Abstract
Background
Oral mucositis is probably the most debilitating complication that can arise in treating a patient with head and neck cancer. Little is known about the impacts of oral microbiota on the initiation and progression of mucositis.
Methods
Based on 16S rRNA gene sequencing, dynamic changes in oral bacterial profile as well as correlations between the severity of mucositis and bacterial shifts during radiotherapy were investigated.
Findings
Our results revealed that bacterial community structure altered progressively during radiation therapy, in parallel with a marked increase in the relative abundance of some Gram-negative bacteria. Patients who eventually developed severe mucositis harbored a significantly lower bacterial alpha diversity and higher abundance of Actinobacillus during the phase of erythema – patchy mucositis. Accordingly, a random forest model for predicting exacerbation of mucositis was generated, which achieved a high predictive accuracy (AUC) of 0.89.
Interpretation
Oral microbiota changes correlate with the progression and aggravation of radiotherapy-induced mucositis in patients with nasopharyngeal carcinoma. Microbiota-based strategies can be used for the early prediction and prevention of the incidence of severe mucositis during radiotherapy.
Keywords: Oral mucositis, Microbiota, Nasopharyngeal carcinoma, Radiotherapy
Highlights
-
•
The oral microbiota of NPC patients are different to that of healthy persons.
-
•
The biodiversity of oral microbiota changed significantly as mucositis progressed during radiotherapy.
-
•
Oral microbiota represents a potential strategy for predicting the aggravation of severe mucositis during radiotherapy.
Oral or oropharyngeal mucositis is the most common side effect of radiation therapy for head and neck cancers. In this prospective cohort study, we found that changes in an oral microbial community correlated with the progression and aggravation of radiotherapy-induced mucositis in the patients with nasopharyngeal carcinoma; and patients who eventually developed severe mucositis transiently harbored a notably higher proportion of Actinobacillus during a mild phase of mucositis, which may potentially play a role in the aggravation of severe mucosal lesions. Moreover, our findings also showed that microbiota-based strategies can be used for the early prediction of the incidence of severe mucositis during radiotherapy.
1. Introduction
Oral/Oropharyngeal mucositis (OM) is probably the most common and debilitating complication that can occur during and shortly after radiotherapy and/or chemotherapy for patients with head-and-neck cancer (HNC). It is typically characterized by: erythema, edema, mucosal ulceration and pseudomembrane formation in the oral cavity and oropharynx. Patients with OM lesions frequently complain of mild to severe pain and pharyngeal dysphagia. Currently, there are no effective mucoprotective strategies for the management of OM. Severe mucositis can not only affect a patients' quality of life, but also increases the need for narcotic analgesics, total parenteral nutrition, interruption of cancer therapy, prolong hospitalization and increases the risk of local and systemic infection (Trotti et al., 2003, Villa and Sonis, 2015, Stokman et al., 2006).
Recent studies have suggested that the pathogenesis of mucositis involves a cascade of inflammatory events, which chronologically consist of five continuous overlapping phases: initiation, upregulation of inflammation, signaling and amplification, ulceration, and finally wound healing (Sonis, 2004). Although it is hypothesized that genetic factors may correlate with an increased risk of developing mucositis (Venkatesh et al., 2014, Pratesi et al., 2011), the role of microorganisms in promoting inflammation and exacerbating mucositis still remains to be explored.
Using culture-based techniques, several Gram-positive/negative species of bacteria were isolated from the oral cavity of patients undergoing radiochemotherapy in the head and neck region (Napenas et al., 2007, Almstahl et al., 2008, Panghal et al., 2012). Their normal bacterial flora was significantly altered during the course of treatment, which leads naturally to the question as to whether changes in the oropharyngeal microflora may correlate with the onset and aggravation of OM. Unfortunately, there are so far no well-defined patterns with respect to these changes, and no consistent correlations were found between the presence of specific pathogens and the severity of OM. On the other hand, it is estimated that > 700 bacterial species have been detected in the oral cavity, of which > 50% remain to be cultivated (Aas et al., 2005). However, using culture-independent high-throughput 16S rRNA gene sequencing, it has been shown that changes in oral microbiota during radiochemotherapy are far more complicated than previously thought (Vanhoecke et al., 2015, Hu et al., 2013). Nevertheless, and more importantly until now, there has been few analysis made to our knowledge on the correlation between the OM severity and the dynamic changes it may make in bacterial profiles throughout radiochemotherapy.
Hence in this study, the OM severity was assessed and scored longitudinally in a cohort of patients with nasopharyngeal carcinoma (NPC) undergoing radiotherapy with or without chemotherapy, during which the mucosa of patients was inspected regularly and sampled eight times throughout their treatment. Based on 16S rRNA gene sequencing and bioinformatics analysis, we were able to investigate dynamic changes in oropharyngeal bacterial profile and their associated correlation between the severity of mucositis and the bacterial shifts which had occurred.
2. Materials and Methods
2.1. Sample Collection
This prospective cohort study was conducted in newly diagnosed NPC patients undergoing three-dimensional conformal radiation therapy at South China's Nanfang Hospital between May 2012 and August 2013. Ethical approval was granted by the Ethics Committee of China's Southern Medical University (SMU). The devised study protocols were explained to each patient and family members, and those who agreed to participate signed an informed consent form. The patients, as well as their relatives, who presented with poor oral hygiene and/or had developed severe forms of periodontal diseases were excluded from this study; and subjects with prior head-and-neck radiation or who had taken antibiotics within 2 weeks before the study commenced, were also not enrolled in the study. A total of 41 NPC patients (27men; 14 women; age range 22–75 years; average age 47.2) were recruited to take part in this study and their medically healthy relatives matched by sex and age were served as controls.
Before radiation treatment began, all patients received a full-mouth clinical examination and oral hygiene instruction. Surgical procedures were carried out in patients with caries, pulpal diseases and gingivitis, including professional dental cleaning, filling, endodontic treatment, and extraction of non-restorable teeth. For all recruited patients, radiotherapy was administered 5 times per week at 2 Gy per fraction in a total dose of 60–70 Gy within 6–7 weeks, in which 17 patients underwent radiotherapy alone, two patients received induction chemotherapy followed by radiotherapy, and the remainder, some 23 patients were given concurrent chemoradiotherapy followed by adjuvant chemotherapy.
In all patients, oral and oropharyngeal mucositis were scored clinically by the same radiation therapist once every 3 days, according to the Radiation Therapy Oncology Group (RTOG) criteria. To trace dynamic changes of bacterial community through the entire course, a series of microbial samples were regularly taken from 19 of these patients prior to irradiation, after their 5th, 10th, 15th, 20th, 25th, 30th and 35th irradiation, corresponding to 10, 20, 30, 40, 50, 60 and 70 Gy respectively. Samples were collected in the patients presenting with OM by swabbing over mucosal lesions in the retropharyngeal wall with disposable medical sterile swabs, whereas in patients without clinical signs of mucositis, or in the control group, retropharyngeal mucosa was sampled. All samples were frozen immediately at − 20 °C and then stored at − 80 °C until analysis.
2.2. DNA Extraction, 16S rRNA Gene Amplification and Sequencing
Bacterial genomic DNA was extracted from mucosal samples using the DNA MAGNETICS and EXTRACT kit (Shenzhen BioEAsy Biotechnologies Co., Ltd., China), according to the manufacturer's instruction as previously validated (Peng et al., 2013). The barcoded V4F-GAGTGCCAGCMGCCGCGGTAA and V4R-GGACTACHVGGGTWTCTAAT primers were used to amplify bacterial 16S rRNA V4 fragments. The PCR cycle conditions were: 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 57 °C for 30 s, and 72 °C for 30 s, and a final extension at 72 °C for 5 min. The PCR products were purified (QIAquick PCR Purification Kit, Qiagen), and sequenced using the 100-bp paired-end strategy on the Illumina HiSeq 2000 according to the manufacturer's instructions. The raw sequence data for 16S rRNA gene sequencing data sets is available from the European Bioinformatics Institute (http://www.ebi.ac.uk/) at accession number PRJEB15392.
