Abstract
Biofilms play a crucial role in the pathogenicity of Staphylococcus epidermidis, while little is known about whether the essential YycFG two-component signal transduction system (TCS) is involved in biofilm formation. We used antisense RNA (asRNA) to silence the yycFG TCS in order to study its regulatory functions in S. epidermidis. Strain 1457 expressing asRNAyycF exhibited a significant delay (~4–5 h) in entry to log phase, which was partially complemented by overexpressing ssaA. The expression of asRNAyycF and asRNAyycG resulted in a 68 and 50% decrease in biofilm formation at 6 h, respectively, while they had no significant inhibitory effect on 12 h biofilm formation. The expression of asRNAyycF led to a ~5-fold increase in polysaccharide intercellular adhesion (PIA) production, but it did not affect the expression of accumulation-associated protein (Aap) or the release of extracellular DNA. Consistently, quantitative real-time PCR showed that silencing yycF resulted in an increased transcription of biofilm-related genes, including icaA, arlR, sarA, sarX, and sbp. An in silico search of the YycF regulon for the conserved YycF recognition pattern and a modified motif in S. epidermidis, along with additional gel shift and DNase I footprinting assays, showed that arlR, sarA, sarX, and icaA are directly regulated by YycF. Our data suggests that YycFG modulates S. epidermidis biofilm formation in an ica-dependent manner.
Keywords: Staphylococcus epidermidis, biofilm, YycFG, two-component signal transduction system, antisense RNA
Introduction
The coagulase-negative Staphylococcus epidermidis, an opportunistic pathogen, has become the most common source of infections associated with indwelling medical devices (Simon et al., 2005; Gordon et al., 2006). The pathogenicity of S. epidermidis is mainly attributed to biofilm formation, which involves multiple matrix components and regulators (Fey and Olson, 2010; Flemming et al., 2016).
Biofilm formation is a phased process that includes initial adhesion, proliferation, and detachment (Otto, 2009; Mack et al., 2013). The matrix of the three-dimensional structured staphylococcal biofilm is mainly composed of extracellular polymeric substances (EPS), which includes polysaccharide intercellular adhesion (PIA) (O'Gara, 2007) and extracellular DNA (eDNA) (Qin et al., 2007). PIA, the major component of staphylococcal biofilm, is synthesized by proteins encoded by icaADBC, which are negatively regulated by the transcriptional repressor IcaR (Jefferson et al., 2003). In addition, eDNA released from cells plays an important role in biofilm formation. We previously reported that AtlE is one of the major murein hydrolases that mediate eDNA release (Qin et al., 2007). Extracellular proteins including accumulation-associated protein (Aap), biofilm-associated protein (Bap/Bhp), and extracellular matrix-binding protein (Embp) also participate in biofilm formation, especially in ica-independent pathways (Vandecasteele et al., 2003; Lasa and Penades, 2006; Christner et al., 2012). In S. epidermidis, Aap is a major component of protein-dependent biofilm formation (Conrady et al., 2008), and a recent study revealed that a Small basic protein (Sbp) serves as an essential scaffold between the B domains of two Aap molecules during cell aggregation (Decker et al., 2015).
Regulation of staphylococcal biofilm formation involves multiple transcriptional regulators that form a complex network. In addition to transcriptional regulators such as IcaR, SarA (Tormo et al., 2005), SarX (Rowe et al., 2011), and SarZ (Wang et al., 2008) that function as single regulators (Knobloch et al., 2001), TCSs play important roles in S. epidermidis biofilm formation. Our previous work showed that depletion of the ArlRS (Wu et al., 2012) or SrrAB (Wu et al., 2015) TCS impaired biofilm formation in S. epidermidis. (Howell et al., 2003; Botella et al., 2011; Fukushima et al., 2011; Delaune et al., 2012; Dhiman et al., 2014). It was noticed that the YycFG (also known as WalKR and VicKR) TCS is involved in biofilm formation in S. aureus (Dubrac et al., 2007), but its regulatory role in S. epidermidis biofilm formation has not previously been confirmed.
The highly conserved YycFG TCS is of noticeable importance because of its essentiality and regulatory roles in metabolism, cell division, cell wall synthesis, autolysis, and virulence (Howell et al., 2003; Botella et al., 2011; Fukushima et al., 2011; Delaune et al., 2012; Dhiman et al., 2014). YycFG TCSs among Gram-positive bacterial species share similar sequences and operon structures that contain 3–6 genes (Dubrac et al., 2008). In S. epidermidis, the yyc operon includes four genes, yycF, yycG, yycH, and yycI. YycG, the histidine kinase (HK), is anchored to the cell membrane, where it senses and transduces environmental signals. YycH and YycI are involved in the activation of YycG in B. subtilis and S. aureus (Santelli et al., 2007; Szurmant et al., 2008; Cameron et al., 2016). YycF is a typical OmpR family protein, serving as a response regulator (RR). When YycF is activated via phosphorylation by YycG, it binds the promoters of target genes based on a conserved pattern that is based on the recognition of a motif sequence [5′-TGT(A/T)A(A/T/C)-N5-TGT(A/T)A(A/T/C)-3′] by the helix-turn-helix domain of YycF. The pattern was first identified in B. subtilis (Howell et al., 2003) and later in S. aureus (Dubrac and Msadek, 2004) and Streptococcus mutans (Senadheera et al., 2005). YycFG TCS regulates biological processes by controlling the expression of various regulons among species (Dubrac and Msadek, 2008; Winkler and Hoch, 2008).
