Abstract
Poor sleep is increasingly being recognised as an important prognostic parameter of health. For those with suspected sleep disorders, patients are referred to sleep clinics which guide treatment. However, sleep clinics are not always a viable option due to their high cost, a lack of experienced practitioners, lengthy waiting lists and an unrepresentative sleeping environment. A home-based non-contact sleep/wake monitoring system may be used as a guide for treatment potentially stratifying patients by clinical need or highlighting longitudinal changes in sleep and nocturnal patterns. This paper presents the evaluation of an under-mattress sleep monitoring system for non-contact sleep/wake discrimination. A large dataset of sensor data with concomitant sleep/wake state was collected from both younger and older adults participating in a circadian sleep study. A thorough training/testing/validation procedure was configured and optimised feature extraction and sleep/wake discrimination algorithms evaluated both within and across the two cohorts. An accuracy, sensitivity and specificity of 74.3%, 95.5%, and 53.2% is reported over all subjects using an external validation dataset (71.9%, 87.9% and 56%, and 77.5%, 98% and 57% is reported for younger and older subjects respectively). These results compare favourably with similar research, however this system provides an ambient alternative suitable for long term continuous sleep monitoring, particularly amongst vulnerable populations.
Keywords: Sleep, polysomnography, actigraphy, ambient intelligence, feature extraction, machine learning
I. Introduction
Sleep is a fundamental physiological process with important restorative functions. It occurs in all living mammals and generally over a significant portion of each day [1]. Sleep problems have been shown to be detrimental to human health. In humans, short (seven hours or less) and long (nine hours or more) durations of sleep have been shown to be significant predictors of death in prospective population studies [2]. Sleep disturbances may be indicative of poor health and functional deficits, especially in older adults [3], [4]. Total sleep time is reduced in the elderly and this is not due to a reduced need for sleep, but in a diminished ability to sleep [5]. Sleep complaints are commonly reported by over 50% of those aged 65 and older [4]. These complaints include getting less sleep, frequent awakenings, waking up too early, excessive daytime sleepiness, and napping during the day. Decreased quality of life, and higher rates of depression and anxiety are reported in patients with sleeping difficulties [6]. High incidences of balance, ambulatory and visual difficulties (after controlling for medication use) have been reported in older adults with sleep problems [7]. Furthermore, decreased total sleep time (TST), an increased sleep latency (SL), defined as the time taken to fall asleep in bed, and a poor sleep efficiency (SE), defined as the percentage of TST over total time in bed (TIB), are linked to a greater risk for mortality (even when controlling for related covariates) [8]. Additionally, the symptoms of various chronic conditions continue into the night and result in a disturbed sleep; these include movement disorders, neuromuscular diseases, depression, dementia, epilepsy, obesity and circadian rhythm disorders [9].
The gold standard sleep assessment technology is polysomnography (PSG) which records multiple physiological signals (including brain activity, muscle tone, eye movements, heart rate and respiration) during sleep. This is generally performed in a sleep clinic and a trained sleep scorer uses a strictly defined set of rules to classify each 30 second epoch into either wake or a variety of sleep stages including rapid eye movement (REM) and non-REM (NREM) sleep [10]. However the application of these rules is subjective and an inter-rater agreement rate of 82% has been reported using data from multiple subjects and across separate sleep laboratories [11]. Additionally, PSG is intrusive, costly, time consuming and often alienates the patient. Wrist actigraphy is the current ambulatory gold standard sleep monitoring device. It consists of a two axis accelerometer which records the rest/activity patterns of the wearer and converts an activity metric to sleep/wake estimates using a thresholding algorithm and some additional logic. Wrist actigraphy has been shown to estimate nocturnal sleep duration and sleep-wake patterns reliably where PSG is not a suitable alternative [12]. However, a low wake detection capacity is often reported with this device as the device cannot discriminate between quiescent wake and sleep [13]. Wrist actigraphy is dependent on the adherence (and active participation) of the wearer. Sleep diaries are also used (often concomitantly with wrist actigraphy) to estimate sleep duration in normal, institutionalised and pediatric populations. However their validity relies upon the attentiveness of the individual filling the diaries out (in cases where the diaries are filled out by the individual). A trade-off exists for these technologies between accuracy and suitability for long term deployment. This paper proposes an ambient sleep/wake and bed-movement sensor suitable for extended home deployments to invesigate long-term changes in sleeping patterns, especially amongst vulnerable populations such as those with memory impairment. Existing systems require either a worn sensor or a sensing device visible within the bedroom which may result in stigma due to the need for such a technology.
In this contribution, a non-contact unobtrusive under-mattress bed sensor (UMBS) is proposed as a means of: (1) quantifying bed-based movements, and; (2) discriminating between sleep and wake. This contribution builds on previous research where novel algorithms were found to reliably measure breathing rate and body movement from UMBS data [14], and a preliminary study of a cohort of community based older adults [15] that showed high correlation between wrist actigraphy and motion metrics derived from UMBS data. This paper initially details a number of movement features which may be extracted during sleep building on previous research [16]. Subsequently, a classification setup is detailed which optimises the reliability and accuracy of the discrimination of sleep and wake using a large and rigorous research clinic-based dataset. It is proposed that the combination of these data provide a holistic description of sleep using a non-invasive sensor which can be deployed over extended periods in the community.
The remainder of this paper is organised as follows. Section II provides a literature review. Section III introduces the UMBS and feature extraction algorithms. An overview of the study, methodology, and techniques used to extract sleep/wake state from the UMBS data is given in Section IV. Results are described in Section V. A discussion and conclusion are given in Sections VI and VII, respectively.
II. Literature review
Proposed approaches for home-based and/or long term monitoring range from ambient (that is, embedded in the environment), worn, and/or image-based solutions. There is a wide disparity between how these systems are validated including methodological differences, the formulation of the gold standard comparison dataset, the potential use of an external test dataset, and also the inconsistent and often non-overlapping performance measures reported across these systems.
Video-based sleep monitoring solutions have been proposed [17], [18] and while their utility is impressive participants are often uncomfortable with the presence of video recording equipment in the home and especially in the bedroom [19]. Privacy concerns were diminished only when unfavourable alternatives (such as nursing homes) were considered [19]. However, other technologies may provide sufficient utility while retaining privacy.
Passive infra-red (PIR) based monitoring systems [20] have been developed and a high accuracy reported, but a number of potential usability issues remain including the varying location of heaters between environments and the presence and types of bed sheets shielding sensors from the minute movements of the individual.
Radar based technologies [21] report high accuracies in detecting movement compared to wrist actigraphy. Further work reported a high accuracy in sleep/wake discrimination (87% sensitivity, 50% specificity) across a test dataset of 113 nights [22].
Load-cell movement detection sensors [23], [24] have been shown to have a high capacity to detect respiration and movement although this may depend on the orientation of the individual.
Bed monitoring systems using pressure pads placed on top of the mattress, underneath the bed sheets, have been developed which detect the ballistocardiogram and detect the heart rate and respiratory rate accurately [25], [26]. Under mattress sensors [27]–[31] have been shown to measure respiration or both respiration and heart rate effectively. A pressure sensitive e-textile bed sheet reported high accuracy in sleep wake discrimination (70% precision, 71% recall) with 7 nights of data [32]. Cardiorespiratory and movement-based signals from a respiratory inductance plethysmography band, electrocardiogram and a pressure sensitive bed were used to discriminate between sleep and wake: 92% accuracy, wake/REM/NREM: 81% and all sleep stages: 69% using 85 nights of sleep [33]. Actigraphic and respiratory effort-derived features and a linear discriminant classifier reported an accuracy of 95.7% using 15 nights of data [34]. Activity and cardiovascular data were used to generate a sleep/wake accuracy of 78.3% using 20 nights of data and a support vector machine and hidden markov model-based system [35].
Pillow based sensors [36] have been proposed as solutions to domestic non-contact sleep monitoring by measuring heart and respiration rate. These systems are particularly suited to non-contact long term sleep monitoring as they do not require specialist expertise to install.
Home-based sleep monitoring has not been forgotten in the burgeoning wave of Internet of Things and Quantified Self devices including the Withings Aura [37], Fitbit [38], Xiaomi Mi Band [39] and smartphone applications like Sleep Cycle Alarm Clock [40]. A comparison of these and similar commercial personal health monitoring devices against global sleep measures (via PSG) in healthy populations has found no significant differences between them in terms of the their ability to estimate total sleep time [41]. However, it is reasonable to assume that a simple in/out of bed sensor may provide similar accuracy levels in terms of total sleep time. An epoch-by-epoch comparison is necessary to provide a more rigorous evaluation of the performance of these systems. Furthermore, healthy adult users were found to be unreliable for any devices which required the user to initiate a sleep tracking setting [41]. Therefore, when moving to long-term sleep monitoring, particularly in populations with potential memory issues, an ambient technology requiring no user interaction is desirable.