2.3. Bioinformatics and Statistic Analysis
The Illumina sequencing quality report indicates that the quality of sequences is relatively high before PE80bp (pair-end 80 bp), while there is a sharp decrease after that. As a result, we trimmed the raw sequences to 80 bp for each end. Sequences were then de-multiplexed, barcode-primer trimmed and filtered if they contain ambiguous bases or mismatches in the primer regions according to BIPES protocol (Zhou et al., 2011). We then used 30Ns to concatenate the two single-ended sequences for the following analyses sequences, because our PE sequences are not able to span the V4 regions of the 16S rRNA gene. All the tools we have used in this study have been validated to be able to handle gapped sequences. UCHIME (implemented in USEARCH, version 6.1) was used to remove chimeras using the de novo mode with default parameters (Edgar et al., 2011). We normalized all samples at the level of 3000 sequences to avoid any uneven sequencing effort between samples. USEARCH was then used to do de novo clustering for the sequences using the default parameters, with the identity cutoff set to 0.97. Multiple alignments of representative sequences were performed using PyNAST. The Green genes core set (Version: 13.8) was used as the template file (Lozupone et al., 2011). The RDP classifier was used to classify these representative sequences into specific taxa using the default database (Wang et al., 2007). PD whole tree and Shannon index were applied to evaluate alpha diversity and the UniFrac distance was used to analyze the beta-diversity (Lozupone and Knight, 2005). All of the analyses, from chimera checking to alpha and beta diversity calculation, were performed using QIIME (1.8.0) (Caporaso et al., 2010). Statistical analysis of the relative abundance of the genera and the diversity indices and estimators were performed using R (v3.2.2). We used LEfSe (linear discriminant analysis effect size) to determine differential features between groups (Segata et al., 2011). LEfSe is an algorithm for high-dimensional biomarker discovery as it identifies genomic features characterizing differences between two or more biological conditions. LEfSe determines the features most likely to explain differences between classes by coupling standard tests for statistical significance with additional tests encoding biological consistency and effect size (Segata et al., 2011). The threshold on the logarithmic LDA score for discriminative features was set to 2.0. Random Forests models were trained by “randomForest” package with default parameters (ntree = 5000, mtry = square root of p, where p is the number of input features) in R using OTU data. The performance of the model was assessed with a ten-fold cross-validation approach and measured by area under the ROC.
3. Results
3.1. The Oropharyngeal Microbiota of NPC Patients are Different to that of Healthy Individuals
In this study, retropharyngeal mucosal samples were taken from patients before the commencement of their radiation treatment and medically healthy controls. Demographic characteristics and health-related lifestyle factors of our NPC patients and control subjects are summarized in Table 1. At baseline, both groups were similar in terms of age, gender, smoking habits and alcohol consumption, because controls were matched to patients based on these factors prior to the analyses. After filtering the low-quality reads and removing chimeras, all samples were normalized to 3000 sequences to avoid the deviation caused by effects of different sequencing depths. Alpha diversity was then calculated within QIIME using the Shannon index and phylogenetic diversity (PD whole tree) (Fig. 1a–b). While the Shannon index accounts for both abundance and evenness of the species present, the value of PD whole tree index represents their evolutionary or genetic distances within the community. Our results showed that the values of both alpha diversity indices were statistically significantly lower in NPC patients than those in the controls (Wilcoxon rank-sum test, P < 0.05), suggesting that the healthy controls harbored a greater diversity of oropharyngeal bacteria than the NPC patients.
Table 1.
Baseline characteristics of nasopharyngeal carcinoma patients and control subjects.
| NPC (n = 41) | CON (n = 49) | P value | |
|---|---|---|---|
| Age (years, mean ± SD) | 47.0 ± 12.9 | 48.3 ± 11.7 | 0.51 |
| Gender, male, n (%) | 27 (65.9) | 37 (75.5) | 0.35 |
| Smoking, n (%) | 14 (34.1) | 9 (18.4) | 0.14 |
| Drinking, n (%) | 9 (22.0) | 11 (22.4) | 0.99 |
Percentages are shown in parentheses as appropriate.
NPC, nasopharyngeal carcinoma; CON, controls.
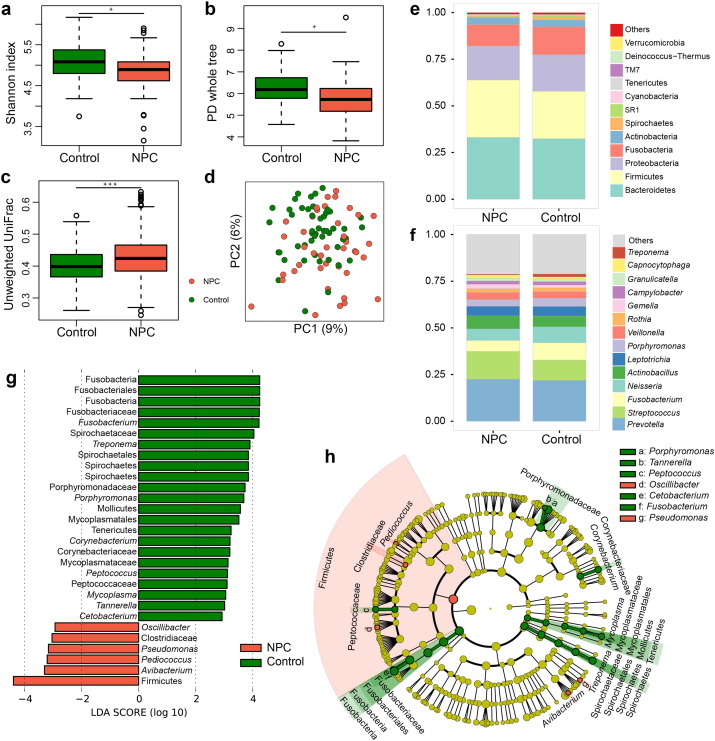
Fig. 1.
Comparison of the oropharyngeal bacterial profile between the healthy controls and NPC patients before the commencement of radiotherapy. The data represent 49 healthy controls (green) and 41 NPC patients (red). (a–b) Box plots of alpha-diversity indices (Shannon and PD whole tree) comparing controls' and patients' samples. Healthy controls harbored significantly greater bacterial diversity than patients. (c) Box plot of the unweighted UniFrac distances comparing inter-variation of controls and NPC patients (d) An unweighted UniFrac-based PCoA plot reveals that controls' bacterial communities cluster separately from patients' bacterial communities, which are more similar to each other. Each circle represents a single patient or a control. (e–f) Average relative abundances of predominant bacterial taxa at the phylum and genus level in controls' and patients' samples. (g–h) Linear Discriminant Analysis Effect Size (LEfSe) identifies the most differentially abundant taxons between the two groups. Control-enriched taxa are indicated with a positive LDA score, and taxa enriched in NPC patients have a negative score. Only taxa meeting an LDA significant threshold > 2 are shown. Wilcoxon rank-sum test was used to determine statistical significance. *P < 0.05; ***P < 0.001.
To compare the phylogenetic differences in bacterial community structure between the two groups, a principal coordinates analysis (PCoA) was performed in QIIME, which revealed some overlaps between the two communities. However the control samples seemed much more tightly clustered than those exhibited in the patients' samples (Fig. 1d), suggesting that the healthy controls shared more similar bacterial communities than those of our NPC patients, as demonstrated by the fact that the control samples exhibited significantly lower average unweighted UniFrac values than those of patients' (Fig. 1c). Moreover, beta diversity was also estimated based on weighted Unifrac distance matrices and Bray-Curtis dissimilarities, which showed similar results as seen in Fig. S1a–d (Wilcoxon rank-sum test, P < 0.001).
Fig. S1.