Since YycFG TCS is an essential element, creating a gene deletion mutant using homologous recombination was not possible. Therefore, antisense RNA (asRNA), which is able to silence target gene expression by stimulating sequence-specific mRNA degradation (Wagner and Simons, 1994; Bai et al., 2012), was used to investigate the functions of YycFG. We investigated the impacts of asRNAyycF or asRNAyycG on bacterial growth and biofilm formation in S. epidermidis. In addition, by carrying out an in silico search for the conserved and modified motif patterns in the YycF regulon of S. epidermidis, we identified YycFG target genes involved in energy production, translation, and cell wall metabolism, as well as biofilm formation. The role of YycF in the regulation of biofilm-related genes was confirmed. This study extends our understanding of the regulatory mechanisms involved in S. epidermidis biofilm formation, in which YycFG TCS plays an important role.
Materials and methods
Bacterial strains and culturing mediums
In this study, the S. epidermidis strain 1457 (SE1457) (Mack et al., 1992) was used as a wild type strain for gene silencing. Escherichia coli DC10B (Monk et al., 2012) was used to isolate shuttle plasmids for directly transforming S. epidermidis via electroporation (Lofblom et al., 2007). Lysogeny broth (LB) medium (1% tryptone, 0.5% yeast extract, and 0.5% NaCl) was used for the cultivation of E. coli. Basic medium (BM) (1% tryptone, 0.5% yeast extract, 0.5% glucose, 0.1% K2HPO4, and 0.5% NaCl) and tryptic soy broth (TSB) (Oxoid, UK) were used for S. epidermidis cultivation and biofilm formation. B2 media (1% tryptone, 2.5% yeast extract, 0.5% glucose, 0.1% K2HPO4, and 2.5% NaCl) was use for bacteria recovery after electroporation. Antibiotics were added at the following concentrations: chloramphenicol at 10 μg/ml and ampicillin at 100 μg/ml. Anhydrotetracycline (ATc, Sigma, USA) was used at a concentration of 250 ng/ml for induction of asRNA.
Construction of asRNA plasmids
To construct an asRNA expression vector, the paired termini 7 (PT7) segment (that can form a hairpin structure) was amplified using PCR with the primers prdtmn-f and prdtmn-r from plasmid pHN678 (Nakashima et al., 2006), digested with KpnI and SacI (Thermo Scientific, USA), and inserted into the ATc-inducible shuttle plasmid pALC2073 (Bateman et al., 2001). The resulting vector was named pMX6 (Figure 1A).
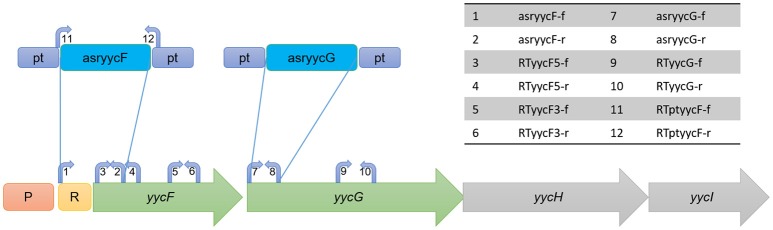
Figure 1.
Representation of structure of yyc operon and design of asRNA as well as primers. The locations of the primers are indicated at the approximate locations on each gene. P, promoter of yyc operon; R, ribosome binding site.
An expression plasmid of yycF asRNA (named pMXyycF) was constructed by first amplifying the predicted Shine-Dalgarno (SD) sequence plus ~100 nt downstream of the start codon of yycF and then inserting the fragment in the reverse direction between EagI and BglII sites downstream of the ATc-inducible promoter in pMX6. As for generating expression plasmids of yycG and ssaA asRNA, a ~120 nt sequence downstream from the start codon of each gene was amplified. All the asRNA expression plasmids were checked using DNA sequencing.
An overexpression plasmid of ssaA (named pMXyycF-ssaA) was constructed. The strong sarA P1 promoter of S. aureus strain 2395 was fused with the coding sequence of S. epidermidis ssaA1 using PCR, and inserted at the SacI site of pMXyycF.
The primers used in this study are listed in Table 1.
Table 1.
Primers used in this study.
| Primer | Sequence (5′ → 3′) |
|---|---|
| FOR CONSTRUCTION OF asRNA PLASMID | |
| prdtmn-f | GGGGTACCTGGAATTGTGAGCGGATAAC |
| prdtmn-r | CGAGCTCCATGGGGTCAGGTGGGAC |
| asryycF-f | GAAGATCTGAACGGAAGAACTACACTTT |
| asryycF-r | AGCGGCCGCGCAATATACGTCGTAACC |
| asryycG-f | GAAGATCTATGAAGTGGCTTAAACAACTAC |
| asryycG-r | AGCGGCCGATTCCTTTTCTAAACTATTCG |
| asrssaA-f | GAAGATCTATGAAAAAAATCGCTACAGCTAC |
| asrssaA-r | AGCGGCCGGTGTAAGTGTAGCTATATGAATAAGGG |
| FOR OVEREXPRESSION OF ssaA1 (serp1880) | |
| saraP1-f | CGGAGCTCCCTGATATTTTTGACTAAACCAAATGCT |
| saraP1-r | GATTTTTTTCATTGTTAAATATCCTCCTAAAAAGATGCATCTTGCTCGATAC |
| ssaA1-f | GTATCGAGCAAGATGCATCTTTTTAGGAGGATATTTAACAATGAAAAAAATC |
