Abstract
Flowering is the result of the coordination between genetic information and environmental cues. Gene regulatory networks have evolved in plants in order to measure diurnal and seasonal variation of day length (or photoperiod), thus aligning the reproductive phase with the most favorable season of the year. The capacity of plants to discriminate distinct photoperiods classifies them into long and short day species, depending on the conditions that induce flowering. Plants of tropical origin and adapted to short day lengths include rice, maize, and sorghum, whereas wheat and barley were originally domesticated in the Fertile Crescent and are considered long day species. In these and other crops, day length measurement mechanisms have been artificially modified during domestication and breeding to adapt plants to novel areas, to the extent that a wide diversity of responses exists within any given species. Notwithstanding the ample natural and artificial variation of day length responses, some of the basic molecular elements governing photoperiodic flowering are widely conserved. However, as our understanding of the underlying mechanisms improves, it becomes evident that specific regulators exist in many lineages that are not shared by others, while apparently conserved components can be recruited to novel functions during evolution.
Keywords: photoperiod, florigen, cereals, flowering, gene regulatory network
Introduction
Several plant species measure day length to start specific developmental switches, e.g., the transition to reproductive growth, during the most appropriate times of the year. Seasonal variation of day length provides a fundamental parameter to synchronize developmental changes, because it is not subject to fluctuations like other environmental cues such as temperature.
Plants can be categorized as long day (LD) or short day (SD) species, depending on the photoperiod most effective at triggering reproductive growth. When day length exceeds a specific critical threshold, flowering is promoted in LD plants, whereas SD plants flower in response to reduction of day length below a critical threshold. Such thresholds are characteristic of each species and largely determined by the region where the species originated and first adapted. Plants growing at low tropical latitudes tend to flower in response to exposure to long nights, whereas species adapted to higher latitudes promote flowering during seasons characterized by LD, indicative of the warm days of spring and summer. Plants adapted to temperate regions that germinate before winter, often also need to satisfy a vernalization requirement (exposure to low non-freezing temperatures for several weeks) to become competent to respond to photoperiodic induction. Additionally, many plants can promote flowering even after long exposures to non-inductive photoperiodic conditions, indicating a facultative response to day length and the existence of floral promoting stimuli that can bypass the requirement for specific conditions. Therefore, plant interactions with its growth environment can be complex, and gene networks have evolved that respond to changing seasonal parameters.
In crop species, responses to day length have been extensively manipulated, creating varieties that can grow, flower and set seeds at latitudes outside of the range occupied by the wild progenitor. Artificial adaptation to broad latitudinal ranges has been a key step during domestication of several species, allowing cultivation and diversification in many regions of the globe. Natural genetic variation has offered the substrate for human selection and remarkably, many domestication loci encode orthologous genes in distantly related species providing a molecular perspective to look at conservation and evolution of pathways regulating flowering.
Here, we will summarize recent advances in understanding photoperiodic flowering regulation in crop species, focusing on cereals. Starting with the tenets established using Arabidopsis as model system, we will discuss how conserved and unique elements have been deployed to evolve flowering networks of LD and SD plants and how they control production of a florigenic systemic signal in leaves.
Arabidopsis Contributed to Develop the Basic Tenets of Photoperiodic Flowering
Photoperiodic flowering has been mostly studied using the dicot Arabidopsis, through which core genetic and molecular mechanisms at the base of the process have been characterized (Song et al., 2015). Arabidopsis might not be fully representative of all plant species but it provides a conceptual framework that can be implemented in other species and also used to discuss evolution of distinct mechanisms typical of distantly related plants (Table 1).
Table 1.
List of genes controlling photoperiodic flowering.