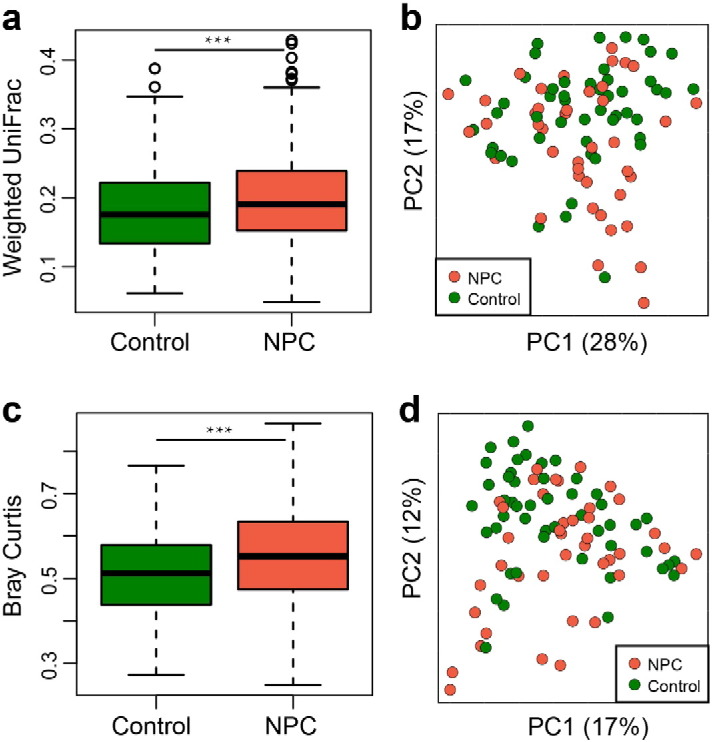
Comparison of the oropharyngeal bacterial beta-diversity between healthy controls and NPC patients before radiotherapy. PCoA plot of bacterial community structure based on weighted UniFrac distance (a–b) and Bray Curtis (c–d). Each circle represents a single patient (red) or a control (green). Wilcoxon rank-sum test was used to determine statistical significance. ***P < 0.001
In this study, the oropharyngeal bacterial community was mainly dominated by a total of 193 genera belonging to 12 major phyla in both patients and healthy individuals: Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria, Actinobacteria, Spirochaetes, Cyanobacteria, Verrucomicrobia, Acidobacteria, TM7, Deinococcus-Thermus and SR1. The predominate 14 top genera which constituted 78.8% of the total sequences were Prevotella, Streptococcus, Fusobacterium, Neisseria, Actinobacillus, Leptotrichia, Porphyromonas, Veillonella, Rothia, Gemella, Campylobacter, Granulicatella, Capnocytophaga and Treponema (Fig. 1e–f). Although these bacterial genera could be detected in both groups, the relative abundance of each genus varied quite widely among them. To further identify which bacterial taxa were distinct between the two groups, LEfSe analysis was applied. Based on LDA effect size, Avibacterium, Pediococcus, Pseudomonas, Oscillibacter, and unclassified Firmicutes and Clostridiaceae were significantly enriched in NPC patients, while Fusobacteria, Porphyromonas, Treponema, Peptococcus, Tannerella and Corynebacterium, etc. were mostly related to the healthy individuals (Fig. 1g–h).
3.2. Association Between Cumulative Radiation Dose, Mucositis Severity and Microbial Community
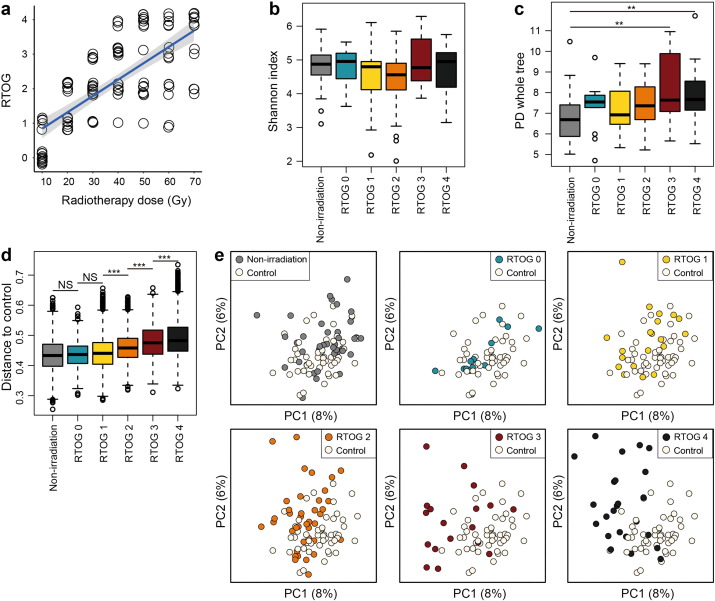
The mucosa of patients' oral cavity and oropharynx was inspected and scored clinically according to the RTOG criteria once every three days during the entire course of their radiotherapy. In this study, all patients experienced at least some degree of OM throughout the treatment, and as mentioned above, a series of microbial samples were regularly taken from 19 patients at eight different time-points, in which: only one patient developed RTOG Grade 1 OM (RTOG 1); five patients suffered from Grade 2 OM; two patients experienced Grade 3 OM; and 11 patients developed Grade 4 OM. Furthermore, it was observed that erythema (RTOG 1) started normally by the end of the first week (10 Gy), and peaked by the end of the second week of radiotherapy (20 Gy). This was followed by patchy mucositis (RTOG 2) that occurred mainly in the second and third week (20–30 Gy), which later progressed to confluent fibrinous mucositis (RTOG 3) during the third to fourth week (30–40 Gy), and then to mucosal ulceration and necrosis (RTOG 4) by the end of fifth week (50 Gy), which then peaked around the sixth to seventh week (60–70 Gy). The scatter plot with trend line is presented in Fig. 2a, indicating a significant positive correlation between accumulated radiation dose and mucositis severity (RTOG Grade) (Spearman's rank correlation, r = 0.73, P < 0.001).
Fig. 2.
Association between cumulative radiation dose, mucositis severity and microbial community diversity. (a) Scatterplot with trend line showing a significant positive correlation between accumulated radiation dose and mucositis severity based on RTOG criteria. (b) Box plot of Shannon diversity index showing no statistically significant differences among non-irradiation (grey) and RTOG groups (RTOG 0–4: blue, yellow, orange, red and black, respectively). (c) Box plot of PD whole tree index also showing no statistically significant differences among the above mentioned groups, except between the non-irradiation and RTOG 3, as well as between the non-irradiation and RTOG 4. (d) Box plot illustrating the unweighted UniFrac distances of different RTOG levels to controls. Higher box indicates higher dissimilarity in microbial community compared to controls. (e) PCoA plots comparing bacterial community structure of oropharyngeal samples between control (white) and different RTOG groups reveals noteworthy overlaps among the Control (white), RTOG 0 and RTOG 1 sample clusters. However, as mucositis progressed to peak severity (from RTOG 2 to RTOG 4), RTOG groups samples became more and more distinguishable from control and dispersed across PC1. Each circle represents a single sample, colored by group. Wilcoxon rank-sum test was used to determine statistical significance. NS, no significance; **P < 0.01; ***P < 0.001.
In all these 19 patients, a total number of 2,035,805 high-quality sequences were obtained after filtering the low-quality reads and removing chimeras, with an average number of 13,134 sequences per sample. Each sample was rarefied to 3000 tags for a consistent sequencing depth. To investigate whether patients' oropharyngeal bacterial profiles were influenced by, and in accordance with the changes in mucositis severity or vice versa, samples were divided into control (healthy individuals), non-irradiation (before radiation treatment) and five RTOG groups: RTOG 0 (samples from patients with mucositis of Grade 0), RTOG 1, 2, 3, and 4. The Shannon diversity index showed no statistically significant differences among the non-irradiation and five RTOG groups. Similar to the Shannon index, the PD whole tree index exhibited the same trend, except for significantly higher index values for the RTOG 3 and RTOG 4 samples, compared to those of non-irradiation samples (Fig. 2b–c). On the other hand, the beta-diversity analysis of unweighted UniFrac distances of all above mentioned patients groups versus controls revealed no significant differences among the non-irradiation, RTOG 0 and RTOG 1 groups. However, as the oral mucosal lesion progressed from Grade 1 to Grade 4 mucositis, their bacterial UniFrac distances to the control group increased significantly per grade level (Fig. 2d) (Wilcoxon rank-sum test, P < 0.001, corrected by FDR).