| ssaA1-r | CGGAGCTCCAGGTATTTGACTATGTTATACAAGTTTTATATG |
| FOR PROTEIN EXPRESSION | |
| REyycF-f | CGGATCCATGGCTAGAAAAGTTGTTGT |
| REyycF-r | CCCTCGAGCTAATCATGTTGTTGGAGGA |
| FOR DETECTION OF eDNA | |
| gyrA-f | CCTTATGAAACTCGGAGATGG |
| gyrA-r | TCAGTAGTAGTAGATTGTTGCG |
| leuA-f | GTGAACGGTATTGGTGAAAGAG |
| leuA-r | GTGGTCCTTCCTTACATATAAAGC |
| lysA-f | TGACAATGGGAGGTACAAGC |
| lysA-r | TGGTCTTCATCGTAAACAATCG |
| serp0306-f | ATGCCACATCCACGAAAGA |
| serp0306-r | TGTAACTGACAATGCCCAATC |
| FOR DETECTION OF GENE EXPRESSION BY REAL-TIME PCR | |
| RTgyrB-f | GCTAATGCCTCGTCAATAC |
| RTgyrB-r | TATGGTGCTGGACAGATAC |
| RTyycF5-f | GTTGTTGTAGTTGACGATGA |
| RTyycF5-r | ACGACATACTTCCATACCAT |
| RTyycF3-f | CCTGGTAGAGATGGTATGG |
| RTyycF3-r | AATAATGACGGCGTAAGTTC |
| RTptyycF-f | AATTTCAGGAGGAATTAACCAT |
| RTptyycF-r | TCATCGTCAACTACAACAAC |
| RTyycG-f | GCTTGGTGTCCTTAACTTAG |
| RTyycG-r | CGACTTGTTGTTGTTCTGT |
| RTyycH-f | ATTAGCCAACCATCCTGAT |
| RTyycH-r | TTGCCTTGTCGTCCATAT |
| RTyycI-f | ACAGCGATGGATGATATACA |
| RTyycI-r | CAGTTAGGTTGGAGTAATTGAA |
| RTrspA-f | GGCGATGTTATTGATGGTAA |
| RTrspA-r | CTACTGACACAACTTCTTCTG |
| RTssaA-f | GGAGTCCAGATCGTGTAA |
| RTssaA-r | TTGTGATTGCGTGTTGTT |
| RTaap-f | CGAGGAATTACAATCATCACA |
| RTaap-r | CGTAGTTGGCGGTATATCTA |
| RTicaR-f | GGAGCACTAGATAATTGAACAT |
| RTicaR-r | CATTGACGGACTTTACCAG |
| RTicaA-f | TCAAGCGAAGTCAATCTCT |
| RTicaA-r | AACAGCAATATCCTCAGTAATC |
| RTarlR-f | CTGTTGATATAGAGAATGATGGAA |
| RTarlR-r | TGATGATAATTGGAGTAGTTGTT |
| RTsarA-f | GTAATGAACACGATGAAAGAACT |
| RTsarA-r | GCTTCTGTGATACGGTTGT |
| RTsarX-f | CTGGCTACAGGAGAGTTAG |
| RTsarX-r | CAACATCTTCAAATAAAGCATCA |
| RTsbp-f | AAGATAGGCGAATCAATGAAG |
| RTsbp-r | GCGTGTAACATTTCCTCTT |
| FOR PROMOTER AMPLIFICATION FOR EMSA | |
| ProqoxB-f | TTACAAACCCCGTCATATCT |
| ProqoxB-r | TGCCAAATAATAGAAGCAAAG |
| PropitR-f | TGATTTTATCGCTCATCATTTT |
| PropitR-r | AAGTGGTTGAGGAACTGATT |
| PromurE-f | CACGATTTTAGTATTGTCTTC |
| PromurE-r | CACGTTCTATTAGATAGTGAT |
| PropstS-f | CATCTATTCATTCAATCAAGT |
| PropstS-r | ATATGTATTTCTTACAGTTCTC |
| ProsceD-f | ATGATAGGAATCATTACGGTT |
| ProsceD-r | CGTGCAGGTTACACTGAAA |
| ProssaA1-f | CACATTGCTATGTTAATTATTAT |
| ProssaA1-r | CGACAAGCCATACTCTAAC |
| ProrpsA-f | AAACTAAATTGACCATCACTT |
| ProrpsA-r | TATGCCTCCTTATACACTAC |
| ProarlR-f | ATAATGCTAGAGGGACTTTTT |
| ProarlR-r | CACCTCACGCTACATCTTA |
| Proica-f | TTCTAAAATCTCCCCCTTATTCAATTTTCT |
| Proica-r | TTTTTCACCTACCTTTCGTTAGTTAGGTTGT |
| Proaap-f | ATTATTCAAATGCTTGTAGT |
| Proaap-r | ATTATACCTCCCATGTATTT |
| prosarA-f | GAATATAGCAAATGCTACAT |
| ProsarA-R | TAATGAAACCTCCCTATTTATAT |
| ProsarX-f | GGTCAATTCTCACCAAGAG |
| ProsarX-r | CTTTCCCTCAGTCTTTTATG |
Detection of bacterial growth and biofilm formation
Bacterial strains were cultured at 37°C and growth curves were determined by measuring the optical density (OD) values at 600 nm, at 1 h intervals.
The biofilm formation of the bacteria was detected using a semi-quantitative microtiter plate assay with 96-well polystyrene plates (Corning, USA). Overnight cultures were diluted 1:200 in 200 μl TSB and cultured at 37°C for 6 or 12 h, with or without the addition of 250 ng/ml ATc. The planktonic culture was removed for detection of cell density at OD600. The biofilms on the bottom of wells were washed with phosphate-buffered saline (PBS), and fixed with 99% methanol. The fixed biofilms were stained with 2% (wt/vol) crystal violet, resolved with acetic acid (30%), and detected at 570 nm with a spectrophotometer (Beckman Coulter DTX880, Beckman Instruments, USA).
Detection of bacterial primary attachment
The primary attachment of bacterial strains was detected according to a protocol developed by Heilmann et al. (1996). Briefly, an S. epidermidis strain with the plasmid pMX6 or pMXyycF was cultured with or without the induction of 250 ng/ml ATc at 37°C until the OD600 reached 0.6. After centrifugation, bacterial cells were resuspended in BM and the volume was adjusted until OD600 reached 0.1. The samples were inoculated into a 6-well polystyrene petri dish (BD, USA) and incubated at 30°C for 30 min. After being washed with PBS five times, the attached bacterial cells were observed using a light microscope (Nikon, Japan) and photographed.