III. Under Mattress Bed Sensor
The UMBS system is composed of an under mattress pressure sensing grid, a data collection system and algorithms to extract respiration and body movement information, or features. The process of collecting and extracting these features is described below.
A. UMBS and Data Collection
The UMBS consists of a grid of 24 fibre optic pressure sensors, or tactels, integrated into a lightweight 1 cm × 90 cm × 23 cm foam mat (Tactex Controls Inc., Canada) and evenly distributed in a 3 × 8 grid as shown in Figure 1. Relative pressure is recorded by measuring the amount of light passing between an emitter and receiver woven into a semi-permeable substrate. Changes in pressure applied to the substrate results in varying amounts of light passing between the fibre optic emitter and receiver. When no pressure is being applied to a tactel, the foam is not compressed and light can pass freely from the source to a receiver. When pressure is applied, the porous nature of the foam is reduced which restricts the amount of light that travels between the emitter and receiver. The light signal is transduced into an electrical signal at a rate of 20Hz and subsequently fed into an analog to digital converter reporting tactel values ranging from 0 (lowest pressure applied, maximum light transmission) to 2047 (full pressure applied, no light transmission). The tactels are optically isolated from one another. Tactel values were polled using customised software via a Universal Serial Bus (USB) serial port connection resulting in a non-constant sampling rate of mean 15.28Hz (range: 9.1Hz – 21.74Hz) [14]. This inconsistent sampling rate was due to a simplex polling protocol and a non-constant delay between data requests and replies. Linear interpolation was employed to provide a constant sampling rate of 10Hz for later analysis.
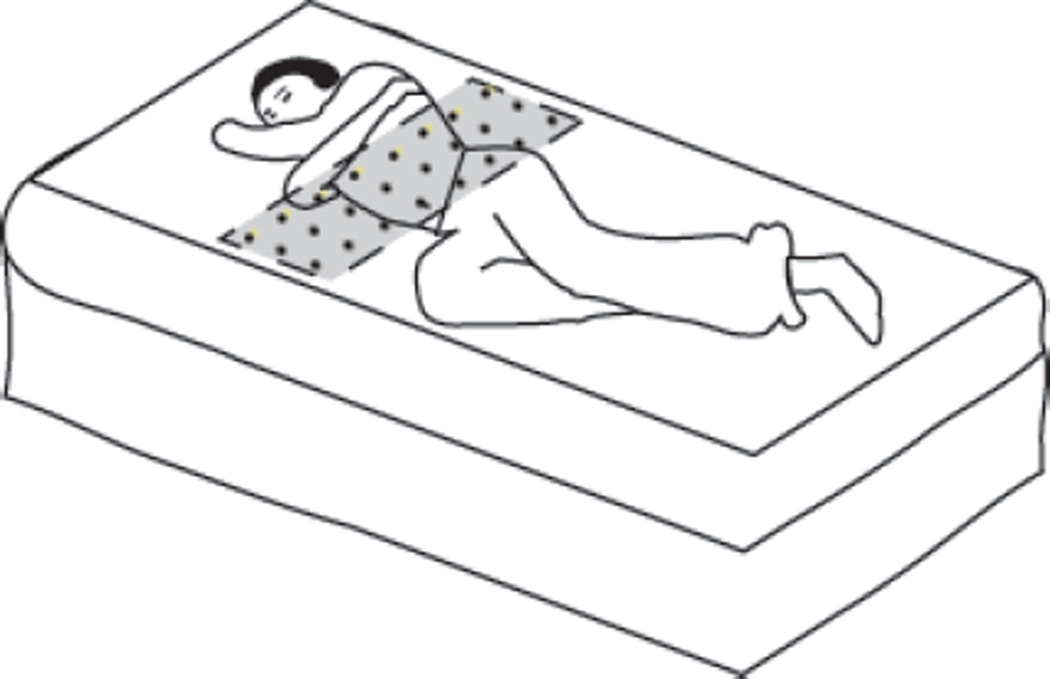
Figure 1.
Position of the UMBS relative to the mattress and subject.
The UMBS pressure sensing grid is placed beneath the upper torso of the subject as shown in Figure 1. When the subject is not lying over the UMBS, tactel values remain low (less than 500 units) and vary with a range of less than 5 units. When subjects lie on the mattress (a bed entry event), their body weight results in an increase in downward pressure on the bed and resultantly onto some of the tactels in the UMBS. When they exit the bed, the tactel values return to their baseline values. Because a person’s weight is not distributed evenly across the mattress, but rather is localised to their position in the bed, some tactels do not experience any change in pressure even when the person is lying on the mattress, while others are saturated as the pressure applied exceeds the dynamic range of the tactels (0–2047 units). Tactels which are neither unaffected nor saturated by a subject’s weight typically contain micro-movements (or cyclic variations) emanating from the individual while lying in the bed.
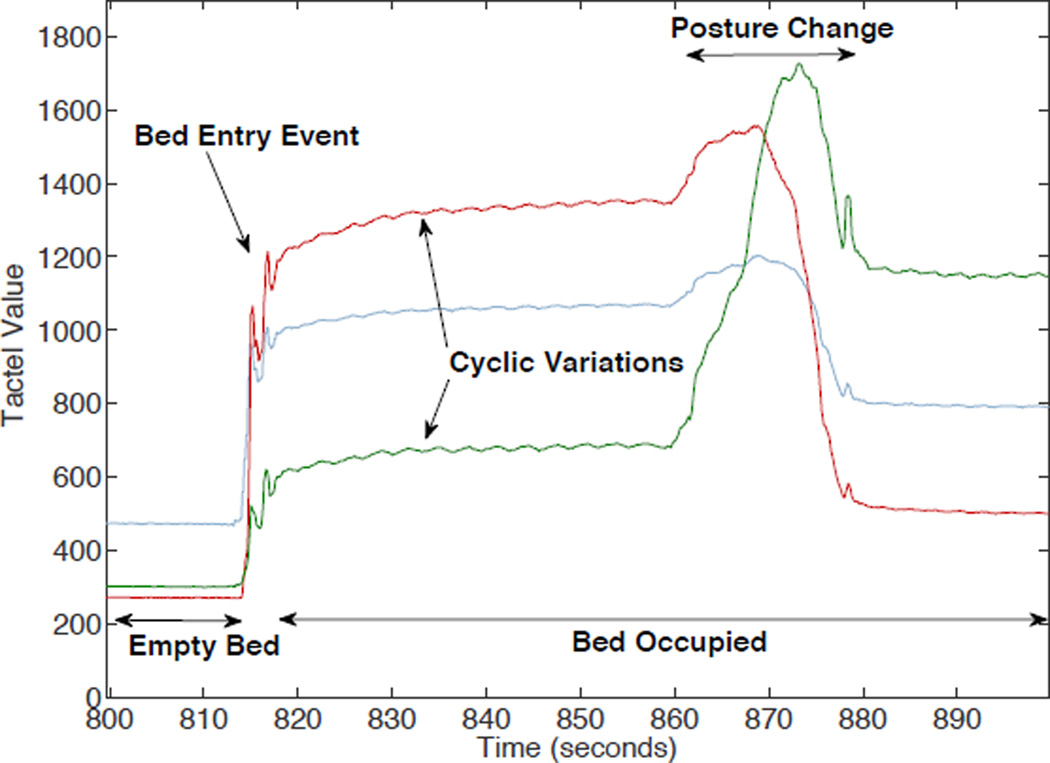
The evolution of the pressure signal for three unsaturated tactels over a 100 second interval around a bed entry event is given in Figure 2. Tactel values are initially low (less than 500 units) and raise upon a bed entry event. Small cyclic variations in pressure can be observed in two of the three tactels which correspond to respiration and in some cases heart rate related movement [14]. Over extended periods of time, body posture changes, or macro-movements, can also be identified.
Figure 2.
Example UMBS data from three active tactels during bed entry and posture change event.
For a more thorough description of the UMBS, including a description of the extraction of respiration rate and body movement, the reader is referred to previous work [14], [16]. Other work elsewhere is examining the use of the UMBS for in-bed mobility monitoring during postural transitions [42].