| Arabidopsis | Rice | Maize | Sorghum | Wheat | Barley |
|---|---|---|---|---|---|
| PhyB (AT2G18790) | OsPhyB (LOC_Os03g19590) | ZmPhyB1 (GRMZM2G124532) ZmPhyB2 (GRMZM2G092174) | SbPhyB (Sb01g037340) | TaPhyB (AY888046) | HvPhyB (DQ201142) |
| n.p. | Ehd1 (LOC_Os10g32600) | ZmEhd1 (GRMZM2G479110) | SbEhd1 (Sb01g019980) | n.f. | n.f. |
| n.p. | Ehd2/RID1/OsID1 (LOC_Os10g28330) | ID1 (GRMZM2G011357) | SbID (Sb01g021480) | n.f. | HvID1 (AK361456) |
| n.p. | Ghd7 (LOC_Os07g15770) | ZmCCT (GRMZM5G868285) | SbGhd7 (Sb06g000570) | n.f. | n.f. |
| NF-YB2 (AT5G47640) NF-YB3 (AT4G14540) | Ghd8 (LOC_Os08g07740) | ZmGhd8 (GRMZM2G444073) | SbGhd8 (Sb07g004740) | TaNFYB-A6 (Traes_2AL_AE22E725E) TaNFYB-B6 (Traes_2BL_3237AA694) TaNFYB-D6 (Traes_2DL_DA577AF57) | HvNF-YB7 (MLOC_57782) |
| GI (AT1G22770) | OsGI (LOC_Os01g08700) | gigz1A/gi1 (GRMZM2G107101) gigz1B/gi2 (GRMZM5G844173) | SbGI (Sb03g003650) | TaGI1 (AF543844) TaGI2 (AY679114) TaGI3 (AY679115) | HvGI (AY740523) |
| FKF1 (AT1G68050) | OsFKF1 (LOC_Os11g34460) | ZmFKF1a (GRMZM2G107945) ZmFKF1b (GRMZM2G106363) | SbFKF1 (Sb05g021030) | TaFKF1 (DQ923399) | HvFKF1 (FJ913271) |
| CDF1 (At5g62430) CDF2 (At5g39660) CDF3 (At3g47500) CDF5 (At1g69570) | OsDOF12 (LOC_Os03g07360) | n.f. | n.f. | n.f. | n.f. |
| PRR3 (AT5G60100) PRR7 (AT5G02810) | PRR37 (LOC_Os07g49460) | ZmPRR37 (GRMZM2G033962 and GRMZM2G005732) | SbPRR37 (Sb06g014570) | Ppd-D1 (AB646976) | HvPpd-H1 (AAY42109) |
| CO (AT5G15840) | Hd1 (LOC_Os06g16370) | conz1 (GRMZM2G405368) | SbCO (Sb10g10050) | TaHd1-1 (AB094487) TaHd1-2 (AB094488) TaHd1-3 (AB094489) | HvCO1 (AF490468) HvCO2 (AF490470) |
| FT (AT1G65480) | Hd3a (LOC_Os06g06320) RFT1 (LOC_Os06g06300) | ZCN8 (GRMZM2G179264) ZCN12 (GRMZM2G103666) | SbFT (Sb10g003940) SbCN8 (Sb09g025760) SbCN12 (Sb03g034580) SbFT12 (Sb06g012260) | HvFT1 (DQ100327) | TaFT (DQ890162) |
Genes on the same row share sequence homology. Each locus corresponds to a unique gene identifier. n.f., not found in public databases; n.p., not present in Arabidopsis.
Flowering of Arabidopsis is promoted under LD. The circadian clock is responsible for the rhythmic expression of several factors implicated in environmental responses. Among them, the GIGANTEA (GI) and FLAVIN BINDING KELCH REPEAT F-BOX PROTEIN 1 (FKF1) proteins are expressed at the end of the light phase and interact in a light-dependent fashion (Sawa et al., 2007). The resulting complex targets a group of CYCLING DOF FACTORs (CDFs) for proteasome-mediated degradation (Fornara et al., 2009). The CDFs encode transcriptional repressors that limit expression of the CONSTANS (CO) zinc finger transcription factor, a central regulator within the photoperiodic flowering pathway (Putterill et al., 1995). Besides the major GI-FKF1-CDFs module, several additional mechanisms contribute to CO expression at the transcriptional and post-transcriptional level, including regulation by transcription factors (Ito et al., 2012), alternative splicing (Gil et al., 2016), photoreceptors (Valverde et al., 2004; Song et al., 2014), as well as ambient temperature signals (Fernández et al., 2016), hormonal signals (Wang et al., 2016) and post-translational modifications (Sarid-Krebs et al., 2015). However, central to the current model for photoperiodic flowering, the most prominent feature of CO is its light-dependent stability (Valverde et al., 2004; Song et al., 2012). During the night and the morning, CO protein is unstable and quickly degraded (Jang et al., 2008; Song et al., 2014; Lazaro et al., 2015). Consequently, its expression is shaped to be highest under LD, during the light phase. At this time of the diurnal cycle, CO protein, acting in the companion cells of the phloem, can directly promote expression of FLOWERING LOCUS T (FT), component of the systemic florigenic signal (An et al., 2004; Corbesier et al., 2007; Mathieu et al., 2007).
The effects of CO protein on the levels and rhythmicity of FT mRNA abundance are mediated by several classes of protein interactors that include transcription factors and transcriptional co-regulators, photoreceptors, histone-like proteins, and ubiquitin ligases (see Brambilla and Fornara, 2016 and references therein). Therefore, the photoperiodic flowering pathway, despite being largely interconnected with other regulatory pathways, can be simplified into a linear molecular cascade, whose major output is the FT protein (Figure 1).
FIGURE 1.