Detection of extracellular polymeric substances (EPS)
For detecting PIA production, S. epidermidis strains were cultivated to the exponential phase until the OD600 reached 0.6. The bacteria were centrifuged for 5 min at 4°C and resuspended in 0.5 M Ethylenediaminetetraacetic acid (EDTA, pH 8.0). After boiling for 5 min, the samples were centrifuged (13,000 g), and the supernatant was treated with 20 mg/ml proteinase K (Merck, Germany) for 3 h at 37°C. Proteinase K was then inactivated by raising the temperature to 100°C for 5 min. Five-fold serial dilutions of the cell extracts were transferred to a nitrocellulose membrane (Merck, Darmstadt, Germany) with a 96-well dot blot device (Biometra GmbH, Germany). The air-dried nitrocellulose membrane was blocked with 5% (wt/vol) skim milk, incubated with 3.2 mg/ml horseradish peroxidase (HRP)-labeled wheat germ agglutinin (WGA-HRP conjugate, Lectinotest Laboratory, Ukraine) for 1 h (Al Laham et al., 2007), and then 4-chloride-1-naphthol (Sigma, USA) was added as the substrate for the chromogenic detection of HRP activity.
For eDNA detection, the bacteria were cultured until the OD600 reached 0.6. After centrifugation, the supernatant was filtered using 0.22-μm syringe filters to remove the bacterial cell debris. The eDNA was extracted using phenol-chloroform-isoamyl that was diluted 1:10 in Tris-EDTA buffer, and it was quantified using quantitative polymerase chain reaction (qPCR) with SYBR Premix Ex-Taq (Takara, Japan) and primers for gyrA, leuA, and lysA (listed in Table 1).
Detection of Aap by western blotting
The presence of Aap was assessed by western blotting with a monoclonal antibody, MAB18B6, that we developed to target S. epidermidis Aap (Hu et al., 2011). The bacteria were cultivated until the OD600 reached 0.6, collected by centrifugation, and treated with 0.1 mg/ml lysostaphin (Sigma, USA). The samples were centrifuged (20,000 g) at 4°C for 30 min. The supernatants were separated using 7% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and electro-transferred onto a 0.45-μm polyvinylidene fluoride (PVDF) membrane (Millipore, USA). The membrane was treated with 10 ng/ml MAB18B6 and HRP-conjugated goat anti-mouse IgG (Santa Cruz, USA) as a secondary antibody, and then visualized using an enhanced chemiluminescence western blotting system (Thermo Scientific).
Detection of bacterial autolysis
Triton X-100 was used to induce the autolysis of the S. epidermidis strains, which was detected by following a protocol developed by Dubrac et al. (2007). In brief, the bacteria were cultured until the OD600 reached 0.6, and centrifuged (13,000 g) for 5 min at 4°C. The pellets were washed with distilled water, resuspended in 50 mM Tris-HCl (pH 7.2) with 0.05% (vol/vol) Triton X-100, and incubated at 37°C with shaking for 4 h. The bacterial cell autolysis was determined by measuring the OD600 absorbance every 30 min.
Observation of bacteria morphology using transmission electron microscopy
The S. epidermidis strains were cultured in TSB medium until the OD600 reached 0.6, centrifuged and resuspended in 2.5% glutaraldehyde in Dulbecco's PBS. After fixation in osmium tetroxide, the samples were dehydrated with increasing alcohol concentrations and transferred onto an electron microscope grid covered with a carbon-coated Formvar film. The bacteria were stained with 1% (w/v) uranyl acetate-lead acetate and examined with an S-520 electron microscope (Hitachi, Japan).
Purification of recombinant YycF
For the gel shift and DNase I footprinting assays, an YycF recombinant expression plasmid (named pETMG-yycF) was constructed. The yycF gene was amplified by PCR with the primers REyycF-f and REyycF-r (Table 1), and inserted into a pETMG plasmid at BamHI and XhoI sites (Hu et al., 2011). After transformation into BL21 (DE3), the bacteria were cultured in LB medium at 37°C for 4 h and incubated for another 4 h at 30°C with 1 mM isopropyl-1-thio-β-D-galactopyranoside. The cells were disrupted using sonication in lysis buffer (50 mM Tris-Cl and 300 mM NaCl, pH 8.0), and they were then centrifuged at 15,000 g for 30 min. The recombinant polyhistidine-tagged GB1-YycF protein in the supernatants was purified using affinity chromatography with an Ni-nitrilotriacetic acid column (Qiagen, Germany) and further purified using Superdex 75 gel filtration columns (GE Healthcare, USA).
Gel shift assay
The gel shift assay was carried out using a protocol developed by Hellman and Fried (2007). The upstream regions of genes were amplified by PCR with the primers listed in Table 1, while a fragment located in the yycF coding region was used as the negative control. Different concentrations of r-YycF were mixed with 20 nM DNA fragments in a binding buffer (10 mM Tris, 25 mM KCl, 1 mM EDTA, 2.5 mM MgCl2, and 5% vol/vol glycerol, pH 8.0). After incubation at 37°C for 30 min, the samples were loaded onto 10% polyacrylamide native gels (with a 29:1 acrylamide: bisacrylamide ratio) for electrophoresis in a Tris-acetate-EDTA buffer at 10 V/cm for about 1.5 h. The gels were stained with Gel-Red (Biotium, USA) and detected using a Gel Doc EZ system (Bio-Rad, USA).