B. Feature Extraction Algorithms
Four types of movement features were derived from the UMBS data: respiration rate, statistical, temporal and spatial (as described in Table I). A brief description of the feature extraction algorithms is given below:
TABLE I.
UMBS Movement Features
| Feature | Description |
|---|---|
| Respiration Rate | The number of breathing cycles in the surrounding 5 minute window. |
| Statistical (Act) | The sum of differences between all pressure values over all samples within each data window. |
| Temporal (TMF) | The mean of the absolute differences between all pressure tactel values over the UMBS for each sample |
| Spatial (SMF) | The horizontal difference in the central body position. |
1) Respiration
A method of extracting respiration rates using the UMBS has been developed and validated against a strain gauge [14]. In this work, a custom respiration rate detection algorithm yielded a mean difference of 0.12 breaths per five minutes and a mean percentage error (MPE) of 0.16% when the sensor was placed beneath the mattress. The algorithm removes the effect of slow postural changes and unwanted spectral noise on the UMBS using filtering, fuses the data from active signals and employs an optimised peak detection algorithm to detect respiration rate. Results are reported on the basis of a 5 minute window to ensure a relatively high number of respiration cycles occur.
2) Statistical Movement Features
Defining the difference between successive tactel values as
| (1) |
where N is the number of samples in an epoch, xij is the value of the jth tactel at the ith sample instant, and δij = 0 for i=1 and 1 ≤ j ≤ 24, an overall UMBS motion metric can be computed as
| (2) |
This metric, which is similar in form to the activity motion count calculated by wrist actigraphy, is computed over a predefined epoch length of 60 seconds.
3) Temporal Movement Features
The mean of δij over all tactels for each time instant i, denoted TMF(i), was used as a temporal movement feature to track the magnitude of motion registered by each tactel over time. TMF is a representation of the total applied force without consideration of the number of tactels experiencing motion. Hence,
| (3) |
Based on an empirical evaluation, determined by experimentally gathered movement and non-movement data, the minimal threshold for movement detection, as measured using TMF, was determined to be 4. In this investigation, background non-movement activity (such as breathing, etc.) generally occurred at TMF less than 1 (where 94% of values below a TMF of 4 occurred during non-movement times). An illustrative example of movement and non-movement using both the UMBS data and the TMF metric is included as supplementary material.
4) Spatial Movement Features
A continuous description of spatial displacement (resultant from postural changes) in bed was derived by examining the pressure applied, and changes therein, to the individual UMBS tactels. This spatial movement feature (SMF) measures the change in position of the subject over time, but does not measure movement where no change in position occurs (TMF should reflect this). Tactels recording instantaneous body posture were defined as those registering a value of greater than 500 s tactels registered less than 500 when in their resting state (i.e. when unloaded). A centre of pressure (COP) metric was derived as the central point in the lateral direction through which pressure was applied. In order to increase the accuracy of COP measurement, linear interpolation was employed between neighbouring real tactels and virtual tactels created to yield a total of 71 equally spaced measurement points across the mat. The centre of pressure metric was then computed as
| (4) |
where statusx,y is a boolean value indicating whether or not pressure is being applied at point (x, y), that is
| (5) |
and Posx is a x-coordinate of the current measurement point (x, y). COPx was not deemed to provide useful information when examined independently, however changes in COPx over time provide a spatial measurement of in-bed movement. The first derivative of the COPx data was used to quantify such movement, that is:
| (6) |
where SMF1 is 0.
IV. Methods
This section describes how data was collected during the clinical studies, details the features extracted from the data, gives an overview of the classifiers used and how they were applied, and how the performance of the system was measured.
A. Study Protocol
Data were collected as part of two research protocols carried out by the Division of Sleep and Circadian Disorders in the Intensive Physiological Monitoring Unit of the Center for Clinical Investigation at the Brigham and Womens’ Hospital, Boston, MA, USA. Full ethical approval was granted by the Partners Health Care Human Research Committee. UMBS data were collected using the system described in Section III. PSG data was collected for each scheduled sleep episode, including EEG (C3, C4), EOG, EMG (submentalis), and ECG using a Vitaport Digital Sleep Recorder (Temec Instruments, Kerkrade, Netherlands) and scored according to the Rechtschaffen and Kales guidelines [10] in 30s epochs. Signals were sampled at 256 Hz, low-pass filtered at 30Hz and high-pass filtered at 0.159Hz (a time constant of 1.0 seconds), and stored at 128 Hz. Two 30 second PSG epochs were concatenated and taken as one 60 second epoch (where there was no change between sleep and wake) for the remainder of this analysis. A consistent lab time was used to ensure concomitant PSG and UMBS time. The electrodes were applied approximately two hours prior to the scheduled lights out and recording began approximately one hour prior to the scheduled lights out. Data during the lights out period was used in this analysis. The PSG data were scored according to the standard criterion [10] into one of seven states: wake, Non-REM sleep (stage 1, stage 2, stage 3 and stage 4), REM sleep or movement. All sleep stages were defined as sleep in this analysis. Movement epochs related to periods in which motion artefacts obscured more than half (≥15 sec) of the epoch and was re-scored as wake in later analysis. Thus, all epochs analysed in this study were either scored as sleep or wake.
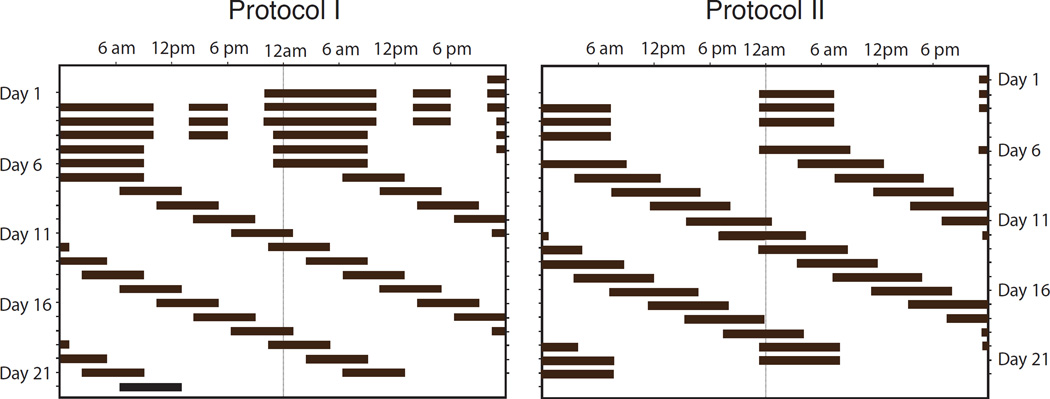
The UMBS study was placed within a broader research program where sleep and waking were scheduled to occur in controlled laboratory conditions, in some cases on a non-24-hour schedule, in order to better understand circadian control of sleep and waking in healthy adults. Each participant in this protocol spent multiple days in the laboratory in which their sleep episodes were scheduled by the experimenters. The subject was instructed to sleep, or attempt to sleep, during lights off episodes, and was required to remain in bed in the dark until scheduled lights on. This process of scheduling the sleep and wake times was external to this analysis. An example of the schedules used in the two experiments is shown in Figure 3. The subjects were continually monitored by a team of specialist healthcare clinicians and technicians throughout both sleep and wake episodes. Data was collected under two protocols. For both protocols, data from the initial 21 days were used for analysis, however the studies ran beyond this duration.
Figure 3.
Double raster plot for both protocols (black represents scheduled periods of sleep).
a) Protocol I
In this protocol, the natural nocturnal sleep length was extended to twelve hours during the initial three days of the study (see Figure 3). A midday nap was scheduled, however sleep during these naps were not included in the present analysis. The study began with extended sleep times and naps in order to ensure the subject had no short term sleep debt. Over the next three days, the scheduled sleep opportunity was ten hours. Over the remaining thirteen days the day length was extended to a 28 hour schedule, while the sleep length was reduced to six and a half hours per 28 hour day. This protocol meant that subjects progressively accumulated sleep loss. As such, the wake time during scheduled sleep episodes diminished as the protocol continued.
b) Protocol II
Baseline data were collected during the initial 3 8-hour nights of the study (see Figure 3). Subsequently the subjects were kept awake for a period of forty hours. Subjects were then scheduled to sleep for 9.33 hours. The day length was then modified to a 28 hour schedule with 9.33 hours of sleep per 28 hour schedule. The subjects were scheduled to be in bed for 8 hours at the same time as the baseline nights for the final two nights of the research study.