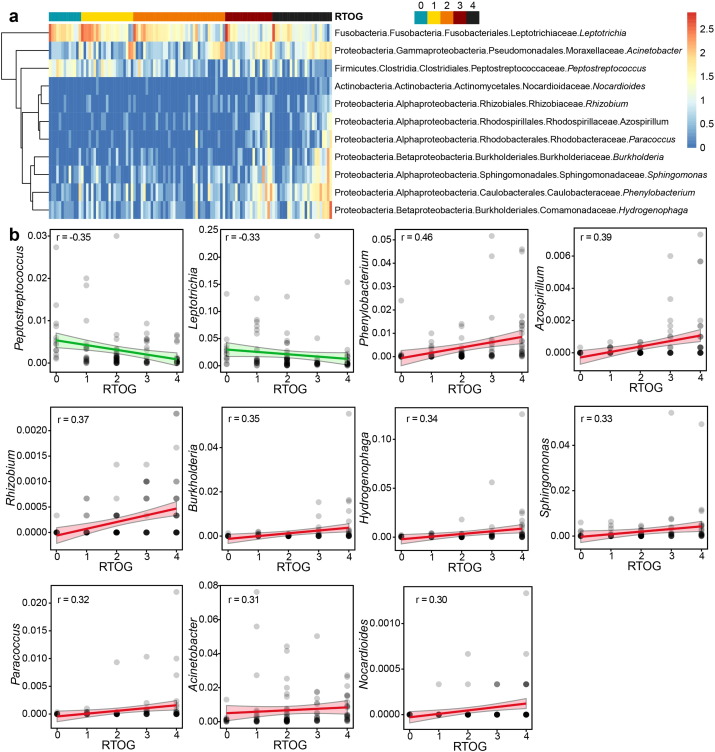
To visualize the microbial dissimilarity among these groups, the unweighted Unifrac distances were then calculated and used for PCoA in the QIIME pipeline. This application revealed a broad overlapping between healthy controls and patients with RTOG 0 as well as RTOG 1 OM (Fig. 2e), suggesting that healthy individuals and patients with RTOG 0–1 OM on the whole shared some similar bacterial communities. However, as the degree of mucositis severity increased, the bacterial communities in the RTOG 2, 3, and 4 gradually became more distinguishable from that in the control along the principal component axis, indicating that the severity of mucositis may indeed be associated with changes in bacterial community structure. Spearman's rank correlation was then employed for the correlation analysis between mucositis severity and changes in abundance of oropharyngeal bacteria at the genus level (Fig. 3a–b), which revealed that the relative abundances of nine genera: Phenylobacterium, Acinetobacter, Burkholderia, Sphingomonas, Azospirillum, Rhizobium, Hydrogenophaga, Paracoccus and Nocardioides were found to be significantly positively associated with an increased RTOG grade, whereas two genera Leptotrichia and Peptostreptococcus were significantly negatively associated with it (FDR < 0.05). Also of note is that a series of samples derived from patients receiving radiotherapy alone (three patients) were also compared with those of radiochemotherapy (16 patients) in order to investigate whether the chemotherapy may profoundly influence the changes in bacterial profiles during radiotherapy. We found that there were no significant differences in alpha diversity (Shannon and PD whole tree) between these two treatment groups and also no remarkable differences between them after RTOG correction as well (Fig. S2a–d). Moreover, among these three patients undergoing radiation therapy alone, two of them developed severe mucositis, whereas 11 of the 16 patients who received radiochemotherapy eventually experienced severe mucositis. The statistical analysis based on Fisher's exact test showed there were also no significant differences regarding the proportion of patients presenting with severe mucositis between these two treatment groups, which may be to some extent due to the smaller sample size in this study. Therefore, further studies with larger sample size are still needed to confirm these results.
Fig. 3.
Association of increased mucositis severity with significant changes in abundance of oropharyngeal bacteria. (a) Heatmap based on Spearman correlations between the relative abundance of 11 bacterial genera and mucositis severity according to RTOG criteria. Leptotrichia and Peptostreptococcus were significantly negatively associated with increased RTOG Grades (RTOG 0–4: blue, yellow, orange, red and black, respectively), whereas other nine genera including Phenylobacterium, Acinetobacter, Burkholderia, Sphingomonas, Azospirillum, Rhizobium, Hydrogenophaga, Paracoccus and Nocardioides were positively associated with it. The relative abundances of 11 bacterial genera were converted to log10 values and represented by a colour gradient for each sample. (b) Scatter plots with trend line showing significant negative or positive correlations between the RTOG Grades and relative abundance of 11 bacterial genera respectively (FDR P < 0.05).
Fig. S2.
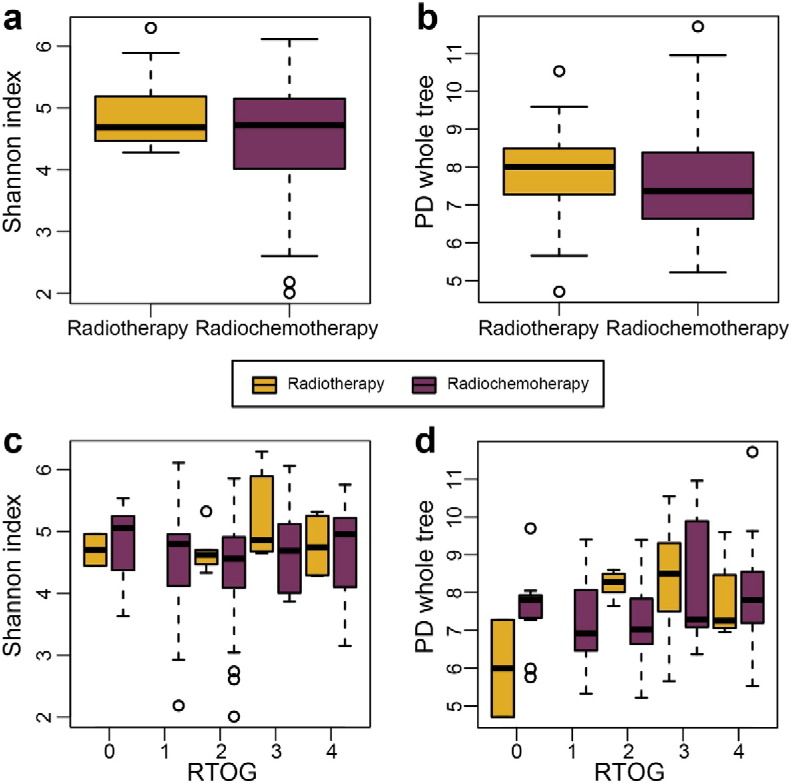
Comparison of the oropharyngeal bacterial alpha-diversity between patients who received radiotherapy alone and those who received radiochemotherapy. Box plots of alpha-diversity indices (Shannon and PD whole tree) showing no statistically significant differences between patients treated with radiotherapy alone and radiochemotherapy (a–b) and also no notable differences between them after RTOG correction as well (c–d).
3.3. Changes in Relative Abundance of Bacterial Genera are Highly Personalized During Radiation Therapy
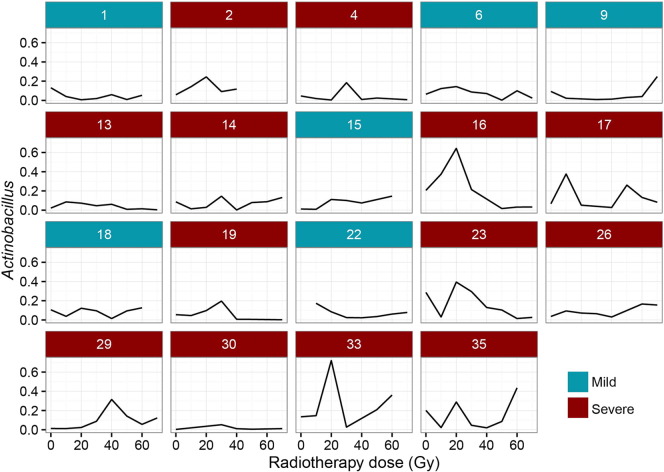
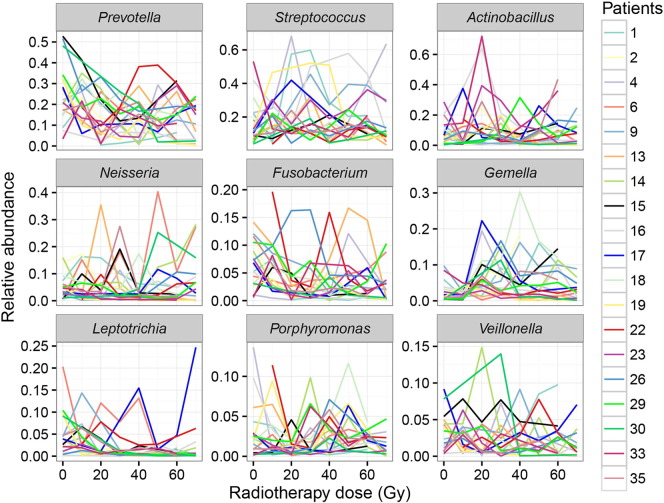
Among the 14 predominate genera identified in patients before irradiation, nine genera including Prevotella, Streptococcus, Actinobacillus, Neisseria, Fusobacterium, Gemella, Leptotrichia, Porphyromonas and Veillonella, due to their higher abundance, were selected for analysis of any intra- and interindividual variations at different dose levels (10–70 Gy). It is noted that the relative abundance of each genus had fluctuated irregularly in each patient during the course of radiotherapy, and this fluctuation as shown in Figs. 4 and S3, tended to be highly personalized and unpredictable. Thus it was difficult to create a general pattern for the interpretation and prediction of variations of these genera associated with increased radiation dose.
Fig. 4.
Illustration of variations in relative abundance of Actinobacillus in each patient. The data represent six patients with mild to moderate mucositis belonging to the mild (blue) subgroup and 13 with severe mucositis belonging to the severe (red) subgroup. A marked, transient peak in the relative abundance of Actinobacillus was often found in patients of the severe subgroup after receiving a cumulative dose of 20 or 30 Gy (corresponding to the second or third week), whereas its abundance remained relatively stable or even decreased in the mild subgroup in the same time frame.