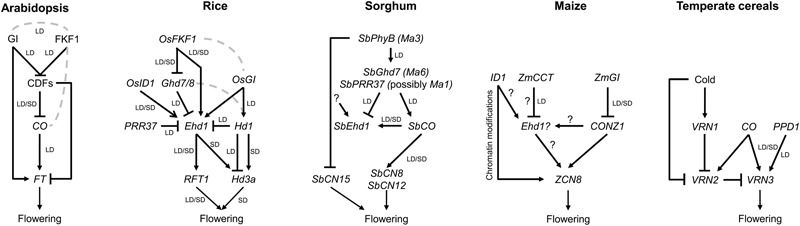
Simplified genetic modules controlling production of the florigens in leaves. (Arabidopsis) The photoperiodic flowering pathway can be simplified into a linear cascade comprising the GI and FKF1 protein complex, targeting CDFs transcriptional repressors for proteasome-mediated degradation. Released CO transcription leads to CO protein accumulation during the evenings of LD and induction of FT expression. Among the CDFs, CDF1 at least can associate directly with FT to repress its expression. (Rice) OsFKF1 and OsGI promote flowering via transcriptional regulation of distinct target genes. However, their protein products can also interact. The Ehd1 and Hd1 proteins promote flowering by activating Hd3a and RFT1 expression under SD. However, under LD, Hd1 switches its function to repress Hd3a transcription. Hd3a transcription is sensitive to induction mediated by Ehd1 under both LD and SD, to the extent that ehd1 rft1 double mutants cannot flower. Conversely, transcription of RFT1 can be activated also under LD, by an unknown mechanism that eventually allows flowering also under unfavorable conditions. (Sorghum) The SbPhyB protein (Ma3) represses flowering by promoting expression of SbGhd7 (Ma6) and SbPRR37 under LD. The coincidence between SbPRR37 and Ma1 is under debate. However, sorghum lines bearing ma3 recessive mutations can flower early also under SD. The SbCO protein is a constitutive activator of flowering, differently from rice Hd1. SbPRR37 can promote SbCO transcription at dawn. (Maize) A higher degree of polygenic control of flowering has been observed in maize compared to other species. However, homologs of flowering genes have been cloned and some mutants characterized. In this diagram, we speculate about the existence of an Ehd1-like function, possibly creating a topology similar to that of other SD species. Major discrete regulators are encoded by ID1 and ZmCCT. (Temperate cereals) Exposure to cold is necessary to reduce VRN2 levels in leaves. Vernalized plants can respond to LDs that promote expression of VRN3/FT via CO homologs and, most importantly, through PPD1, encoding a CCT-domain protein similar to PRR37. Arrows indicate transcriptional activation; flat-end arrows indicate transcriptional repression. Dashed lines indicate that the protein products can interact. Question marks are speculative, and indicate the possible existence of unknown factors with specific functions on gene expression. LD, long day; SD, short day.
Rewiring Photoperiodic Networks in Rice Modifies Day Length Responses
Rice flowering is accelerated by exposure to SD. Seasonal and diurnal time measurements are mediated by a circadian clock that shares components with that of Arabidopsis, and when mutated results in altered sensitivity to the length of the day (Izawa et al., 2011; Matsubara et al., 2012). Homologs of GI, FKF1, the CDFs, CO, and FT exist in rice and have been partly linked in a cascade that resembles the photoperiodic pathway of Arabidopsis (Shrestha et al., 2014) (Table 1). The OsGI and OsFKF1 proteins can interact with each other and with a CDF protein, OsDOF12, similarly to their Arabidopsis homologs (Li et al., 2009; Han et al., 2015). However, mutations in OsFKF1 delay flowering under any photoperiod tested, whereas osgi mutants are late flowering under SD, while having only mild effects under LD (Hayama et al., 2003; Izawa et al., 2011). The phenotypic effects of the two mutations are therefore different. Overexpression of OsDOF12 increases transcription of Heading Date 3a (Hd3a), a homolog of FT, under LD while having no impact on transcription of Heading date 1 (Hd1), a homolog of CO. Thus, the function of OsDOF12 is opposite to that of Arabidopsis CDFs, effectively promoting flowering (Li et al., 2009). It is still unclear whether the interaction between OsGI and OsFKF1 is dependent upon the photoperiod, or if it is necessary for the degradation of OsDOF12 or other DOF proteins. These data indicate that a similar arrangement of regulators exists upstream of Hd3a, but that their molecular function or day length-dependency is very different from Arabidopsis. Both the DOF-CO and the GI-FKF1 modules are evolutionarily ancient as indicated by data from the unicellular alga Chlamydomonas reinhardtii and the liverwort Marchantia polymorpha, where they control phase transition (Kubota et al., 2014; Lucas-Reina et al., 2015). However, evolution has likely re-shaped the function of the dimer several times, readjusting it depending on the species.
Cloning of Hd1 indicated that it encodes a homolog of CO (Yano et al., 2000). However, the Hd1 protein not only promotes flowering under SD but also represses it under LD. Mutations in Hd1 result in accelerated flowering under LD and have been extensively introgressed in varieties cultivated at high latitudes (Izawa et al., 2002; Hayama et al., 2003; Gao et al., 2014; Gómez-Ariza et al., 2015; Goretti et al., 2017). A second important flowering QTL, Early Heading Date 1 (Ehd1) was later cloned and shown to encode a B-type response regulator (Doi et al., 2004). Ehd1 integrates circadian and light inputs and is required to promote flowering under both LD and SD (Itoh et al., 2010), and to modulate it also in response to abiotic stress, including water deficit (Galbiati et al., 2016; Zhang et al., 2016). Under SD, Ehd1 induces flowering mainly by promoting Hd3a expression, and this function is not shared with dicot species (Zhao et al., 2015). Under LD, expression of Ehd1 is limited by several repressors that delay flowering, including Grain Number Plant Height and Heading Date 7 (Ghd7), Hd1, and Pseudo Response Regulator 37 (PRR37) (Gao et al., 2014; Gómez-Ariza et al., 2015). The Hd1 and Ghd7 proteins interact forming a repressor dimer and at least the Ghd7 protein can directly bind the promoter of Ehd1 (Nemoto et al., 2016). Thus, genetic and molecular evidences indicate how a conserved inductive cascade has been repurposed and integrated with unique components to create a novel network topology (Figure 1).