B. Subject Details
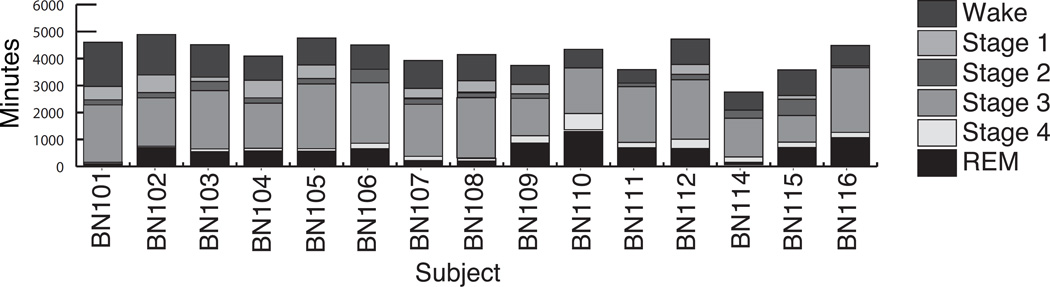
Data, collected from 30 people, were split into 5 datasets for this analysis as detailed in Table II. Subjects were both healthy younger and older adults and were split between protocol I and protocol II. Datasets A (8 younger adults) and B (7 older adults) contained at least 13 sleep records each. These data were used to train and validate the classification algorithms. The total distribution of sleep stages per subject over the sleep episodes is given in Figure 4. Datasets C (8 younger adults) and E (1 older adult) contained data from one and three sleep records respectively and were used to test the performance of the optimal classification algorithm on unseen subjects. The cohort in dataset D did not have any corresponding PSG data and was used to assess the correlation in the underlying data. The mean and standard deviation of the sleep efficiency over all sleep episodes where data was collected for each subject is also given in Table II
TABLE II.
Data sets For Classifier Training and Validation.
| UMBS Subcode |
Study | Age yrs |
Sex m/f |
Records scored |
Sleep efficiency mean ± std. (%) |
|---|---|---|---|---|---|
| Data Set A: Healthy Younger Adults; For classifier training and validation | |||||
| BN101 | Protocol II | 19 | M | 13 | 1.87 ± 1.19 |
| BN102 | Protocol II | 23 | M | 13 | 13.14 ± 7.32 |
| BN103 | Protocol II | 21 | F | 13 | 9.47 ± 6.26 |
| BN104 | Protocol II | 24 | M | 13 | 14.75 ± 10.31 |
| BN105 | Protocol II | 19 | F | 13 | 10.09 ± 5.26 |
| BN106 | Protocol II | 21 | M | 13 | 15.12 ± 3.53 |
| BN107 | Protocol I | 20 | F | 13 | 9.51 ± 7.74 |
| BN108 | Protocol I | 24 | F | 13 | 3.93 ± 2.23 |
| Data Set B: Healthy Older Adults; For classifier training and validation | |||||
| BN109 | Protocol I | 56 | F | 14 | 22.02 ± 10.97 |
| BN110 | Protocol I | 70 | M | 14 | 33.04 ± 13.59 |
| BN111 | Protocol I | 56 | M | 14 | 21.0 ± 12.40 |
| BN112 | Protocol I | 58 | F | 14 | 15.52 ± 8.89 |
| BN114 | Protocol I | 64 | M | 14 | 5.63 ± 4.87 |
| BN115 | Protocol I | 55 | M | 14 | 13.48 ± 17.47 |
| BN116 | Protocol I | 60 | F | 14 | 25.29 ± 12.36 |
| Data Set C: Healthy Younger Adults; For classifier testing | |||||
| BN117 | Protocol I | 19 | F | 1 | 7.26 |
| BN118 | Protocol I | 18 | F | 1 | 16.32 |
| BN119 | Protocol I | 27 | M | 1 | 22.32 |
| BN120 | Protocol I | 24 | F | 1 | 19.87 |
| BN1211 | Protocol II | 19 | F | 1 | 2.70 |
| BN1221 | Protocol II | 24 | M | 1 | 2.26 |
| BN1231 | Protocol II | 23 | M | 1 | 1.04 |
| BN124 | Protocol II | 27 | M | 1 | 14.13 |
| Data Set D: Healthy Younger Adults; used For investigative purposes | |||||
| BN1252 | Protocol II | 23 | F | 0 | - |
| BN1262 | Protocol II | 22 | F | 0 | - |
| BN1272 | Protocol II | 21 | F | 0 | - |
| BN1282 | Protocol II | 19 | M | 0 | - |
| BN1292 | Protocol II | 21 | F | 0 | - |
| BN1302 | Protocol II | 24 | M | 0 | - |
| Data Set E: Healthy Older adult; For classifier testing | |||||
| BN113 | Protocol I | 55 | F | 3 | 41.56 ± 10.35 |
Subjects had less than twenty 60 second epochs of wake.
Subjects sleep records were not scored, however their UMBS data were used to generate independent coefficients.
Figure 4.
Distribution of sleep stages per subject over all sleep episodes. BN101-BN108 belong to the younger adult cohort, while BN109-116 are part of the older adult cohort.
C. Features
13 UMBS features, taken from the temporal (TMF), spatial (SMF) and statistical (Act) metrics and the UMBS respiration rate estimate, were used in this analysis (as given in Table III). A large number of features were found to be correlated with each other (see Table IV) using dataset D.
TABLE III.
UMBS-Derived Features.
| No. | Feature Type | Feature Description |
|---|---|---|
| 1 | Respiration | Number of Respiratory Peaks |
| (see Section III-B1) | ||
| 2 | Spatial Movement | Feature Standard Deviation |
| 3 | SMF (Eqn. 6) | Maximum |
| 4 | Mean | |
| 5 | Time Greater Than 0 | |
| 6 | Num. Distinct Movements | |
| 7 | Temporal Movement Feature | Standard Deviation |
| 8 | TMF2 (Eqn. 3) | Maximum |
| 9 | Mean | |
| 10 | Median | |
| 11 | Time Greater Than 4 | |
| 12 | Num. Distinct Movements | |
| 13 | Statistical | Log of Act |
| (See Equation 2) | ||
TABLE IV.
Correlation (r) Between the UMBS Features Averaged over the 6 Subjects in Data Set D.
| Resp. Peaks |
SMF | TMF | Statistical Act |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| St. Dev.1 | Max | Mean | TMT2 | NCM3 | St. Dev.1 | Max | Mean | Median | TMT2 | NDM3 | ||||
| Resp. | Peaks | 1.000 | ||||||||||||
| SMF | St. Dev.1 | −0.145 | 1.000 | |||||||||||
| Max | −0.159 | 0.810 | 1.000 | |||||||||||
| Mean | −0.107 | 0.923 | 0.569 | 1.000 | ||||||||||
| TMT2 | −0.094 | 0.854 | 0.517 | 0.942 | 1.000 | |||||||||
| NCM3 | −0.093 | 0.847 | 0.509 | 0.937 | 0.997 | 1.000 | ||||||||
| TMF | St. Dev.1 | −0.137 | 0.376 | 0.606 | 0.174 | 0.140 | 0.138 | 1.000 | ||||||
| Max | −0.144 | 0.353 | 0.577 | 0.161 | 0.131 | 0.130 | 0.979 | 1.000 | ||||||
| Mean | −0.120 | 0.372 | 0.584 | 0.182 | 0.137 | 0.134 | 0.958 | 0.909 | 1.000 | |||||
| Median | 0.020 | 0.156 | 0.259 | 0.070 | 0.027 | 0.025 | 0.529 | 0.484 | 0.700 | 1.000 | ||||
| TMT2 | −0.157 | 0.412 | 0.618 | 0.215 | 0.170 | 0.171 | 0.897 | 0.842 | 0.936 | 0.593 | 1.000 | |||
| NCM3 | −0.141 | 0.390 | 0.539 | 0.231 | 0.178 | 0.179 | 0.661 | 0.622 | 0.708 | 0.494 | 0.851 | 1.000 | ||
| Stat. | Act | −0.124 | 0.274 | 0.463 | 0.112 | 0.076 | 0.078 | 0.638 | 0.627 | 0.623 | 0.420 | 0.663 | 0.647 | 1.000 |
Standard Deviation (St. Dev.)