Fig. S3.
Illustration of inter- and intraindividual differences in relative abundance of nine predominant genera respectively in 19 NPC patients under seven different cumulative radiation doses. The abscissas represent cumulative radiation doses and the ordinates indicate relative abundances of each genus. Each colored broken line represents dynamic changes in the relative abundance of one specific genus (Prevotella, Streptococcus, Actinobacillus, Neisseria, Fusobacterium, Gemella, Leptotrichia, Porphyromonas or Veillonella) in one patient during radiotherapy. Based on irregular fluctuations in abundances of each genus shown in the graph above, there is no consistent pattern of variations found either in one patient or among patients.
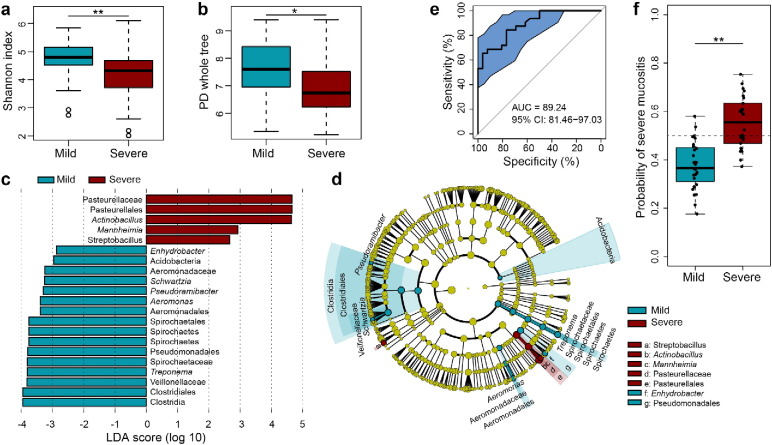
Of particular interest, however, was the fact that although 13 of the 19 patients developed RTOG Grade 3–4 OM in this study, there were still six patients who, after receiving about the same cumulative radiation dose, only experienced Grade 1–2 OM eventually during the entire course. To investigate whether the aggravation of mucositis was associated with the changes in bacterial diversity during their treatment or not, these 19 patients were divided into two subgroups: a mild subgroup (the patients experienced only Grade 1 to 2 mucositis), and a severe subgroup (patients which experienced Grade 3 to 4 mucositis). The biodiversity of bacterial communities was then compared between the two subgroups. These results showed there were in fact no significant differences in alpha diversity between the two groups either before or shortly after irradiation (RTOG 0) (Figs. S4a–b and S5a–b). However what is worthy of note was that a significantly higher relative abundance of Streptococcus was found in the severe subgroup during the phase of RTOG 0 OM (Fig. S5c). As radiation accumulated, mucosal lesions became more apparent in both groups. During the phase of RTOG 1–2 OM, surprisingly, samples from the severe subgroup showed a significantly lower alpha diversity than those from the mild (Fig. 5a–b), suggesting that the latter had a more diverse bacterial community at this stage in their development than the former. What's more, LEfSe analysis showed that the severe subgroup also particularly harbored a significantly higher abundance of some specific non-motile, gram-negative rod-shaped facultative anaerobic bacteria, including the three genera Actinobacillus, Mannheimia and Streptobacillus, and unclassified Pasteurellales and pasteurellaceae also being abundantly present during this phase (Fig. 5c–d). Notably, the proportion of Actinobacillus peaked around the end of second week or third week, corresponding to 20 or 30 Gy, in most patients of this severe subgroup (Fig. 4).
Fig. S4.
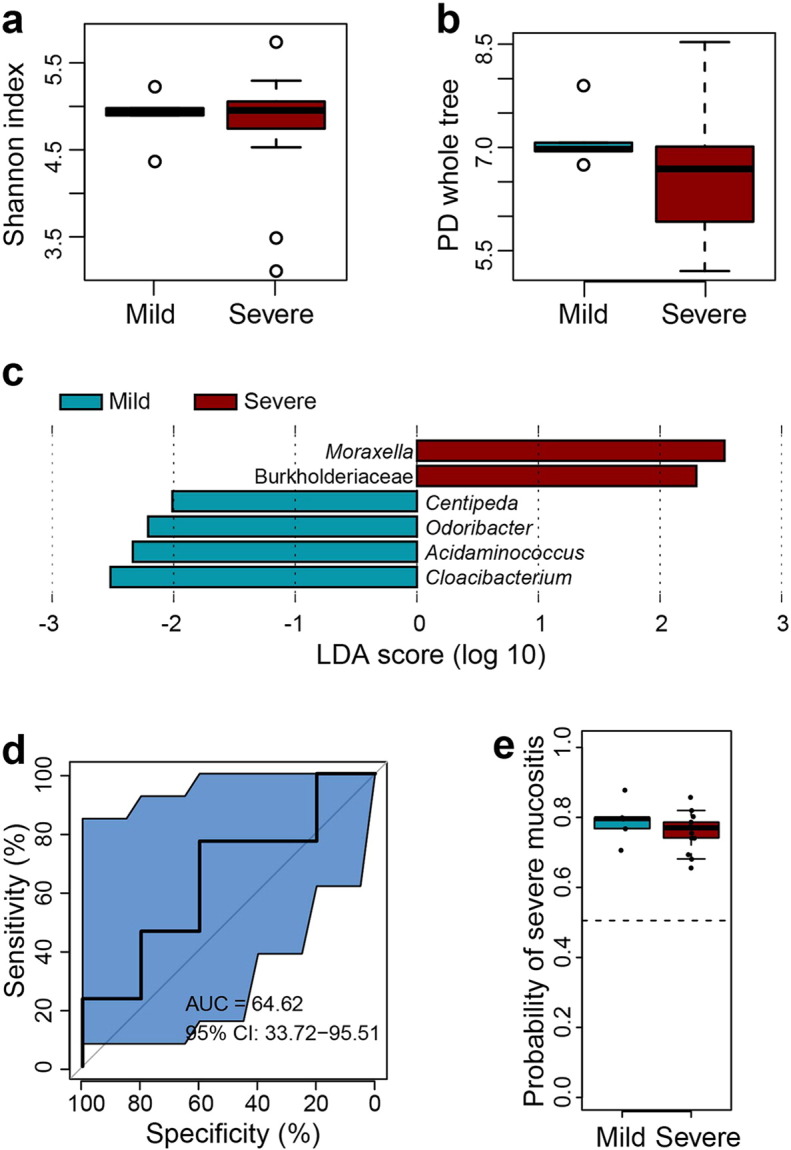
Comparison of bacterial diversity of oropharyngeal samples from the mild and severe subgroups prior to radiotherapy. (a–b) Box plots of alpha-diversity indices (Shannon and PD whole tree) showing no statistically significant differences between non-irradiated samples from patients of the mild (blue) subgroup who only experienced mild mucositis throughout the course of treatment and from patients of severe (red) subgroups who eventually developed severe mucositis (Wilcoxon rank-sum test, P > 0.05). (c) LEfSe analysis identifies the most differentially abundant taxons between the two subgroups. Severe-patients-enriched taxa are indicated with a positive LDA score, and taxa enriched in mild patients have a negative score. Only taxa meeting an LDA significant threshold > 2 are shown. (d) The ROC curves of supervised classification using all the oropharyngeal samples from the mild and severe subgroups prior to radiotherapy to predict their final OM. (e) Box plot showing predicted probabilities of developing severe or mild mucositis based on non-irradiated samples microbiota from the severe and mild subgroups. There is no significant difference between the two subgroups (Wilcoxon rank-sum test, P > 0.05).
Fig. S5.