DNase I footprinting assay
The DNase I footprinting assay was performed by following a protocol developed by Wang et al. (2012). Briefly, the promoters of the arlR, ica, and pitR genes were cloned into a pUC18B-T vector (Shanghai Biotechnology Corporation, China), and the plasmids were used as the template for preparation of fluorescein amidite (FAM)-labeled probes with the primers M13F and M13R (both FAM-labeled). The FAM-labeled probes were purified using Wizard SV Gel and a PCR Clean-Up System (Promega, Southampton, UK), and quantified using NanoDrop 2000C (Thermo Scientific). For the DNase I footprinting assay, 200 ng probes were incubated with different amounts of r-YycF in 40 μl of binding buffer at 30°C for 30 min. Subsequently, 10 μl DNase I (0.01 unit) (Promega, UK) and 100 nmol CaCl2 were added, incubated for 1 min at 25°C, and stopped using 140 μl DNase I stopping solution (200 mM unbuffered sodium acetate, 30 mM EDTA, and 0.15% SDS). The DNA samples extracted with phenol/chloroform and precipitated with ethanol, and the pellets were dissolved in 35 μl MilliQ water. The samples were loaded onto a device to carry out capillary electrophoresis, and data were collected using the GeneScan-500 LIZ dye Size Standard (Applied Biosystems, USA).
RNA extraction and quantitative real-time (qRT)-PCR
For RNA extraction, the S. epidermidis strains were cultured at 37°C with shaking. For the detection of asRNA expression and the gene silencing efficiency, the bacteria were cultured for 6 or 12 h. For detecting the expression levels of biofilm-related genes, the bacteria were cultivated until the OD600 reached 0.6. The cell pellets were washed with ice-cold normal saline and then homogenized using 0.1-mm Zirconia-silica beads in a Mini-BeadBeater (Biospec, Bartlesville, USA) at a speed of 3,600 rpm for 40 s following cooling on ice for 1 min. This homogenization and cooling cycle was repeated five times, then the samples were centrifuged at 15,000 rpm and the bacterial RNA in the supernatant was purified using an RNeasy Mini kit (Qiagen) and quantified using an ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, USA). RNA samples that had a 260/280 ratio between 2.0 and 2.2 were reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad) following the manufacturer's protocol. The mRNA levels were quantified by using qRT-PCR with SYBR green PCR reagents (Takara, Japan) and the primers listed in Table 1, with the housekeeping gene gyrB being used as an endogenous control. The amplification efficiency of all primer pairs were determined according to the standard curve with four magnitude of templates. The specificity of primer pairs was determined with melting curve. All the qRT-PCR experiments were carried out in triplicate and the relative gene expression data were analyzed using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Sequence analysis
A comparison of the protein sequence of YycF among various bacterial species was carried out using Clustal X 2.0 (http://www.clustal.org). The YycF regulon in the S. epidermidis RP62A genome was predicted using a bioinformatics analysis with a custom-made script on Perl and an online relational database (http://genolist.pasteur.fr).
Motif-based sequence analysis was performed online using Motif Discovery from the MEME (Multiple Em for Motif Elicitation) suite (http://meme-suite.org/).
Results
Silencing of YycFG TCS by asRNA
Since YycFG TCS is essential, we used asRNA technology to individually silence the expression of yycF and yycG. First, we constructed an ATc-inducible asRNA-expressing plasmid, pMX6, which contained a paired termini sequence for the formation of a hairpin structure that mediated the asRNA transcriptional termination (Supplementary Figure 1). For silencing yycF and yycG, the asRNA plasmids pMXyycF and pMXyycG were constructed. The asRNAyycF was designed to target a sequence from the Shine-Dalgarno site to the 103rd nt of the yycF coding sequence, while the segment of asRNAyycG was designed to target a sequence from the start codon to the 129th nt of yycG to avoid interference with yycF expression (Figure 1).
After transformation of the pMXyycF plasmid into SE1457, the expression levels of asRNAyycF and yycF mRNA were quantified using qRT-PCR. In the bacteria that were incubated with 250 ng/ml ATc for 6 h, the transcription of asRNAyycF was ~20-fold higher than that without ATc induction, but it decreased by ~50% at 12 h (Figure 2A). Correspondingly, the level of yycF mRNA when ATc was present was reduced by over 99% at 6 h compared to when ATc was not present, and it was ~75% lower at 12 h (Figure 2B). The similar time dependent trend was observed by detection of asRNAyycG and yycG mRNA during induction of asRNAyycG (Supplementary Figure 2).
Figure 2.
Detection of asRNAyycF and its effects on yycF mRNA. SE1457 and its transformants with different plasmids were grown in BM for 6 or 12 h, with or without addition of ATc to 250 ng/ml. Total RNA was extracted and expression levels of asRNA (A) and mRNAs (B) were examined by qRT-PCR.
Effect of asRNAyycF or asRNAyycG on bacterial growth and cell morphology
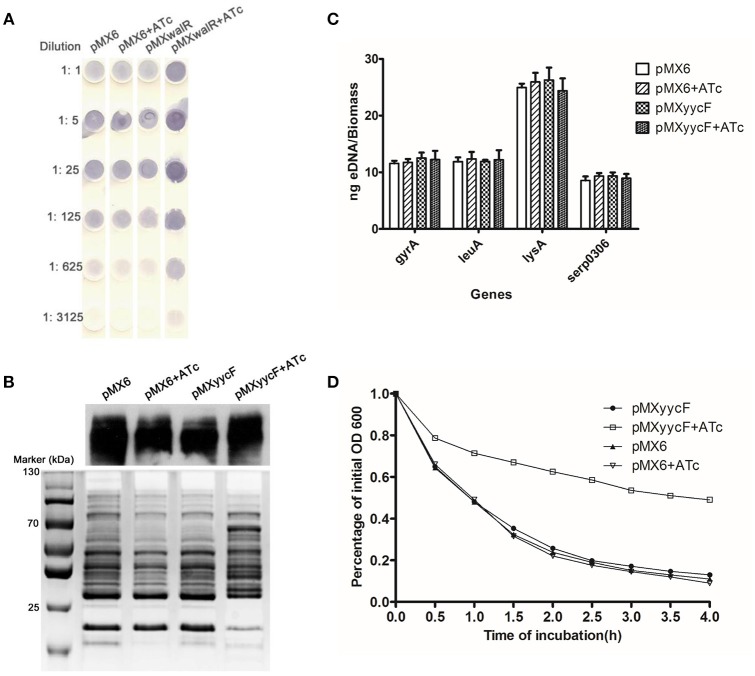
The individual effects of asRNAyycF and asRNAyycG on bacterial growth were investigated. Under induction with 250 ng/ml ATc, the entrance into the log phase of SE1457 expressing asRNAyycF was significantly delayed (~4–5 h) compared with non-ATc induction. However, the effect of asRNAyycG on bacterial growth was weaker than the effect of asRNAyycF (Figure 3A). Meanwhile, the addition of ATc did not affect the growth of the control strain, SE1457 containing the pMX6 vector.