As with all photoperiodic response networks, the major outputs of the regulatory cascade include Hd3a and its paralog RICE FLOWERING LOCUS T 1 (RFT1). Both proteins encode mobile leaf-borne systemic signals, but whereas Hd3a is required only under SD to induce flowering, the RFT1 protein is expressed and can promote flowering under both SD and LD (Komiya et al., 2008, 2009; Zhao et al., 2015). Thus, the facultative response of rice is based on a system comprising two florigens subject to differential regulation. The molecular basis of this differential sensitivity to the photoperiod is still poorly understood.
Mechanisms of Photoperiodic Flowering in Other Short Day Monocots Including Sorghum and Maize
Sorghum (Sorghum bicolor) is a SD plant evolved in Africa, in the Sudan region. Six major QTLs controlling flowering time and termed Maturity loci (Ma1–Ma6) have been detected in sorghum. Almost all QTLs have been identified as photoperiodic flowering regulators and their study is demonstrating the strong homology occurring between the sorghum and rice pathways (Wolabu and Tadege, 2016) (Table 1).
Cloning of the Ma3 locus showed that it encodes SbPhyB, a light receptor which can mediate light signaling and flowering repression (Childs et al., 1997). When SbPhyB is mutated, sorghum becomes insensitive to the photoperiod and flowers early compared to the wild type both under LD and SD (Yang et al., 2014a). One of the functions of SbPhyB is to promote the transcription of SbPRR37 (possibly Ma1) and SbGhd7 (Ma6). These genes encode flowering repressors that limit mRNA expression of downstream targets under LD, including Ehd1, SbFT, and SbZCN8 (collinear orthologs of Hd3a and maize ZCN8, respectively) (Murphy et al., 2011). The flowering suppressor role of these sorghum genes reflects the function of rice OsPRR37 and Ghd7, indicating that these components are shared among SD cereals. Recent data suggested that the Ma1 QTL does not correspond to PRR37, but rather to an FT-like gene, SbFT12, that could act as floral suppressor (Cuevas et al., 2016; Wolabu and Tadege, 2016). Additional data will be required to confirm the true identity of the Ma1 gene.
The regulation of SbCO transcription mediated by SbPRR37 has also been investigated. The data suggest that SbPRR37 modulates SbCO expression at dawn, promoting its transcription under LD, whereas under SD SbCO expression seems not to depend upon SbPRR37 (Murphy et al., 2011). SbCO can activate florigen production under both SD and LD conditions through the activation of SbEhd1, SbCN8, and SbCN12 (Yang et al., 2014b). The role of sorghum SbCO as constitutive floral activator is therefore different from that of rice Hd1, implicating a different regulatory mechanism.
Thirteen different FT-like genes have been identified in the sorghum genome, three of which (SbFT1/SbCN15, SbFT8/SbCN12, and SbFT10/SbCN8) could promote flowering when constitutively expressed in Arabidopsis (Yang et al., 2014b; Wolabu et al., 2016). The transcripts of SbCN8, SbCN12, and SbCN15 peak at dawn but show distinct sensitivities to SbCO mutations. Whereas the transcripts of SbCN8 and SbCN12 are strongly reduced in the Sbco mutant background under LD, SbCN15 shows only a phase shift, suggesting different regulation by SbCO (Yang et al., 2014b). The transcriptional patterns of SbCN8, SbCN12, and SbCN15 under different photoperiods and mutant backgrounds could provide in the future valuable data to understand similarities and differences with the dual florigen system of rice.
Maize (Zea mays) was domesticated in central Mexico from Teosinte, which is a SD plant. The first flowering gene cloned in maize was INDETERMINATE 1 (ID1): plants with mutations in this gene delay the floral transition and produce aberrant inflorescences (Colasanti et al., 1998). ID1 encodes a zinc-finger transcription factor expressed in immature leaves which can activate the floral transition and is not under the control of the circadian clock (Wong and Colasanti, 2007). Although the precise function of ID1 in the photoperiodic pathway is still unclear, recent analyses demonstrated that ID1 controls chromatin modifications of loci encoding maize florigens, and that it can regulate flowering through histone methylations (Mascheretti et al., 2015). A rice homolog of ID1, OsEhd2, is required to induce OsEhd1 expression (Matsubara et al., 2008) (Table 1). Although a maize Ehd1 homolog has not yet been found, the high homology between ID1 and OsEhd2 could suggest a similar regulatory mechanism, possibly indicating the existence of a ZmEhd1-like protein subject to similar regulation. Indirect evidence supporting this view is that the CCT-domain transcription factor ZmCCT shows sequence homology with OsGhd7, and encodes a strong LD flowering repressor (Figure 1). Mutations in ZmCCT cause early flowering and have been artificially selected to expand maize cultivation to higher latitudes (Hung et al., 2012).