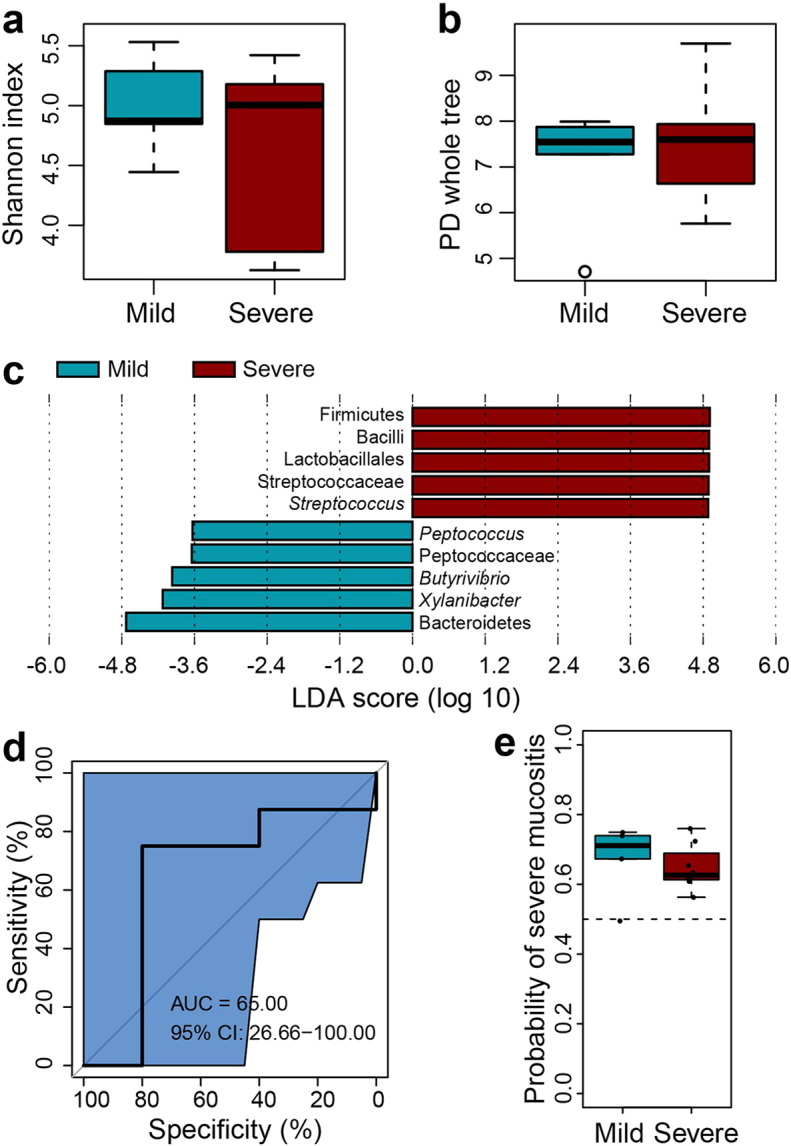
Comparison of bacterial diversity of oropharyngeal samples from the mild and severe subgroups at the phase of RTOG 0 OM. (a–b) Box plots of alpha-diversity indices (Shannon and PD whole tree) showing no statistically significant differences between RTOG 0 samples from patients of the mild (blue) subgroup who only experienced mild mucositis throughout the course of treatment and from patients of severe (red) subgroups who eventually developed severe mucositis (Wilcoxon rank-sum test, P > 0.05). (c) LEfSe analysis identifies the most differentially abundant taxons between the two subgroups. Severe-subgroup-enriched taxa are indicated with a positive LDA score, and taxa enriched in the mild subgroup have a negative score. Only taxa meeting an LDA significant threshold > 2 are shown. (d) The ROC curves of supervised classification using all the oropharyngeal samples from the mild and severe subgroups during the phase of RTOG 0 OM to predict their final OM. (e) Box plot showing predicted probabilities of developing severe or mild mucositis based on RTOG 0 samples microbiota from the severe and mild subgroups. There is no significant difference between the two subgroups (Wilcoxon rank-sum test, P > 0.05)
Fig. 5.
Comparison of bacterial diversity of oropharyngeal samples from the mild and severe subgroups at the phase of RTOG 1–2 OM and disease state classification based on RTOG 1–2 microbiota profiles. (a–b) Box plots of alpha-diversity indices (Shannon and PD whole tree) comparing the mild (blue) and severe (red) subgroups' samples. During the phase of RTOG 1–2 OM, patients who eventually developed severe mucositis (severe subgroup) harbored significantly lower bacterial diversity than patients who only experienced mild mucositis (mild subgroup) throughout the course of treatment. (c–d) LEfSe analysis identifies the most differentially abundant taxons between the two subgroups. Severe-patients-enriched taxa are indicated with a positive LDA score, and taxa enriched in mild patients have a negative score. Only taxa meeting an LDA significant threshold > 2 are shown. (e) The ROC curves of supervised classification using all the oropharyngeal samples from the mild and severe subgroups presenting with RTOG1–2 OM to predict their final outcome in OM. (f) Box plot showing predicted probabilities of developing severe or mild mucositis based on RTOG 1–2 samples microbiota from the severe and mild subgroups. The difference in probabilities is statistically significant. Wilcoxon rank-sum test was used to determine statistical significance. *P < 0.05; **P < 0.01.
3.4. Predictive Modeling of Aggravation of OM Using Oral Microbiota
As this project progressed, a hypothesis was proposed of the existence of an obvious discrimination of oral microbiota between the two subgroups prior to the exacerbation of severe mucositis, and this may reflect significant differences in the extent of microbiota dysbiosis between them at this phase in their treatment, which in turn may ultimately lead to different disease states, by either becoming a stable (RTOG1–2) or a further exacerbated condition. To test this hypothesis, a random forest model was developed using the OTU data of RTOG 1–2 OM from both subgroups. The performance of model was evaluated by a 10-fold cross-validation approach, and the predictive power was scored in a receiver-operating characteristic (ROC) analysis. The area under the curve (AUC) was 0.89 (Fig. 5e,f), indicating that the oral bacterial community is related to the progression and aggravation of mucositis, and that the oral microbiota has the potential to predict mucositis outcomes. However, the AUC was only 0.64 and 0.65 respectively by using the OTU data of non-irradiated and RTOG 0 OM samples as training set, which suggested that this advanced outcome can not be identified by oral microbiota at an earlier stage of radiation-induced mucositis (Figs. S4d–e and S5d–e).
4. Discussion
Nasopharyngeal carcinoma is one of the most common epithelial malignancies in Southern China (Guo et al., 2014). A number of theories regarding its pathogenesis have been proposed over the years (Mcdermott et al., 2001, Tsao et al., 2014). However, previously the possible role of oral and oropharyngeal bacteria in the development of NPC was rarely investigated. Thus it was of interest that we found healthy controls harbored more diverse and shared more similar bacterial communities than NPC patients. It is commonly believed that a stable and diverse microbiota is essential for several host physiological processes and mucosal immune functions, since in the oral cavity and oropharynx, the normal bacterial flora can prevent colonization by exogenous pathogens and overgrowth of indigenous pathobionts, whereas an altered composition of commensal flora, termed dysbiosis, could lead to immune dysregulation and increase a person's risk of developing several diseases (Tomkovich and Jobin, 2016).
Moreover, compared to healthy individuals, our NPC patients harbored significantly greater abundances of Pseudomonas, Pediococcus and Oscillibacter in the oropharynx, all of which have been shown to be associated with human infections. Of particular note is the genus Pseudomonas that contains several nitrate- and/or nitrite-reducing bacteria, such as P. aeruginosa and P. fluorescens. These oral species can, in particular, endogenously synthesize N-nitroso compounds that are more commonly found in food samples from NPC high-risk areas (Feng et al., 2007) and represent a major class of important chemical carcinogens. In fact, the colonization of the nasopharynx by microflora that contain abundant nitrate-reducing microorganisms has been suggested to be a risk factor for NPC in Maghrebian populations (Charriere et al., 1991). Notwithstanding, it is questionable whether such changes in oropharyngeal bacterial profile are a cause or consequence of NPC. However to postulate further on this, is beyond the scope of this study.
Radiotherapy remains the cornerstone of treatment for NPC. A systematic literature review reported that OM is one of the most frequent toxicities for HNC, with the mean incidence rate being 80% (Rodriguez-Caballero et al., 2012). Virtually, all patients in this study experienced some degree of mucositis. It has been shown that radiochemotherapy compromises the mouth's defense mechanisms and causes distinct shifts in the oral microbiota (Vanhoecke et al., 2015). To analyze these complex changes, patients' retropharyngeal mucosa was longitudinally sampled in this study using disposable medical sterile swabs, because the mucosal swab sampling, compared to mouthwash, dental plaque and saliva sampling, may more accurately and factually reflect the dynamic changes in mucosal microbiota. Actually, once a pseudomembrane (biofilm) has formed in the oral cavity during treatment, rinsing the mouth out with liquid (mouthwash), although less invasive than mucosal sampling, can only remove planktonic bacteria but not the bacteria living within the biofilm. This may partially explain that the use of topical antimicrobial agents in different forms (solution, lozenge, paste or gel) failed to prevent the occurrence of severe mucositis in many previous experiments (Wijers et al., 2001, Stokman et al., 2003, Donnelly et al., 2003), because microorganisms growing in a biofilm show much greater resistance to antimicrobial agents than their free-living counterparts.