Figure 3.
Effects of asRNA on growth and morphology. (A) Expression of yycF or yycG asRNA on bacterial growth. S. epidermidis 1457 with plasmids were grown in BM medium at 37°C, and growth was monitored every hour by measuring the turbidity of the cultures at OD600. (B) Effects of overexpression of ssaA on growth inhibition by asRNAyycF. The initial inoculation of each strain was 1:1,000 to optimize the effect of the asRNA. Similar results were obtained in three independent experiments. ATc, anhydrotetracycline (added to a final concentration of 250 ng/ml). (C) Transmission electron microscopy of effects from silencing of yycF and YycF target genes. SE 1457 strains were incubated in BM containing 10 μg/ml CM and 250 ng/ml ATc until an OD 600 of 0.6–0.8 was reached. From cells in pMX6, pMXyycF, and pMXssaA (first column on the left), cells with abnormal appearances from pMXyycF and pMXssaA were shown in the right 3 columns, while none was found in pMX6, which is the plasmid control. The white patches inside of some normal cells were probably due to insufficient penetration of EP612 resin into cell walls of gram positive bacteria.
In S. aureus, the indispensability of the YycFG TCS can be circumvented by overexpressing two autolysin genes, ssaA or lytM, found in its regulon, which has been shown to restore normal cell division under YycFG starvation (Delaune et al., 2011). By carrying out a genomic search for ssaA and lytM, it was found that the SE1457 genome possesses two ssaA genes, ssaA1 (serp1880) and ssaA2 (serp2136), but it does not contain lytM. The two ssaA genes share the same coding sequence with a different promoter sequence.
Two more plasmids, pMXssaA and pMXyycF-ssaA, were constructed and transformed into SE1457 to study the role of SsaA in S. epidermidis. The pMXssaA plasmid was used to express asRNAssaA and silence the two ssaA genes, while the pMXyycF-ssaA plasmid was used to constitutively overexpress SsaA and bring about the inducible expression of asRNAyycF. There was a significant decrease in bacterial growth due to asRNAssaA, while the overexpression of SsaA partially prevented the growth inhibition effects of asRNAyycF. The pMXyycF-ssaA transformant entered log phase 2 h earlier than the SE1457::pMXyycF strain with ATc induction (Figure 3B).
The effect of yycF or ssaA silencing on bacterial cell morphology was observed with a transmission electron microscope. Silencing of either yycF or ssaA led to abnormal morphology, including cell enlargement, distorted shapes, and misplaced division septa. The disruption of the cell envelope resulted in cell death, with leaking of cytosol into the medium (Figure 3C). Meanwhile, none of the morphological changes were observed in the pMX6 transformant with ATc induction.
Effect of asRNAyycF or asRNAyycG on biofilm formation
We investigated the individual effects of asRNAyycF and asRNAyycG on biofilm formation in vitro. After ATc (250 ng/ml) induction for 6 h, asRNAyycF and asRNAyycG resulted in a decrease in biofilm formation of 68 and 50%, respectively, compared with the control without ATc (Figure 4A). Meanwhile, the inhibition of bacterial growth by asRNAyycF (76%) was also greater than that caused by asRNAyycG (51%) (Figure 4B). At 12 h, no significant inhibition of biofilm formation by asRNAyycF or asRNAyycG was observed but the growth inhibition by asRNA was still remarkable (Figure 4B).
Figure 4.
Effects of yycF and asRNAyycG on biofilm formation. The strains were resuspended in TSB (1:200 dilution) and incubated in 96-well plates for 6 or 12 h. (A) Biofilm formation and (B) growth were measured by detecting the absorbance at 570 and 600 nm, respectively. (C) Primary attachment of the S. epidermidis 1457 strains to polystyrene surfaces. Overnight cultures grown to an OD600 of 0.6 were adjusted to an OD600 of 0.1 in PBS and inoculated into 6-well plates (2 ml/well). After 2 h at 37°C, the primary attachment at the bottom of the plates was observed under microscopy using a 40-fold objective lens.
Since yycF silencing showed more significant impacts on growth and biofilm formation than yycG silencing, we focused on asRNAyycF in the subsequent analyses. Also, to overcome the interference to biofilm formation caused by growth inhibition, we detected the effects of asRNAyycF on primary attachment, biofilm matrix production and biofilm gene expression with cultures at the same density (OD600 = 0.6), based on the consideration that cell density probably is a more important indicator of growth state than incubation time. By normalization of cell numbers, the effect of asRNAyycF on primary attachment to polymer surfaces was assessed. After incubation in a 6-well polystyrene petri dish at 30°C for 30 min, SE1457 expressing asRNAyycF formed more and much larger cell clusters compared with the control strains with or without ATc. The density of the attached bacterial cells in the areas without cell clusters was similar among all the strains (Figure 4C).