Two GI homologs are present in maize, GIGANTEA1 (GI1) and GIGANTEA2 (GI2) (Miller et al., 2008). In Arabidopsis and rice, GI is under circadian clock control and regulates the expression of several genes important for the floral induction. In maize, gi1 mutations cause early flowering under LD conditions. Transcriptional analysis of these mutants demonstrated that GI1 is necessary to repress transcription of CONZ1 (homolog of OsHd1) and ZCN8 (homolog of Hd3a), both of which displayed increased expression in the gi1 background (Bendix et al., 2013). These data demonstrate that ZmGI function is similar to OsGI which can repress flowering under LD conditions, a function opposite to that of AtGI (Hayama et al., 2003). Whether mutations in CONZ1 influence flowering is unknown, but the data suggest it to be downstream of GI1, and possibly upstream of ZCN8 as positive regulator of flowering (Miller et al., 2008).
From the analysis of 15 maize FT-like genes, ZCN8 was identified as the strongest candidate for the maize florigen (Meng et al., 2011). ZCN8 encodes a homolog of FT that delays flowering if silenced, and can complement ft mutants when expressed in Arabidopsis (Lazakis et al., 2011). The regulation of ZCN8 is similar to that of another putative maize florigen, ZCN7, and is under the control of chromatin modifications governed by ID1 (Mascheretti et al., 2015). However, whether ZCN7 satisfies the criteria of a florigenic protein is still to be clarified.
Flowering Mechanisms in Long Day Temperate Cereals
Differently from rice, sorghum, and maize, the temperate cereals wheat (Triticum spp.) and barley (Hordeum vulgare) were domesticated in the Eastern Mediterranean region, in areas characterized by the alternation of cold and warm seasons. These cereals have evolved mechanisms to prevent flowering when temperatures are low, to protect the meristem from cold damage. Flowering is promoted after exposure to vernalizing conditions, when plants resume growth in the spring. During domestication, some cultivars of these species have lost sensitivity to vernalization and, depending on the response to cold, they could be classified as winter or spring types. Winter-types have an obligate vernalization requirement. Such response is controlled by the VERNALIZATION (VRN) loci (Ream et al., 2012). VRN1 is a MADS-box floral promoter homologous to FRUITFULL (FUL) and APETALA1 (AP1) of Arabidopsis, whereas VRN2 is a floral repressor sharing sequence similarity to Ghd7 of rice (Table 1). Under low temperatures, the expression of VRN1 is induced and the protein directly binds to the promoter of VRN2 to reduce its expression during vernalization (Trevaskis, 2006; Deng et al., 2015). Dominant mutations in VRN1 or recessive mutations in VRN2 confer a spring growth habit, and have been exploited by breeders to expand cultivation areas (Yan et al., 2004; Fu et al., 2005; Loukoianov, 2005).
Downregulation of VRN2 is required to induce VRN3 expression during the floral transition. VRN3 proteins (designated as TaFT and HvFT in wheat and barley, respectively) are homologs of the Arabidopsis and rice florigens, and move to the apical meristem to promote flowering upon exposure to warm temperatures and LD (Yan et al., 2006; Li and Dubcovsky, 2008). Thus, cold signals coordinate VRN expression to activate flowering and long-distance florigenic signaling only when a vernalization requirement has been satisfied.
As soon as VRN2 levels decrease, exposure to LDs is required to promote flowering. Temperate cereals flower earlier under LDs, whereas exposure to SDs delays flowering. The PHOTOPERIOD 1 (Ppd1) gene has been described as the major factor controlling sensitivity to day length in wheat and barley (Turner et al., 2005; Beales et al., 2007). Mutations in PPD1 delay flowering under LD and reduce VRN3/FT expression. PPD1 proteins are homologous to PRR37 proteins of rice and sorghum, both of which repress flowering under LD. The functional divergence of PRR37 proteins observed among LD temperate and SD tropical cereals deserves further attention, as it might be at the base of their distinct photoperiodic requirements.
Homologs of CO and Hd1 have been identified in wheat and barley (Campoli and Von Korff, 2014). The TaHd1-1 gene could complement a rice hd1 mutant, suggesting functional conservation of protein function in a heterologous system (Nemoto et al., 2003). In barley, studies based on overexpression have provided important clues to the position of Hd1 homologs in flowering regulatory networks. Overexpression of HvCO1 and HvCO2 promoted flowering under both LD and SD, but plants retained sensitivity to the photoperiod, because of independent control of HvFT1 by PPD1 (Campoli et al., 2012). Thus, barley flowering depends on two parallel pathways controlling FT expression (Figure 1). Interestingly, overexpression of HvCO2 was recently shown to increase expression of VRN2 under LD and SD in a winter variety (Mulki and von Korff, 2016). Despite such increase of the VRN2 repressor, overexpression of HvCO2 could still promote flowering, likely through a VRN2-independent pathway. The data might suggest that HvCO2 mediates a floral repressive function through VRN2, to limit FT expression. Whether barley orthologs of Hd1 display dual functions similarly to rice Hd1 awaits further testing. The use of mutant resources and possibly of edited alleles might help to address this issue.