It has been shown that chemotherapy was associated with a decrease in intestinal microbial diversity, and that this decrease coincided in time with the development of severe mucositis (Van Vliet et al., 2010, Van Vliet et al., 2009). However in this study we did not find any significant changes in bacterial diversity among the non-irradiated samples, RTOG 0 and RTOG 1 (erythema). This suggests therefore that at least the initiation of erythematous lesion may indeed be due to a direct cytotoxic effect of radiation or chemotherapeutic drugs; but not related to an alteration in oral microbiota. Sonis (2009), on the other hand proposed that bacteria, at least quantitatively, could not drive the development of mucosa ulceration, since their work suggests that ulceration preceded the increase in bacterial population, even though its peak severity coincided with peak bacterial loads. This raises a question of whether shifts in bacterial community structure may play a role in the aggravation of ulcers. In our study we found the relative abundance of several Gram-negative bacteria increased significantly as mucositis progressed to peak severity. Most of them were facultative anaerobes and belong to the phylum Proteobacteria, some of which are known opportunistic pathogens capable of causing a wide variety of nosocomial infections and are associated with ulcerative mucosal and/or skin lesions (Zhu et al., 2010, Broniek et al., 2014, Lestin et al., 2008, Song et al., 2011, Soares et al., 2011). Therefore the increase in the proportion of the Gram-negative bacteria that act as “modifiers” may facilitate the aggravation of mucosal inflammation through the activation of host pattern recognition receptors (e.g. TLR, NOD) by bacterial components (e.g. LPS, fimbriae, membrane vesicles, etc.) and the formation of pro-inflammatory cytokines such as TNF-α, IL-1, and IL-6 as well as PGE2 and MMPs (Vasconcelos et al., 2016).
Surprisingly, only six of our 19 patients in this study experienced mild to moderate mucositis eventually, while the rest developed severe mucositis. This led to the question, as to what differences existed between the mild and severe subgroups with respect to bacterial profile. We found patients from the severe subgroup had a significantly higher abundance of Streptococcus before erythema became visible. This Gram-positive genus consists of approximately 50 species, including species forming a major part of oral commensals and many important human pathogens (Innings et al., 2005). It has been shown that the level of Streptococcus mitis increased significantly in NPC patients after irradiation, and in patients with breast cancer after chemotherapy (Tong et al., 2003, Napenas et al., 2010). This Streptococcus mitis, together with some other oral Streptococci, are mucin-degrading bacteria which play a pivotal role in the breakdown of mucus in the human oral cavity (van der Hoeven and Camp, 1994). When the degradation proceeds, the barrier function of epithelium is compromised, potentially causing pathogen translocation into the lamina propria and subsequent recruitment of inflammatory cells (Deplancke and Gaskins, 2001). Therefore it appears that a higher abundance of Streptococci might predispose patients to oral mucus dysfunction in the very early stage of mucositis, thus potentially disturbing the ecological balance of the oral microenvironment to a greater extent in the severe subgroup prior to erythema arising. From the occurrence of visible erythema to the beginning of severe mucositis (RTOG 3), we found that the patients in the mild subgroup harbored significantly more diverse bacteria and a lower abundance of Actinobacillus than those in the severe category. It is commonly believed that a more diverse oral bacterial community may contribute a lot to the protection of the oral ecosystem from overgrowth of indigenous pathobionts, such as Actinobacillus spp. observed in this study. These species were usually found to serve as both commensals of the oropharynx and opportunistic pathogens often involved in the pathogenesis of meningitis, sinusitis, pleural empyema, bronchopneumonia in patients with their associated underlying diseases (Friis-Moller et al., 2001). Due to its higher abundance shown in the server group and its commensal/pathogen life-style, Actinobacillus spp. may profoundly affect oropharyngeal microbial homeostasis, and be one of associated factors that predispose patients to severe mucositis. Thus, additional research is still needed to determine the species of Actinobacillus and germ-free and gnotobiotic animals can provide invaluable experimental tools for further investigation of host-microbe interactions, since colonization of one or more defined species can be achieved.
Moreover, as to the existence of a remarkable difference in microbiota which preceded the manifestation of severe mucositis, a random forest model was built into this study to examine whether microbiota composition could identify their pre-aggravation status. It is worth mentioning that this predictive model has been shown to be a suitable model for the prediction of aggravation of mucositis (RTOG 3–4) in patients presenting with mild to moderate mucositis during the treatment. However, we failed to build models in predicting such aggravation prior to irradiation or before the visual manifestation of erythema, which may be essentially due to non-significant differences in microbiota between the two subgroups and or limited samples derived from these groups within these two stages as well. Both factors may in turn affect the accuracy of the modeling and final results of prediction. Therefore, larger and prospective cohort studies are further required to verify and validate this predictive model.
In conclusion, our results showed that changes in oral microbial community correlated with the progression and aggravation of radiotherapy-induced mucositis in the NPC patients and microbiota-based strategies can be used for the early prediction and prevention of the incidence of severe mucositis during radiotherapy.
The following are the supplementary data related to this article.
Funding Sources
This study is supported by the National Natural Science Foundation of China (NSFC31570497, 31322003, 31270152). The funding sources had no role in the study design; in the collection, analysis and interpretation of data; in the writing of the manuscript; or in the decision to submit the paper for publication.
Conflicts of Interest
The authors declare no conflict of interest.
Author Contributions
X.X.Z, Y.H, and H.W.Z conceived the idea, X.X.Z, Y.L.C, and H.Y.L performed the experiment, X.J.Y, H.M.Z, H.F.S, Y.H and H.W.Z analyzed the data, X.J.Y wrote the manuscript, X.J.Y, Y.H and H.W.Z revised the manuscript.
Acknowledgements
We highly appreciated the language editing of A. Evans.
References
- Aas J.A., Paster B.J., Stokes L.N., Olsen I., Dewhirst F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almstahl A., Wikstrom M., Fagerberg-Mohlin B. Microflora in oral ecosystems in subjects with radiation-induced hyposalivation. Oral Dis. 2008;14:541–549. doi: 10.1111/j.1601-0825.2007.01416.x. [DOI] [PubMed] [Google Scholar]
- Broniek G., Langwinska-Wosko E., Szaflik J., Wroblewska M. Acinetobacter junii as an aetiological agent of corneal ulcer. Infection. 2014;42:1051–1053. doi: 10.1007/s15010-014-0647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., Mcdonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charriere M., Poirier S., Calmels S., De Montclos H., Dubreuil C., Poizat R., Hamdi Cherif M., de The G. Microflora of the nasopharynx in Caucasian and Maghrebian subjects with and without nasopharyngeal carcinoma. IARC Sci. Publ. 1991:158–161. [PubMed] [Google Scholar]
- Deplancke B., Gaskins H.R. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 2001;73:1131S–1141S. doi: 10.1093/ajcn/73.6.1131S. [DOI] [PubMed] [Google Scholar]
- Donnelly J.P., Bellm L.A., Epstein J.B., Sonis S.T., Symonds R.P. Antimicrobial therapy to prevent or treat oral mucositis. Lancet Infect. Dis. 2003;3:405–412. doi: 10.1016/s1473-3099(03)00668-6. [DOI] [PubMed] [Google Scholar]
- Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B.J., Jalbout M., Ayoub W.B., Khyatti M., Dahmoul S., Ayad M., Maachi F., Bedadra W., Abdoun M., Mesli S., Hamdi-Cherif M., Boualga K., Bouaouina N., Chouchane L., Benider A., Ben Ayed F., Goldgar D., Corbex M. Dietary risk factors for nasopharyngeal carcinoma in Maghrebian countries. Int. J. Cancer. 2007;121:1550–1555. doi: 10.1002/ijc.22813. [DOI] [PubMed] [Google Scholar]