Total Movement Time (TMT)
Number of Distinct Movements (NDM)
Bold relates to correlations which are statically significant (p ≤ 0.05)
D. Classifier
A number of classifiers were investigated for discriminating between sleep and wake using the features derived from the UMBS. In addition to classical Linear and Quadratic Discriminant Analysis (LDA and QDA), the nonparametric k-Nearest Neighbour (kNN) classifier, and the nonlinear Artificial Neural Network (ANN) and Support Vector Machine (SVM) classifiers were tested to see if they provided superior performance to LDA. These are now briefly introduced and details given on how their internal configurations were selected and optimised. The software implementation employed for each classifier was that provided in MATLAB version 7.12.0 (R2011a). Default parameters and training algorithms were used, unless otherwise specified.
a) Discriminant Classifiers
Discriminant classifiers, configured using training data, divide an n-dimensional input (or feature) space into subspaces which optimally discriminate between classes on the assumption that the conditional probability distribution for each class is multivariate normal. Decision boundaries between classes are linear if class covariance matrices are assumed to be the same and quadratic otherwise, yielding linear discriminant analysis (LDA) and quadratic discriminant analysis (QDA), respectively [45]. An attractive feature of LDA and QDA is that they have relatively low complexity and are parameter free.
b) k-Nearest Neighbour (kNN) classifier
The k-Nearest Neighbour classifier is a simple, non-linear classification method which generally results in relatively high accuracy. It is a non-parametric, memory-based algorithm which classifies new samples based on the classification of the k most similar samples in a reference training dataset. Here, the Euclidian distance between samples was used as a similarity metric and the majority class of the k nearest neighbours taken as the sample classification. The optimum number of nearest neighbours k was determined by cross-validation from a candidate set k=1, 5, 9 based on a preliminary investigation that showed that the optimal k value was less than 10. The restricted search was undertaken to reduce the computational overhead.
c) Artificial Neural Network (ANN) Classifier
An ANN is an interconnected network of nodes, or neurons, loosely modelled on the interconnection and operation of neurons in the brain. By employing appropriate learning strategies to adjust network parameters ANNs can learn to approximate arbitrary non-linear input-output relationships between variables. For the classification task considered here, a single hidden layer multilayer perceptron (MLP) neural network with tanh activation functions was used to perform the classification task. Early stopping using cross-validation was employed to determine the optimum number of iterations in order to avoid over-fitting. To allow for the potential of getting stuck in a poor local minimum training was repeated 5 times with different random parameter initializations and the best performing network selected. There are several options for optimising the topology of a neural network, for example varying the number of hidden layer neurons or even the number of hidden layers. Here, a single hidden layer topology was used and the number of hidden layer units was selected as either 20 or 40. More extensive optimization of network topology was not undertaken due to the computational overhead involved, and instead early stopping was employed to ensure good generalization performance. For further information on ANNs the interested reader is referred to Bishop [46].
d) Support Vector Machines (SVM)
Support Vector Machines generate boundaries that maximally separate classes. Boundaries can be linear or nonlinear. Non-linear boundaries are achieved by employing the so called kernel trick, where the feature space is mapped into a high dimensional space using a kernel function, K(x, y) [47], [48]. Determining linear boundaries in this high dimensional space corresponds to defining non-linear boundaries in the original feature space. Many functions can be used for the mapping kernel. In this work the radial basis function (RBF) kernel, a popular choice for continuous feature spaces, is selected. Using this configuration SVMs have two hyperparameters, the kernel width, σ, and the box constraint, C. The box constraint is a parameter which controls the extent to which misclassifications are allowed to occur on the training data and effectively determines the smoothness of the boundaries between classes. The optimal hyperparameters for each classifier were determined by performing a grid search and selecting the values that yielded the best cross-validation error. Following an initial investigation, and mindful of the computational complexity of SVM training, the search grid range was set as 0.01 to 4 for σ with a grid resolution of 0.5 and 0.01 to 20 for C with a grid resolution of 1 unit.
E. Performance Measures
The performance of a binary classifier can be quantified in terms of its ability to correctly predict the actual class (positive or negative) for all samples under investigation. When a class is incorrectly predicted it results in either a false negative (FN) or false positive (FP), while correctly classified samples are either true positives (TP) or true negatives (TN). Using these definitions the accuracy of a classifier can be defined as
| (7) |
However, seeking to maximise this metric can be sub-optimal if the data be largely skewed toward either class. In order to take account of this bias, the specificity and sensitivity (also referred to as recall) metrics, defined as:
| (8) |
| (9) |
are often reported instead. These metrics quantify the proportion of correctly classified negative and positive samples, respectively. Alternatively, the classifier performance can be expressed in terms of the F-Score,
| (10) |
which is the harmonic mean of the recall (eqt. 9) and the precision of the classier, where precision is defined as:
| (11) |
The use of the harmonic mean ensures that the F-score is heavily skewed toward the lower value and this addresses the situation where there is a large difference between precision and recall, which can arise with unbalanced class sizes.
F. Classifier Training and Validation Preprocessing Procedure
Datasets A and B were split into subsets of sleep and wake epochs on a per subject basis. 400 epochs of sleep and 400 epochs of wake, randomly chosen from each person, were grouped into a single training/validation dataset. For two of the younger subjects (BN101 and BN108) and one older adult (BN113), random sampling with replacement was used to select the wake epochs as there were less than 400 wake epochs available (95, 198 and 196 wake epochs available respectively as shown in Figure 4). These data were then grouped into three sets: younger adults, older adults and all subjects.
The number of samples available to train the classifiers for the younger, older and all subjects groups are given in Table V. For the SVM, a reduced number of samples was used for the training procedure (as per Table V) due to the significantly increased computational overhead. Due to the low number of data points available, Monte Carlo cross-validation was used to avoid data related bias and ensure consistency of results. The training/validation task, as described above, was repeated 5 times for each classifier with randomly chosen subsets of samples in the training and validation datasets.
TABLE V.
Sample sizes for all classifiers.
| Classifier | No. Samples | |||||
|---|---|---|---|---|---|---|
| Younger | Older | All | ||||
| Tr | Va | Tr | Va | Tr | Va | |
| SVM | 320 | 6080 | 280 | 5320 | 300 | 11700 |
| Others | 4266 | 2134 | 3732 | 1868 | 8000 | 4000 |
No. of Training Samples(Tr), No. of Validation Samples (Va)
G. Independent Classifier Testing Cohort
Each classifier was applied to respective independent data of 5 younger adults (1 night each), an older adults (3 nights) and a combination of these (datasets C and E as per Table II).
For the younger adult cohort, fifty samples of sleep and fifty samples of wake were randomly selected from each of the five subjects in dataset C and compiled into the younger adult testing dataset. Random sampling with replacement was used to select the fifty wake epochs due to the low number of wake epochs available in this dataset. A much lower number of samples was used here compared to the training and validation stage as the number of wake epochs was below 50 for subjects BN117 and BN120 (23 and 25 epochs respectively). The subjects were only awake for 13% of the recordings. Random sampling without replacement was used to select fifty of the sleep epochs in this dataset. The subsets of chosen sleep and wake epochs from all subjects were combined to create a set of 500 chosen epochs.
For the older adult cohort, 200 samples of sleep and 200 samples of wake were randomly selected for each subject over the three nights and compiled into the older adult testing dataset. Random sampling without replacement was used to select the sleep and wake epochs in this dataset as the dataset contained over 41% wake epochs. The subsets of chosen sleep and wake epochs from all sleeping episodes were combined to create a set of 400 chosen epochs.
The data from one sleeping episode from one younger subject (BN119) and one older subject (BN113) were compiled into a testing set for older and younger subjects. The data from the younger subject contained 113 wake epochs and 472 sleep epochs. The data from the older subject contained 264 wake epochs and 233 sleep epochs. 200 samples were randomly selected, without replacement where possible, to create a dataset of 400 sleep epochs and 400 wake epochs. This dataset contained an equal contribution of data from younger and older adults.
V. UMBS Sleep/Wake Classification Results
In this section, results from applying each classifier to the features with the aim of discriminating sleep from wake epochs to training/validation UMBS data from each cohort, using Monte Carlo cross-validation, are given. Finally, the optimally trained classifier is selected and applied to an independent testing dataset to provide impartial results.