Concluding Remarks
The examples discussed above illustrate the flexibility of photoperiodic flowering networks and how adaptation to distinct environments modifies their topology. Major changes include the integration of vernalization modules in some networks and the recruitment of non-shared regulators, such as Ehd1 and Ghd7, in others. A common theme appears to be the requirement for upstream master regulators to control expression of FT-like genes, but their number and relative contributions to heading time broadly varies between species. While in Arabidopsis, CO acts as central and primary regulator of FT, CO homologs in crops are coupled to parallel pathways largely sharing the workload, and FT expression often strongly depends on additional regulators.
Efforts will be needed in the future to isolate all components of the networks in crop species, many of which are still to be cloned. Quantification of transcripts offers a rapid way of determining relationships between genes, but provides only limited information on protein expression or biochemical function. Finally, molecular networks are starting to be built, based on protein–protein or protein–DNA interactions especially in rice. Expanding these efforts toward other crops will prove necessary.
Author Contributions
FF and VB organized the manuscript and wrote the Arabidopsis and rice sections. MC wrote the maize and sorghum section and prepared Figure 1. JG-A wrote the temperate cereals section. FF revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by an ERC Starting Grant #260963, and by a grant from the Italian Ministry of Education and Research (MIUR) #20153NM8RM to FF.
References
- An H., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., et al. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131 3615–3626. 10.1242/dev.01231 [DOI] [PubMed] [Google Scholar]
- Beales J., Turner A., Griffiths S., Snape J. W., Laurie D. A. (2007). A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 115 721–733. 10.1007/s00122-007-0603-4 [DOI] [PubMed] [Google Scholar]
- Bendix C., Mendoza J. M., Stanley D. N., Meeley R., Harmon F. G. (2013). The circadian clock-associated gene gigantea1 affects maize developmental transitions. Plant Cell Environ. 36 1379–1390. 10.1111/pce.12067 [DOI] [PubMed] [Google Scholar]
- Brambilla V., Fornara F. (2016). Y flowering? Regulation and activity of CONSTANS and CCT-domain proteins in Arabidopsis and crop species. Biochim. Biophys. Acta 10.1016/j.bbagrm.2016.10.009 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Campoli C., Drosse B., Searle I., Coupland G., von Korff M. (2012). Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J. 69 868–880. 10.1111/j.1365-313X.2011.04839.x [DOI] [PubMed] [Google Scholar]
- Campoli C., Von Korff M. (2014). Genetic control of reproductive development in temperate cereals. Adv. Bot. Res. 72 131–158. 10.1016/B978-0-12-417162-6.00005-5 [DOI] [Google Scholar]
- Childs K. L., Miller F. R., Cordonnier-Pratt M. M., Pratt L. H., Morgan P. W., Mullet J. E. (1997). The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol. 113 611–619. 10.1104/pp.113.2.611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J., Yuan Z., Sundaresan V. (1998). The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93 593–603. 10.1016/S0092-8674(00)81188-5 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316 1030–1033. 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- Cuevas H. E., Zhou C., Tang H., Khadke P. P., Das S., Lin Y.-R., et al. (2016). The evolution of photoperiod-insensitive flowering in sorghum, a genomic model for panicoid grasses. Mol. Biol. Evol. 33 2417–2428. 10.1093/molbev/msw120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Casao M. C., Wang P., Sato K., Hayes P. M., Finnegan E. J., et al. (2015). Direct links between the vernalization response and other key traits of cereal crops. Nat. Commun. 6:5882 10.1038/ncomms6882 [DOI] [PubMed] [Google Scholar]
- Doi K., Izawa T., Fuse T., Yamanouchi U., Kubo T., Shimatani Z., et al. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18 926–936. 10.1101/gad.1189604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández V., Takahashi Y., Le Gourrierec J., Coupland G. (2016). Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J. 86 426–440. 10.1111/tpj.13183 [DOI] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K. C. S., Gissot L., Sauerbrunn N., Rühl M., Jarillo J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17 75–86. 10.1016/j.devcel.2009.06.015 [DOI] [PubMed] [Google Scholar]
- Fu D., Szûcs P., Yan L., Helguera M., Skinner J. S., von Zitzewitz J., et al. (2005). Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol. Genet. Genomics 273 54–65. 10.1007/s00438-004-1095-4 [DOI] [PubMed] [Google Scholar]
- Galbiati F., Chiozzotto R., Locatelli F., Spada A., Genga A., Fornara F. (2016). Hd3a, RFT1 and Ehd1 integrate photoperiodic and drought stress signals to delay the floral transition in rice. Plant Cell Environ. 39 1982–1993. 10.1111/pce.12760 [DOI] [PubMed] [Google Scholar]
- Gao H., Jin M., Zheng X.-M., Chen J., Yuan D., Xin Y., et al. (2014). Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. U.S.A. 111 16337–16342. 10.1073/pnas.1418204111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil K.-E., Park M.-J., Lee H.-J., Park Y.-J., Han S.-H., Kwon Y.-J., et al. (2016). Alternative splicing provides a proactive mechanism for the diurnal CONSTANS dynamics in Arabidopsis photoperiodic flowering. Plant J. 89 128–140. 10.1111/tpj.13351 [DOI] [PubMed] [Google Scholar]