- Friis-Moller A., Christensen J.J., Fussing V., Hesselbjerg A., Christiansen J., Bruun B. Clinical significance and taxonomy of Actinobacillus hominis. J. Clin. Microbiol. 2001;39:930–935. doi: 10.1128/JCM.39.3.930-935.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Winkler C.A., Li J., Guan L., Tang M., Liao J., Deng H., de The G., Zeng Y., O'brien S.J. Evaluation and integration of genetic signature for prediction risk of nasopharyngeal carcinoma in Southern China. Biomed. Res. Int. 2014;2014:434072. doi: 10.1155/2014/434072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.J., Shao Z.Y., Wang Q., Jiang Y.T., Ma R., Tang Z.S., Liu Z., Liang J.P., Huang Z.W. Exploring the dynamic core microbiome of plaque microbiota during head-and-neck radiotherapy using pyrosequencing. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innings A., Krabbe M., Ullberg M., Herrmann B. Identification of 43 Streptococcus species by pyrosequencing analysis of the rnpB gene. J. Clin. Microbiol. 2005;43:5983–5991. doi: 10.1128/JCM.43.12.5983-5991.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestin F., Kraak R., Podbielski A. Two cases of keratitis and corneal ulcers caused by Burkholderia gladioli. J. Clin. Microbiol. 2008;46:2445–2449. doi: 10.1128/JCM.02442-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Lladser M.E., Knights D., Stombaugh J., Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdermott A.L., Dutt S.N., Watkinson J.C. The aetiology of nasopharyngeal carcinoma. Clin. Otolaryngol. Allied Sci. 2001;26:82–92. doi: 10.1046/j.1365-2273.2001.00449.x. [DOI] [PubMed] [Google Scholar]
- Napenas J.J., Brennan M.T., Bahrani-Mougeot F.K., Fox P.C., Lockhart P.B. Relationship between mucositis and changes in oral microflora during cancer chemotherapy. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2007;103:48–59. doi: 10.1016/j.tripleo.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Napenas J.J., Brennan M.T., Coleman S., Kent M.L., Noll J., Frenette G., Nussbaum M.L., Mougeot J.L., Paster B.J., Lockhart P.B., Bahrani-Mougeot F.K. Molecular methodology to assess the impact of cancer chemotherapy on the oral bacterial flora: a pilot study. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2010;109:554–560. doi: 10.1016/j.tripleo.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Panghal M., Kaushal V., Kadayan S., Yadav J.P. Incidence and risk factors for infection in oral cancer patients undergoing different treatments protocols. BMC Oral Health. 2012;12:22. doi: 10.1186/1472-6831-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Yu K.Q., Deng G.H., Jiang Y.X., Wang Y., Zhang G.X., Zhou H.W. Comparison of direct boiling method with commercial kits for extracting fecal microbiome DNA by Illumina sequencing of 16S rRNA tags. J. Microbiol. Methods. 2013;95:455–462. doi: 10.1016/j.mimet.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Pratesi N., Mangoni M., Mancini I., Paiar F., Simi L., Livi L., Cassani S., Buglione M., Grisanti S., Almici C., Polli C., Saieva C., Magrini S.M., Biti G., Pazzagli M., Orlando C. Association between single nucleotide polymorphisms in the XRCC1 and RAD51 genes and clinical radiosensitivity in head and neck cancer. Radiother. Oncol. 2011;99:356–361. doi: 10.1016/j.radonc.2011.05.062. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Caballero A., Torres-Lagares D., Robles-Garcia M., Pachon-Ibanez J., Gonzalez-Padilla D., Gutierrez-Perez J.L. Cancer treatment-induced oral mucositis: a critical review. Int. J. Oral Maxillofac. Surg. 2012;41:225–238. doi: 10.1016/j.ijom.2011.10.011. [DOI] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares A.F., Aquino A.R., Carvalho C.H., Nonaka C.F., Almeida D., Pinto L.P. Frequency of oral mucositis and microbiological analysis in children with acute lymphoblastic leukemia treated with 0.12% chlorhexidine gluconate. Braz. Dent. J. 2011;22:312–316. doi: 10.1590/s0103-64402011000400009. [DOI] [PubMed] [Google Scholar]
- Song E., Jaishankar G.B., Saleh H., Jithpratuck W., Sahni R., Krishnaswamy G. Chronic granulomatous disease: a review of the infectious and inflammatory complications. Clin. Mol. Allergy. 2011;9:10. doi: 10.1186/1476-7961-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonis S.T. The pathobiology of mucositis. Nat. Rev. Cancer. 2004;4:277–284. doi: 10.1038/nrc1318. [DOI] [PubMed] [Google Scholar]
- Sonis S.T. Mucositis: the impact, biology and therapeutic opportunities of oral mucositis. Oral Oncol. 2009;45:1015–1020. doi: 10.1016/j.oraloncology.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Stokman M.A., Spijkervet F.K., Burlage F.R., Dijkstra P.U., Manson W.L., De Vries E.G., Roodenburg J.L. Oral mucositis and selective elimination of oral flora in head and neck cancer patients receiving radiotherapy: a double-blind randomised clinical trial. Br. J. Cancer. 2003;88:1012–1016. doi: 10.1038/sj.bjc.6600824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokman M.A., Spijkervet F.K., Boezen H.M., Schouten J.P., Roodenburg J.L., De Vries E.G. Preventive intervention possibilities in radiotherapy- and chemotherapy-induced oral mucositis: results of meta-analyses. J. Dent. Res. 2006;85:690–700. doi: 10.1177/154405910608500802. [DOI] [PubMed] [Google Scholar]
- Tomkovich S., Jobin C. Microbiota and host immune responses: a love-hate relationship. Immunology. 2016;147:1–10. doi: 10.1111/imm.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H.C., Gao X.J., Dong X.Z. Non-mutans streptococci in patients receiving radiotherapy in the head and neck area. Caries Res. 2003;37:261–266. doi: 10.1159/000070868. [DOI] [PubMed] [Google Scholar]
- Trotti A., Bellm L.A., Epstein J.B., Frame D., Fuchs H.J., Gwede C.K., Komaroff E., Nalysnyk L., Zilberberg M.D. Mucositis incidence, severity and associated outcomes in patients with head and neck cancer receiving radiotherapy with or without chemotherapy: a systematic literature review. Radiother. Oncol. 2003;66:253–262. doi: 10.1016/s0167-8140(02)00404-8. [DOI] [PubMed] [Google Scholar]
- Tsao S.W., Yip Y.L., Tsang C.M., Pang P.S., Lau V.M., Zhang G., Lo K.W. Etiological factors of nasopharyngeal carcinoma. Oral Oncol. 2014;50:330–338. doi: 10.1016/j.oraloncology.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Van Der Hoeven J.S., Camp P.J. The use of lectins in monitoring degradation of oligosaccharide chains in mucin by oral streptococci. Caries Res. 1994;28:257–261. doi: 10.1159/000261979. [DOI] [PubMed] [Google Scholar]
- Van Vliet M.J., Tissing W.J., Dun C.A., Meessen N.E., Kamps W.A., De Bont E.S., Harmsen H.J. Chemotherapy treatment in pediatric patients with acute myeloid leukemia receiving antimicrobial prophylaxis leads to a relative increase of colonization with potentially pathogenic bacteria in the gut. Clin. Infect. Dis. 2009;49:262–270. doi: 10.1086/599346. [DOI] [PubMed] [Google Scholar]
- Van Vliet M.J., Harmsen H.J., De Bont E.S., Tissing W.J. The role of intestinal microbiota in the development and severity of chemotherapy-induced mucositis. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoecke B., De Ryck T., Stringer A., Van De Wiele T., Keefe D. Microbiota and their role in the pathogenesis of oral mucositis. Oral Dis. 2015;21:17–30. doi: 10.1111/odi.12224. [DOI] [PubMed] [Google Scholar]
- Vasconcelos R.M., Sanfilippo N., Paster B.J., Kerr A.R., Li Y., Ramalho L., Queiroz E.L., Smith B., Sonis S.T., Corby P.M. Host-microbiome cross-talk in oral mucositis. J. Dent. Res. 2016;95:725–733. doi: 10.1177/0022034516641890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh G.H., Manjunath V.B., Mumbrekar K.D., Negi H., Fernandes D.J., Sharan K., Banerjee S., Bola Sadashiva S.R. Polymorphisms in radio-responsive genes and its association with acute toxicity among head and neck cancer patients. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa A., Sonis S.T. Mucositis: pathobiology and management. Curr. Opin. Oncol. 2015;27:159–164. doi: 10.1097/CCO.0000000000000180. [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijers O.B., Levendag P.C., Harms E.R., Gan-Teng A.M., Schmitz P.I., Hendriks W.D., Wilims E.B., Van Der Est H., Visch L.L. Mucositis reduction by selective elimination of oral flora in irradiated cancers of the head and neck: a placebo-controlled double-blind randomized study. Int. J. Radiat. Oncol. Biol. Phys. 2001;50:343–352. doi: 10.1016/s0360-3016(01)01444-4. [DOI] [PubMed] [Google Scholar]
- Zhou H.W., Li D.F., Tam N.F., Jiang X.T., Zhang H., Sheng H.F., Qin J., Liu X., Zou F. BIPES, a cost-effective high-throughput method for assessing microbial diversity. ISME J. 2011;5:741–749. doi: 10.1038/ismej.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.H., Li F., Xu J.H., Xiang L.H., Kang K.F. Cutaneous infectious granuloma caused by Phenylobacterium in an adult with myelodysplastic syndrome: a first case report. Am. J. Clin. Dermatol. 2010;11:363–366. doi: 10.2165/11533200-000000000-00000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.