For the training/validation procedure, data from 8 younger subjects over 13 scheduled sleeps each and 7 older subjects over 14 scheduled sleeps each were collected. 400 epochs of sleep and 400 epochs of wake were randomly selected from each subject and evenly split to create training and validation data sets for a younger, older and all subjects cohorts. For the testing procedure, data from 5 younger subjects over 1 schedule sleep each and 1 older subject over 3 sleep episodes were used to create an independent test data set of younger cohort and older cohort respectively. 50 epochs of sleep and 50 epochs of wake were selected from each sleep episode from the younger data set. 200 epochs of sleep and 200 epochs of wake was extracted over the 3 sleep episodes from the older subject. An additional all subjects cohort was extracted using data from one sleep episode from one younger and one older subject to ensure the data was not biased to a certain cohort.
The mean performance of the classifiers (F-score) over all repetitions on the validation data is reported in Table VI. Low standard deviations (less than 0.025) were found in the F-score, sensitivity and specificity values.
TABLE VI.
Performance (F-score) of the classifier in discriminating sleep and wake over each cohort
| Younger | Older | All Subjects | |
|---|---|---|---|
| LDA | 0.799 | 0.768 | 0.783 |
| QDA | 0.784 | 0.748 | 0.765 |
| KNN1 | 0.714 | 0.698 | 0.704 |
| KNN5 | 0.762 | 0.734 | 0.745 |
| KNN9 | 0.780 | 0.752 | 0.761 |
| NN20 | 0.800 | 0.767 | 0.784 |
| NN40 | 0.796 | 0.772 | 0.789 |
| SVM | 0.760 | 0.714 | 0.743 |
The optimal classifier was a neural network, however the performance of the LDA classifier (see Table VII) was only slightly lower (by less than 0.006) and within one standard deviation of the Neural Network. The LDA classifier is characterised by very high sensitivity values and adequate specificity values. Additionally, due to the simpler implementation in low-level hardware of an LDA classifier, it was chosen as the optimal classifier for sleep/wake classification. Low standard deviations (less than 0.02) were found in the F-score, sensitivity and specificity values.
TABLE VII.
LDA classifier performance on validation data.
| Cohort | F-score | Sens. | Spec. |
|---|---|---|---|
| Younger | 0.799 | 0.902 | 0.645 |
| Older | 0.768 | 0.856 | 0.627 |
| All Subjects | 0.783 | 0.880 | 0.633 |
Performance results for the optimal classifiers, as applied to their cohorts independent unseen data is given in Table VIII.
TABLE VIII.
LDA classifier performance on their cohorts’ independent unseen data (data sets C and E).
| Cohort/Classifier | F-score | Sens. | Spec. | Acc. |
|---|---|---|---|---|
| Younger Adult | 0.758 | 0.879 | 0.560 | 0.719 |
| Older Adult | 0.813 | 0.980 | 0.570 | 0.775 |
| All Subjects | 0.788 | 0.955 | 0.532 | 0.743 |
VI. DISCUSSION
A systematic and robust study has been presented demonstrating that a non-contact sleep monitoring system offers similar performance to existing non-clinical/home-based validated sleep monitoring solutions in a manner conducive to long term monitoring, especially amongst vulnerable populations such as those with memory impairment. The system was trained and validated using a large dataset and a varied cohort, especially given the constraints of collecting sleep related data. The underpinning feature extraction was optimized using a cross-validation procedure with an independent unseen testing cohort to ensure validity across new data sets when presented to the system.
Results derived on a per-night basis using an independent test data set are given in Table IX. The performance levels of state-of-the-art commercially available regulated sleep monitors are also reported in the table for comparison purposes. As can be seen, the UMBS system provides comparable results. Wrist actigraphy reports slightly higher specificity levels, however this technology requires subjects to wear a watch for the duration of data collection. The UMBS is particularly suitable for sensitive populations, such as people with dementia, for whom unobtrusive ambient technologies are preferred or where wearable options have low adherence rates due to extended data collection periods. Overall, the UMBS can provide an accurate and reliable estimation of clinically important sleep metrics over extended periods in the real-world.
TABLE IX.
Comparison of wrist actigraphy, Biancamed SleepMinder and UMBS for sleep/wake discrimination (mean± st. dev.)
Within this study, some constraints must be noted. Firstly, the subjects partaking in this research were screened for health and potential sleep disorders and therefore they may not be representative of a population requiring sleep monitoring technologies. Additional research evaluating this technology on a clinical population should be carried out. In particular, its suitability for heavier and lighter populations and those suffering with insomnia and breathing problems needs to be assessed. It is noted that the system was designed based on changes in pressure distribution rather than absolute weight, and as such performance is expected to be robust to variation in weight. However, in the case of sleep conditions such as sleep apnea and insomnia performance is likely to degrade in line with other movement based sleep monitoring devices due to the abnormal sleep movement profiles associated with these conditions. Secondly, the sleep schedules in these studies are atypical of natural sleep timing patterns. While the extended day length and sleep restriction were artificially enforced, this should not have any bearing on the relationship between UMBS-derived data and PSG-derived sleep/wake state. The range of sleep timing imposed in the studies also produced sleep that varied in quality from night-to-night, and while artificial, resulted in a wide range of sleep qualities that allowed for more robust testing. As such, this data is adequate for the investigation of the classification of wake and sleep stages from UMBS data. Additionally, sleep efficiencies over all nights were within the expected range for healthy younger and older adults (mean of 9.7 % and 19.4% respectively). Thirdly, the nature of clinical sleep research is artificial by definition and, as such, all data collected in a clinic is non-representative of real-world sleep. For example, participants may have increased wake time due to the PSG recording equipment, and may lie more passively than when in their own homes resulting in an increased durations or formations of quiescent wake. However, this is an inherent constraint for any type of clinical research.
Methods for reducing the dimensionality of the feature space (Principal component analysis [43] and feature subset selection [44]) were investigated, but these did not significantly increase the overall accuracy of the system. As such only results for classification using the full set of input features have been reported.
A variety of linear and non-linear classifiers were investigated for sleep/wake classification. On balance the LDA classifier is recommended as the preferred solution over all cohorts as although its performance is marginally inferior to other classifiers considered (it is within 1% of the best F-score achieved for each cohort), it is the least complex to implement and hence has a much lower computational overhead.
Sleep stage discrimination and wake/REM/NREM discrimination was also investigated, however the system did not perform to an acceptable level. A similar design to that described above with additional classes relating to each sleep stage was used. Similarly to the approach taken in this paper, these data were examined on a per-cohort basis, however using a per-subject design may achieve better performance but would require significant quantities of data. Integrating the timing of the sleep epoch within the sleep episode as a feature may improve the performance of this type of system as increased levels of wake occur at the beginning and end of sleep episodes. This was not considered in the current study, where the focus has been on relating UMBS data directly to sleep/wake state such that is provides a balanced identification of wake episodes occurring throughout a sleep episode.
Often in sleep research, sensitivity and specificity values are reported due to the inherently biased nature of sleep datasets. This paper proposes the use of a harmonic mean performance measure (F-score) which is insensitive to an uneven sample distribution.
Another consideration not generally taken into account in the design and development of home-based sleep/wake monitoring systems is the usability or acceptability of the proposed solution over long durations. Considerations like the perceived benefit of the system, the stigma in the systems use, physical space limitations, the aesthetic design and the visibility of the system to visitors may reduce the practical use/deployment of such a system.
VII. Conclusion
In this paper, a non-contact unobtrusive pressure based technology has been shown to offer similar performance to existing non-clinical/home-based validated sleep monitoring solutions and to also capture body movement information during sleep. The UMBS sleep scoring system is suitable for long term placement in domestic homes and is ideal for the non-intrusive collection of sleep data. The UMBS is especially suited to monitoring vulnerable populations where it is preferential to keep modifications to the sleeping environment at a minimum (such as those with mild cognitive impairment or dementia where sleeping difficulties pervade). The performance and reliability of the UMBS technology is comparable to the current ambulatory sleep/wake monitoring gold standard, wrist actigraphy [13], and an alternative non-contact sleep/wake monitor (BiancaMed SleepMinder [?], [22] (as given in Table IX and calculated over the data for each complete sleeping episode).
Acknowledgments
The authors wish to thank the study participants, the technical staff of the Division of Sleep and Circadian Disorders, and the technical and nursing staff of the Intensive Physiological Monitoring Unit. We are indebted to the Project Leaders of the studies, Drs. Anne-Marie Chang, Sean W. Cain, and Mirjam Y. Münch. The studies were supported by grants P01 AG09975 (to CAC) and R01 HL080978 (to JFD) from the US National Institutes of Health, and were carried out in the Brigham and Women’s Hospital Center for Clinical Investigation, part of the Harvard Catalyst — The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award UL1 TR001102) and financial contributions from Harvard University and its affiliated academic healthcare centers. L. Walsh was supported by a Fulbright Award for Science and Technology.