- Gómez-Ariza J., Galbiati F., Goretti D., Brambilla V., Shrestha R., Pappolla A., et al. (2015). Loss of floral repressor function adapts rice to higher latitudes in Europe. J. Exp. Bot. 66 2027–2039. 10.1093/jxb/erv004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goretti D., Martignago D., Landini M., Brambilla V., Gomez-Ariza J., Gnesutta N., et al. (2017). Transcriptional and post-transcriptional mechanisms limit Heading Date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genet. 1:e1006530 10.1371/journal.pgen.1006530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.-H., Yoo S.-C., Lee B.-D., An G., Paek N.-C. (2015). Rice FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (OsFKF1) promotes flowering independent of photoperiod. Plant Cell Environ. 38 2527–2540. 10.1111/pce.12549 [DOI] [PubMed] [Google Scholar]
- Hayama R., Yokoi S., Tamaki S., Yano M., Shimamoto K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422 719–722. 10.1038/nature01549 [DOI] [PubMed] [Google Scholar]
- Hung H.-Y., Shannon L. M., Tian F., Bradbury P. J., Chen C., Flint-Garcia S. A., et al. (2012). ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl. Acad. Sci. U.S.A. 109 E1913–E1921. 10.1073/pnas.1203189109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Song Y. H., Josephson-Day A. R., Miller R. J., Breton G., Olmstead R. G., et al. (2012). FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109 3582–3587. 10.1073/pnas.1118876109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Nonoue Y., Yano M., Izawa T. (2010). A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 42 635–638. 10.1038/ng.606 [DOI] [PubMed] [Google Scholar]
- Izawa T., Mihara M., Suzuki Y., Gupta M., Itoh H., Nagano A. J., et al. (2011). Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 23 1741–1755. 10.1105/tpc.111.083238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T., Oikawa T., Sugiyama N., Tanisaka T., Yano M., Shimamoto K. (2002). Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16 2006–2020. 10.1101/gad.999202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K. C. S., Wenkel S., Soppe W., Deng X.-W., et al. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27 1277–1288. 10.1038/emboj.2008.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R., Ikegami A., Tamaki S., Yokoi S., Shimamoto K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135 767–774. 10.1242/dev.008631 [DOI] [PubMed] [Google Scholar]
- Komiya R., Yokoi S., Shimamoto K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136 3443–3450. 10.1242/dev.040170 [DOI] [PubMed] [Google Scholar]
- Kubota A., Kita S., Ishizaki K., Nishihama R., Yamato K. T., Kohchi T. (2014). Co-option of a photoperiodic growth-phase transition system during land plant evolution. Nat. Commun. 5 3668 10.1038/ncomms4668 [DOI] [PubMed] [Google Scholar]
- Lazakis C. M., Coneva V., Colasanti J. (2011). ZCN8 encodes a potential orthologue of Arabidopsis FT florigen that integrates both endogenous and photoperiod flowering signals in maize. J. Exp. Bot. 62 4833–4842. 10.1093/jxb/err129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A., Mouriz A., Piñeiro M., Jarillo J. A. (2015). Red light-mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in Arabidopsis. Plant Cell 27 2437–2454. 10.1105/tpc.15.00529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Dubcovsky J. (2008). Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 55 543–554. 10.1111/j.1365-313X.2008.03526.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Yang C., Li X., Gan Q., Zhao X., Zhu L. (2009). Functional characterization of rice OsDof12. Planta 229 1159–1169. 10.1007/s00425-009-0893-7 [DOI] [PubMed] [Google Scholar]
- Loukoianov A. (2005). Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol. 138 2364–2373. 10.1104/pp.105.064287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas-Reina E., Romero-Campero F. J., Romero J. M., Valverde F. (2015). An evolutionarily conserved DOF-CONSTANS module controls plant photoperiodic signaling. Plant Physiol. 168 561–574. 10.1104/pp.15.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascheretti I., Turner K., Brivio R. S., Hand A., Colasanti J., Rossi V. (2015). Florigen-encoding genes of day-neutral and photoperiod-sensitive maize are regulated by different chromatin modifications at the floral transition. Plant Physiol. 168 1351–1363. 10.1104/pp.15.00535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17 1055–1060. 10.1016/j.cub.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Matsubara K., Ogiso-Tanaka E., Hori K., Ebana K., Ando T., Yano M. (2012). Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol. 53 709–716. 10.1093/pcp/pcs028 [DOI] [PubMed] [Google Scholar]
- Matsubara K., Yamanouchi U., Wang Z.-X., Minobe Y., Izawa T., Yano M. (2008). Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol. 148 1425–1435. 10.1104/pp.108.125542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Muszynski M. G., Danilevskaya O. N. (2011). The FT-Like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23 942–960. 10.1105/tpc.110.081406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T. A., Muslin E. H., Dorweiler J. E. (2008). A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 227 1377–1388. 10.1007/s00425-008-0709-1 [DOI] [PubMed] [Google Scholar]