The influence of asRNAyycF on EPS, including PIA, Aap, and eDNA, was explored. The effects of asRNAyycF on PIA production was detected using a semi-quantitative dot-blot assay with a WGA-HRP conjugate. After the addition of 250 ng/ml ATc, the silencing of yycF in SE1457 resulted in a ~5-fold increase in PIA production compared with when there was no ATc induction and with the control plasmid (Figure 5A). The production of a major biofilm associated protein Aap that forms intracellular ligands was also detected. After silencing of yycF, no obvious change in Aap expression (Western blot, Figure 5B) was observed. No significant impact of asRNAyycF on release of eDNA, an important factor that stabilizes the second structure of biofilms, was observed either (by qPCR, Figure 5C). The results combined indicate that YycFG mainly affects production of PIA.
Figure 5.
Effects of asRNAyycF on EPS production, Aap expression, and autolysis. (A) Detection of PIA synthesis by S. epidermidis after silencing yycF. Serial dilutions of the PIA extractions detected using spot assays. The data represent one of three independent experiments. (B) Detection of Aap synthesis after silencing yycF. Aap expression was detected using western blotting with MAb25C11 (1 ng/mL). After separation of the proteins using 7% SDS-PAGE, the gel sections carrying high-molecular-weight proteins (>130 kDa) were excised for the western blot assay, and the remaining gel was stained using Coomassie brilliant blue as the endogenous control. (C) Extracellular DNA quantification. Extracellular DNA was isolated from the supernatants of each culture. Q-PCRs of four chromosomal loci were performed for eDNA quantification. (D) Detection of effects of asRNAyycF on autolysis. Cultures grown to an OD600 of 0.6 were re-adjusted to an OD600 of 1. Autolysis induced by Triton X-100 at 30°C in the presence of 0.1% Triton X-100. The lysis percentage was calculated as follows: [(ODt0 − ODtx /ODt0) × 100%]. Experiments were performed three times independently.
The strain with induced asRNAyycF exhibited a high level of resistance to Triton X-100-induced autolysis (~50% lysis was observed), while the OD600 of the control strains with or without ATc dropped to ~20% (Figure 5D).
We also used qRT-PCR to assess the effects of asRNAyycF on the transcriptional levels of genes involved in biofilm formation (Table 2). The expression of icaA, sbp, arlR, sarA, and sarX clearly increased during asRNAyycF induction (especially icaA) by more than 20-fold, while no significant change in the expression of other genes was observed (Figure 6).
Table 2.
Biofilm associated genes and detection.
| Gene | qRT-PCR (ATc+/ATc–) | EMSA | Biological functions/pathways | References |
|---|---|---|---|---|
| icaA | ↑ | + | PIA synthesis | Rohde et al., 2001 |
| icaR | NC | + | Regulation of icaADBC | Jefferson et al., 2004 |
| sdrG | NC | ND | Cell surface clumping factor | Hartford et al., 2001 |
| clpP | NC | ND | Protease involved in biofilm formation | Michel et al., 2006) |
| spx | NC | − | Regulation of icaADBC | Wang et al., 2010 |
| ygs | NC | − | Stress responder, regulator of PIA synthesis | Wang et al., 2011 |
| aap | NC | + | Surface protein for accumulation | Conrady et al., 2008 |
| embp | NC | − | Surface protein for intracellular adhesion | Christner et al., 2012 |
| sbp | ↑ | ND | Scaffold protein for PIA and Aap | Decker et al., 2015 |
| agrA | NC | − | Quorum sensing regulator | Lauderdale et al., 2009 |
| sigB | NC | − | Regulation of PIA synthesis | Pane-Farre et al., 2006 |
| sarA | ↑ | + | Regulation of PIA synthesis | Tormo et al., 2005 |
| sarX | ↑ | + | Regulation of PIA synthesis | Rowe et al., 2011 |
| sarZ | NC | − | Regulator of biofilm formation and virulence | Wang et al., 2008 |
+, binding by YycF; −, no binding by YycF; NC, no change (cutoff = 2); ND, not detected.
Figure 6.

Effects of asRNAyycF on the expression of biofilm-related genes. The expression of the genes in Table 2 was detected using qRT-PCR, with gyrB as an internal control. The experiment was carried out in triplicate and the expression ratios of the biofilm-related genes are represented as means with standard deviations.
Genome-based prediction of the S. epidermidis YycG/YycF regulon
In silico searches based on conserved motif pattern have been widely used among low-GC Gram positive bacterial species to provide information about the potential target genes that are directly regulated by YycF (Howell et al., 2003; Senadheera et al., 2012; Dhiman et al., 2014). To assess whether the pattern can be applied in S. epidermidis, the amino acid sequence of S. epidermidis YycF was compared with those of B. subtilis str. 168, S. aureus RN4220, and S. mutans UA159 using the Clustal X 2.0 program. The helix-turn-helix domain (180–217th amino acids) of the SE1457 YycF shares 100% identity with that of S. aureus, and it has a difference of one amino acid with the corresponding domain of B. subtilis (Supplementary Figure 3).
Based on the conserved pattern [5′-TGT(A/T)A(A/T/C)-N5-TGT(A/T)A(A/T/C)-3′], an in silico search of the S. epidermidis RP62A genome was carried out to predict the target genes of YycF, especially the biofilm-related target genes. We identified 28 potential binding sites of YycF, which were located in the promoter region (<400 bp upstream of each start codon) of various genes/operons (Table 3). The genes of the putative YycF regulon were divided into several groups, including metabolism (four genes/operons), protein production (three), phosphor transport (three), cell wall synthesis, and lysis (nine), as well as biofilm formation (two). The largest number of genes/operons (including both ssaA genes) were found to belong to the cell wall metabolism group, while only two genes (arlR and sbp) were found to belong to the biofilm formation group.
Table 3.