Addendum I
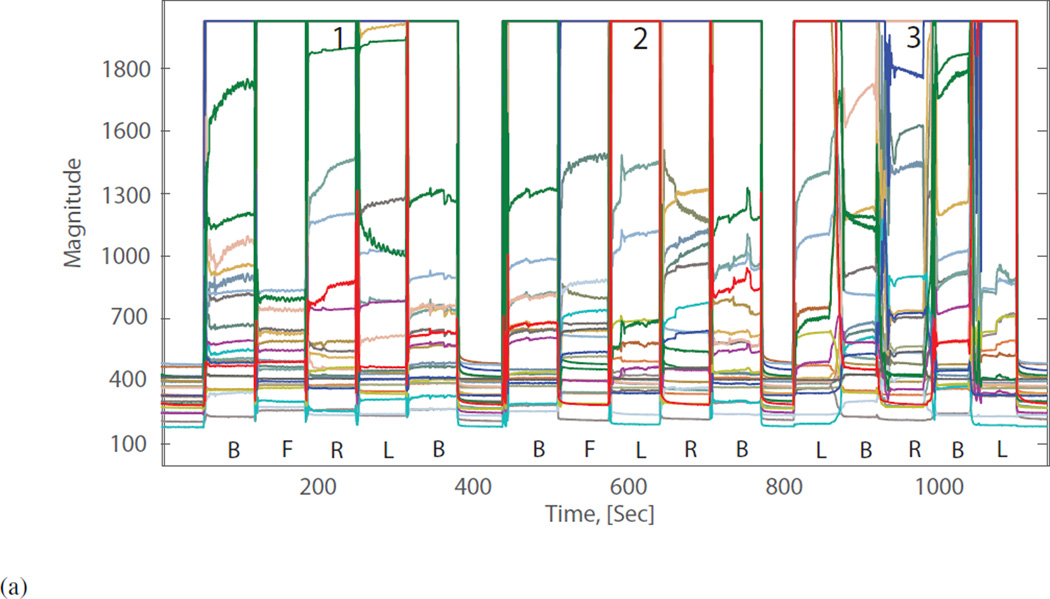
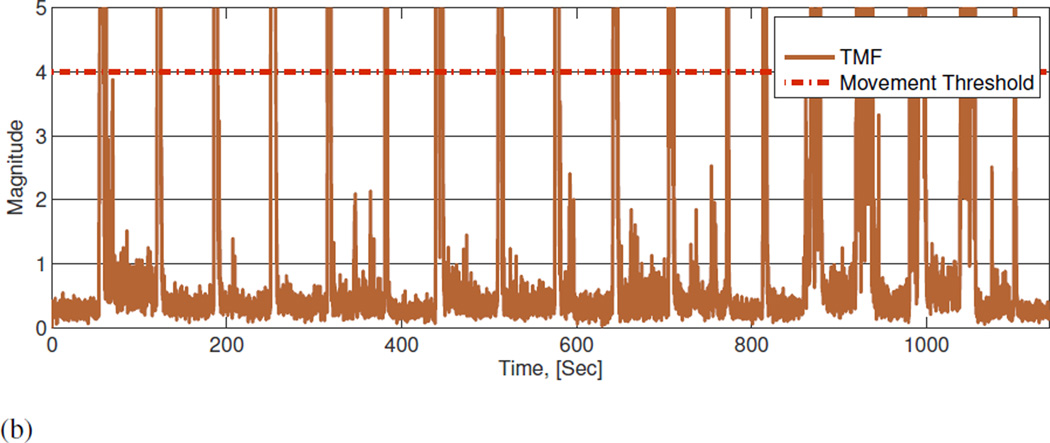
An investigation into UMBS derived movement and non-movement data. Data were collected experimentally to investigate the effect of movement and non-movement on UMBS sensor data. A healthy adult was asked to shift between 4 postures: lying on back (B), lying on front (F), lying on left side (L) and lying on right side (R) under three conditions: 1) rapid transition without lateral movement, 2) rapid transition with lateral movement, and 3) slow rolling movement with lateral movement. UMBS data relating to the various bed entry, bed exits and postural changes are shown in Figure 1A below. The corresponding TMF data is presented in Figure 1B (with the range of value given an upper limit of 5 for this example). It can be determined empirically that all bed entry, bed exit and postural changes can be discriminated from non-movement activity (e.g. micro-movements from breathing) using a TMF threshold of 4.
(a) UMBS data collected while the participant was asked to assume and shift between four typical sleeping postures: lying on their back (B); left side (L); right side (R); and front (F). The subject was asked to change postures under three conditions 1) rapid transition without lateral displacement, 2) rapid transition with lateral displacement and 3) slow rolling transitions (inherently including lateral displacement).
(b) Corresponding temporal movement features with movement threshold (time domain).
Figure 5. UMBS derived temporal feature with movement threshold of 4 from a participant shifting between postures on the UMBS.
Contributor Information
Lorcan Walsh, CASALA, Dundalk Institute of Technology, Co. Louth, Ireland.
Seán McLoone, Energy, Power and Intelligent Control (EPIC) Research Cluster at Queens University Belfast, Northern Ireland, U.K..
Joseph Ronda, Harvard Medical School and the Division of Sleep and Circadian Disorders, Brigham and Women’s Hospital, MA 02115, USA.
Jeanne F. Duffy, Harvard Medical School and the Division of Sleep and Circadian Disorders, Brigham and Women’s Hospital, MA 02115, USA
Charles A. Czeisler, Harvard Medical School and the Division of Sleep and Circadian Disorders, Brigham and Women’s Hospital, MA 02115, USA
References
- 1.Zepelin H. Ch. Mammalian Sleep. Philadelphia: Saunders; 2000. Principles and practice of sleep medicine; pp. 82–92. [Google Scholar]
- 2.Cappuccio FP, et al. Sleep duration and all-cause mortality: a systematic review and meta-analysis of prospective studies. Sleep. 2010 May;33(5):585–592. doi: 10.1093/sleep/33.5.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manabe K, et al. Sleep patterns and mortality among elderly patients in a geriatric hospital. Gerontology. 2000;46(6):318–322. doi: 10.1159/000022184. [DOI] [PubMed] [Google Scholar]
- 4.Miles LE, Dement WC. Sleep and aging. Sleep. 1980;3(3):119–220. [PubMed] [Google Scholar]
- 5.Ancoli-Israel S. Sleep problems in older adults: Putting myths to bed. Geriatrics. 1997;52(1):20–29. [PubMed] [Google Scholar]
- 6.Barbar SI, et al. Sleep disturbances and their correlates in elderly japanese american men residing in Hawai’i. J. Gerontol. A. Biol. Sci. Med. Sci. 2000 Jul;55(7):M406–M411. doi: 10.1093/gerona/55.7.m406. [DOI] [PubMed] [Google Scholar]
- 7.Brassington GS, King AC, Bliwise DL. Sleep problems as a risk factor for falls in a sample of community-dwelling adults aged 64–99 years. J. Am. Geriatr. Soc. 2000 Oct;48(10):1234–1240. doi: 10.1111/j.1532-5415.2000.tb02596.x. [DOI] [PubMed] [Google Scholar]
- 8.Dew MA, et al. Healthy older adults’ sleep predicts all-cause mortality at 4 to 19 years of follow-up. Psychosom. Med. 2003;65(1):63–73. doi: 10.1097/01.psy.0000039756.23250.7c. [DOI] [PubMed] [Google Scholar]
- 9.Happe S. Excessive daytime sleepiness and sleep disturbances in patients with neurological diseases. Drugs. 2003;63(24):2725–2737. doi: 10.2165/00003495-200363240-00003. [DOI] [PubMed] [Google Scholar]
- 10.Rechtschaffen A, Kales A. A manual of standardized terminology, techniques and scoring system for sleep stages of human subjects. Washington, DC: NIH Publication No. 204, US Government Printing Office; 1968. [Google Scholar]
- 11.Danker-Hopfe H, et al. Interrater reliability for sleep scoring according to the rechtschaffen & kales and the new AASM standard. Journal of sleep Research. 2009;18:74. doi: 10.1111/j.1365-2869.2008.00700.x. [DOI] [PubMed] [Google Scholar]
- 12.Kushida CA, et al. Comparison of actigraphic, polysomnographic, and subjective assessment of sleep parameters in sleep-disordered patients. Sleep Med. 2001 Sep;2(5):389–396. doi: 10.1016/s1389-9457(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 13.Paquet J, Kawinska A, Carrier J. Wake detection capacity of actigraphy during sleep. Sleep. 2007;30(10):1362–1369. doi: 10.1093/sleep/30.10.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh L, McLoone S. Non-contant Under-Mattress Sleep Monitoring. Journal of Ambient Intelligence and Smart Environments. 2014;6(4):385–401. [Google Scholar]
- 15.Walsh L, et al. The deployment of a non-intrusive alternative to sleep/wake actigraphy in a home-based study of the elderly. Conf. Proc. IEEE EMBS. 2008:1687–1690. doi: 10.1109/IEMBS.2008.4649500. [DOI] [PubMed] [Google Scholar]
- 16.Walsh L, Moloney E, McLoone S. Identification of nocturnal movements during sleep using the non-contact under mattress bed sensor. Conf. Proc. IEEE EMBS. 2011:1660–1663. doi: 10.1109/IEMBS.2011.6090478. [DOI] [PubMed] [Google Scholar]
- 17.Nakajima K, Matsumoto Y, Tamura T. Development of real-time image sequence analysis for evaluating posture change and respiratory rate of a subject in bed. Phys. Meas. 2001;22:N21–N28. doi: 10.1088/0967-3334/22/3/401. [DOI] [PubMed] [Google Scholar]
