after Surgical Resection of Pancreatic Ductal Adenocarcinoma. Ann Surg Oncol. 2014:1–9. doi: 10.1245/s10434-014-4241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang Y, Zhang R, Suzuki R, Li S, Roife D, Truty MJ, et al. Two-dimensional culture of human pancreatic adenocarcinoma cells results in an irreversible transition from epithelial to mesenchymal phenotype. Lab Invest. 2014 doi: 10.1038/labinvest.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]