- Mulki M. A., von Korff M. (2016). CONSTANS controls floral repression by up-regulating VERNALIZATION2 (VRN-H2) in Barley. Plant Physiol. 170 325–337. 10.1104/pp.15.01350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R. L., Klein R. R., Morishige D. T., Brady J. A., Rooney W. L., Miller F. R., et al. (2011). Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. U.S.A. 108 16469–16474. 10.1073/pnas.1106212108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y., Kisaka M., Fuse T., Yano M., Ogihara Y. (2003). Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J. 36 82–93. 10.1046/j.1365-313X.2003.01859.x [DOI] [PubMed] [Google Scholar]
- Nemoto Y., Nonoue Y., Yano M., Izawa T. (2016). Hd1,a CONSTANS orthlog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 86 221–233. 10.1111/tpj.13168 [DOI] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 847–857. 10.1016/0092-8674(95)90288-0 [DOI] [PubMed] [Google Scholar]
- Ream T. S., Woods D. P., Amasino R. M. (2012). The molecular basis of vernalization in different plant groups. Cold Spring Harb. Symp. Quant. Biol. 77 105–115. 10.1101/sqb.2013.77.014449 [DOI] [PubMed] [Google Scholar]
- Sarid-Krebs L., Panigrahi K. C. S., Fornara F., Takahashi Y., Hayama R., Jang S., et al. (2015). Phosphorylation of CONSTANS and its COP1-dependent degradation during photoperiodic flowering of Arabidopsis. Plant J. 84 451–463. 10.1111/tpj.13022 [DOI] [PubMed] [Google Scholar]
- Sawa M., Nusinow D. A., Kay S. A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318 261–265. 10.1126/science.1146994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R., Gómez-Ariza J., Brambilla V., Fornara F. (2014). Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann. Bot. 114 1445–1458. 10.1093/aob/mcu032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. H., Estrada D. A., Johnson R. S., Kim S. K., Lee S. Y., MacCoss M. J., et al. (2014). Distinct roles of FKF1, gigantea, and zeitlupe proteins in the regulation of constans stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. U.S.A. 111 17672–17677. 10.1073/pnas.1415375111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. H., Shim J. S., Kinmonth-Schultz H. A., Imaizumi T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66 441–464. 10.1146/annurev-arplant-043014-115555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. H., Smith R. W., To B. J., Millar A. J., Imaizumi T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336 1045–1049. 10.1126/science.1219644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B. (2006). HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol. 140 1397–1405. 10.1104/pp.105.073486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A., Beales J., Faure S., Dunford R. P., Laurie D. A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310 1031–1034. 10.1126/science.1117619 [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 1003–1006. 10.1126/science.1091761 [DOI] [PubMed] [Google Scholar]
- Wang H., Pan J., Li Y., Lou D., Hu Y., Yu D. (2016). The DELLA-CONSTANS transcription factor cascade integrates gibberellic acid and photoperiod signaling to regulate flowering. Plant Physiol. 172 479–488. 10.1104/pp.16.00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolabu T. W., Tadege M. (2016). Photoperiod response and floral transition in sorghum. Plant Signal. Behav. 11:e1261232 10.1080/15592324.2016.1261232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolabu T. W., Zhang F., Niu L., Kalve S., Bhatnagar-Mathur P., Muszynski M. G., et al. (2016). Three FLOWERING LOCUS T-like genes function as potential florigens and mediate photoperiod response in sorghum. New Phytol. 210 946–959. 10.1111/nph.13834 [DOI] [PubMed] [Google Scholar]
- Wong A. Y. M., Colasanti J. (2007). Maize floral regulator protein INDETERMINATE1 is localized to developing leaves and is not altered by light or the sink/source transition. J. Exp. Bot. 58 403–414. 10.1093/jxb/erl206 [DOI] [PubMed] [Google Scholar]
- Yan L., Fu D., Li C., Blechl A., Tranquilli G., Bonafede M., et al. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U.S.A. 103 19581–19586. 10.1073/pnas.0607142103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Blechl A., Tranquilli G., Ramakrishna W., SanMiguel P., et al. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303 1640–1644. 10.1126/science.1094305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Murphy R. L., Morishige D. T., Klein P. E., Rooney W. L., Mullet J. E. (2014a). Sorghum phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and SbGHD7, repressors of SbEHD1, SbCN8 and SbCN12. PLoS ONE 9:e105352 10.1371/journal.pone.0105352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Weers B. D., Morishige D. T., Mullet J. E. (2014b). CONSTANS is a photoperiod regulated activator of flowering in sorghum. BMC Plant Biol. 14:148 10.1186/1471-2229-14-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M., Katayose Y., Ashikari M., Yamanouchi U., Monna L., Fuse T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12 2473–2484. 10.1105/tpc.12.12.2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Liu J., Zhao T., Gomez A., Li C., Yu C., et al. (2016). A drought-inducible transcription factor delays reproductive timing in rice. Plant Physiol. 171 334–343. 10.1104/pp.16.01691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Chen H., Ren D., Tang H., Qiu R., Feng J., et al. (2015). Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/ RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). New Phytol. 208 936–948. 10.1111/nph.13503 [DOI] [PubMed] [Google Scholar]