- 18.Okada S, et al. Examination of non-restrictive and non-invasive sleep evaluation technique for children using difference images. Conf. Proc. IEEE EMBS. 2008:3483–3487. doi: 10.1109/IEMBS.2008.4649956. [DOI] [PubMed] [Google Scholar]
- 19.Townsend D, Knoefel F, Goubran R. Privacy versus autonomy: a tradeoff model for smart home monitoring technologies. Conf Proc IEEE EMBS. 2011:4749–4752. doi: 10.1109/IEMBS.2011.6091176. [DOI] [PubMed] [Google Scholar]
- 20.Choi JM, et al. Unobtrusive body movement monitoring during sleep using infrared motion detector and zigbee protocol. Proc. 3rd IEEE/EMBS International Summer School on Medical Devices and Biosensors. 2006:32–33. [Google Scholar]
- 21.de Chazal P, et al. Assessment of sleep/wake patterns using a non-contact biomotion sensor. Conf. Proc. IEEE EMBS. 2008:514–517. doi: 10.1109/IEMBS.2008.4649203. [DOI] [PubMed] [Google Scholar]
- 22.De Chazal P, et al. Sleep/wake measurement using a non-contact biomotion sensor. J. Sleep Res. 2011 Jun;20(2):356–366. doi: 10.1111/j.1365-2869.2010.00876.x. [DOI] [PubMed] [Google Scholar]
- 23.Brink M, Muller CH, Schierz C. Contact-free measurement of heart rate, respiration rate, and body movements during sleep. Behav. Res. Methods. 2006;38(3):511–521. doi: 10.3758/bf03192806. [DOI] [PubMed] [Google Scholar]
- 24.Choi BH, et al. Slow-wave sleep estimation on a load-cell-installed bed: a non-constrained method. Phys. Meas. 2009;30(11):1163–1170. doi: 10.1088/0967-3334/30/11/002. [DOI] [PubMed] [Google Scholar]
- 25.Mack DC, et al. Sleep assessment using a passive ballistocardiography-based system: Preliminary validation. Conf. Proc. IEEE EMBS. 2009:4319–4322. doi: 10.1109/IEMBS.2009.5333805. [DOI] [PubMed] [Google Scholar]
- 26.Jung DW, et al. Estimation Using Unobtrusively Measured Ballistocardiogram. IEEE Trans. on Biomed. Eng. 2014;61(1):131–138. doi: 10.1109/TBME.2013.2278020. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe T, Watanabe K. Noncontact method for sleep stage estimation. IEEE Trans. on Biomed. Eng. 2004;51(10):1735–1748. doi: 10.1109/TBME.2004.828037. [DOI] [PubMed] [Google Scholar]
- 28.Carlson BW, Neelon VJ, Hsiao H. Evaluation of a non-invasive respiratory monitoring system for sleeping subjects. Phys. Meas. 1999;20(1):53–63. doi: 10.1088/0967-3334/20/1/004. [DOI] [PubMed] [Google Scholar]
- 29.Shin JH, et al. Nonconstrained sleep monitoring system and algorithms using air-mattress with balancing tube method. IEEE Trans. Inf. Technol. Biomed. 2010;14(1):147–156. doi: 10.1109/TITB.2009.2034011. [DOI] [PubMed] [Google Scholar]
- 30.Kortelainen JM, et al. Sleep staging based on signals acquired through bed sensor. IEEE Trans. Inf. Technol. Biomed. 2010;14(3):776–785. doi: 10.1109/TITB.2010.2044797. [DOI] [PubMed] [Google Scholar]
- 31.Brueser C, et al. Adaptive beat-to-beat heart rate estimation in ballistocardiograms. IEEE Trans. Inf. Technol. Biomed. 2011;15(5):778–786. doi: 10.1109/TITB.2011.2128337. [DOI] [PubMed] [Google Scholar]
- 32.Samy L, et al. Unobtrusive Sleep Stage Identification Using a Pressure-Sensitive Bed Sheet. IEEE Sensors Journal. 2013;14(7):2092–2101. [Google Scholar]
- 33.Willemen T, et al. An evaluation of cardiorespiratory and movement features with respect to sleep-stage classification. IEEE Journal of Biomedical and Health Informatics. 2014;18(2):661669. doi: 10.1109/JBHI.2013.2276083. [DOI] [PubMed] [Google Scholar]
- 34.Long X, et al. Sleep and wake classification with actigraphy and respiratory effort using dynamic warping. IEEE Journal of Biomedical and Health Informatics. 2014;18(4):12721284. doi: 10.1109/JBHI.2013.2284610. [DOI] [PubMed] [Google Scholar]
- 35.Domingues A, Paiva T, Sanches JM. Hypnogram and sleep parameter computation from activity and cardiovascular data. IEEE Transactions on Biomedical Engineering. 2014;61(6):17111719. doi: 10.1109/TBME.2014.2301462. [DOI] [PubMed] [Google Scholar]
- 36.Zhu X, et al. Real-time monitoring of respiration rhythm and pulse rate during sleep. IEEE Trans. on Biomed. Eng. 2006;53(12):2553–2563. doi: 10.1109/TBME.2006.884641. [DOI] [PubMed] [Google Scholar]
- 37.Withings Aura. Withings Aura [Online] 2014 Sep; Available: http://www.withings.com/us/withings-aura.html. [Google Scholar]
- 38.Fitbit. Fitbit. 2014 Sep; Available: http://www.fitbit.com. [Google Scholar]
- 39.Band Xiaomi Mi. Xiaomi [Online] 2015 Aug; Available: http://www.mi.com/in/miband/ [Google Scholar]
- 40.Sleep Cycle Alarm Clock. Northcube [Online] 2015 Aug; Available: http://www.sleepcycle.com/ [Google Scholar]
- 41.Mantua J, Gravel N, Spencer RMC. Reliability of sleep measures from four personal health monitoring devices compared to researchbased actigraphy and polysomnography. Sensors. 2016;16:646. doi: 10.3390/s16050646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bennett S, et al. In-Bed Mobility Monitoring Using Pressure Sensors. IEEE Trans. on Inst. and Meas. 2015;64(8):2110–2120. [Google Scholar]
- 43.Jolliffe I. Principal Component Analysis. New York: Springer-Verlag; 1986. [Google Scholar]
- 44.Cantu-Paz E, Newsam S, Kamath C. Proceedings, ACM International Conference on Knowledge Discovery and Data Mining. Seattle, WA: 2004. Aug, Feature selection in scientific applications; pp. 788–793. [Google Scholar]
- 45.Hastie T, Tibshirani R, Friedman JH. The Elements of Statistical Learning. Springer-Verlag; 2001. [Google Scholar]
- 46.Bishop CM. Neural networks for pattern recognition. 1995 [Google Scholar]
- 47.Shawe-Taylor J, Christiani N. Support Vector Machines and other kernel-based learning methods. Cambridge University Press; 2000. [Google Scholar]
- 48.Burges CJC. A tutorial on supp ort vector machines for parttern recognition. Data Mining and Knowledge Discovery. 1998;2(2) [Google Scholar]