Genes potentially regulated by YycFG in S. epidermidis.
| Predicted YycF binding sites* | Gene | Gene symbol | Product | Biological functions/pathways | References |
|---|---|---|---|---|---|
| −54 TGTAAATATTGTGTAAT → | SERP_RS03290 | qoxB | Quinol oxidase, subunit II | Terminal oxidases involved in energy production | Winstedt and von Wachenfeldt, 2000 |
| SERP_RS03285 | qoxA | Quinol oxidase, subunit I | |||
| SERP_RS03280 | qoxC | Quinol oxidase, subunit III | |||
| SERP_RS03275 | qoxD | Quinol oxidase, subunit IV | |||
| −159 TGTAAATATATTGTTAT → | SERP_RS02575 | sufB | FeS assembly protein | Iron-sulfur cluster biosynthesis | Fontecave and Ollagnier-de-Choudens, 2008 |
| −175 TGTAAATCTAATGTTAA → | SERP_RS02470 | pgm | Phosphoglycerate mutase | Glucose metabolism | Nukui et al., 2007 |
| −317 TGTAATTATTATGTTAA → | SERP_RS06410 | Chorismate mutase | Shikimate pathway | Kloosterman et al., 2003 | |
| −76 TGTAAAAAAACTGTTAA → | SERP_RS11260 | Acetyl-CoA biotin carboxyl carrier | Fatty acid metabolism | ||
| −105 TGTTAAACTTTTGTTAT → | SERP_RS05210 | rpsA | 30s ribosomal protein S1 | Protein production | Agalarov et al., 2006 |
| −47 GTAACAAAGCATTTACA← | SERP_RS08380 | tRNA-Leu-5 | tRNAs | Protein production | |
| SERP_RS08375 | tRNA-Gly-5 | ||||
| −96 TTAACAAAAAATTTACA← | SERP_RS04810 | pstS | Phosphate ABC transporters | Phosphate transport | Chan and Torriani, 1996 |
| SERP_RS04805 | pstC | ||||
| SERP_RS04800 | pstA | ||||
| SERP_RS04795 | pstB | ||||
| SERP_RS04790 | phoU | ||||
| −229 TGTTAAGAATTTGTAAA → | SERP_RS01725 | pitR | Transcription regulator | Zinc and phosphate transport | Mechler et al., 2015 |
| −121 TTTACACAATTTTTACA← | SERP_RS01730 | pitA | Transporter of Zn2+ in complex with Pi | ||
| −43 TTAACAGTTTTTTTACA← | SERP_RS11255 | phnD | Phosphonate transport system regulatory protein | Phosphonate transport | Gebhard and Cook, 2008 |
| SERP_RS11250 | |||||
| SERP_RS11245 | |||||
| SERP_RS11240 | |||||
| −161 TTAACACTACCTTAACA← | SERP_RS03100 | murE | UDP-N-acetylmuramoy-L-alanyl-D-glutamate-L-lysine ligase | Cell wall synthesis | Gardete et al., 2004; Zaher and Green, 2011 |
| SERP_RS03105 | |||||
| SERP_RS03120 | prfC | peptide-chain-release factor 3 | Protein production accuracy | ||
| −226 ATTACAATAAGATTACA← | SERP_RS02025 | ltaS | Glycerol phosphate lipoteichoic acid synthase | Lipoteichoic acid synthesis | Grundling and Schneewind, 2007b |
| −243 ATTACAAATGTATTACA← | SERP_RS02885 | oatA | O-acetyltransferase | Teichoic acid modification | Bera et al., 2005 |
| −344 TGTTAAGGTAGTGTTAA → | SERP_RS03095 | ltaA | Diacylglycerol glucosyltransferase | Cell wall synthesis | Grundling and Schneewind, 2007a |
| SERP_RS03090 | ypfP | Lipoteichoic synthesis | |||
| −112 TGTAATTGTAGTGTAAA → | SERP_RS08520 | sceD | Lytic Transglycosylases | Cell wall hydrolase | Stapleton et al., 2007 |
| −146 GTTACAAGATAATAACA← | SERP_RS01735 | LysM domain protein | Cell wall hydrolase | ||
| −77 ATTACATCAATATAACA← | SERP_RS02225 | LysM domain autolysin | Cell wall hydrolase | ||
| −269 TTTACATTCATGTAACA← | SERP_RS09425 | ssaA1 | CHAP domain autolysin | Cell wall hydrolase | Lang et al., 2000 |
| −140 ATTACAAATCAATAACA← | |||||
| −252 ATTACAAGAATATAACA← | SERP_RS10600 | ssaA2 | CHAP domain autolysin | Cell wall hydrolase | |
| −146 ATTACAAAATGATAACA← | |||||
| −63 ATAACAAATCATTTACA← | SERP_RS04480 | arlR | DNA-binding response regulator ArlR | Two-component system | Wu et al., 2014 |
| SERP_RS04475 | arlS | Histidine kinase | |||
| −287 TGTTACATGAATGTAAA → | SERP_RS09420 | Schuller et al., 2012 | |||
| SERP_RS09415 | sarY | AraC family transcriptional regulator | Transcription regulation | ||
| SERP_RS09410 | |||||
| −69 TGTAATATTATTGTTAT → | SERP_RS01525 | sbp | Scaffold protein | Biofilm formation | Decker et al., 2015 |
| −248 TGTTATTATCTTGTAAC → | SERP_RS01740 | Conserved hypothetical protein | |||
| −63 TGTAACATTAATGTAAT → | SERP_RS11965 | Conserved hypothetical protein | |||
| −179 TGTAAAGTTGATGTTAT → | SERP_RS01900 | Conserved hypothetical protein |
The numbers indicate the distance between the start codon of putative YycF target gene and the putative binding sites in the promoter region.
Binding of YycF to the predicted target genes
To verify the predicted YycF target genes in S. epidermidis, a gel shift assay was performed. The recombinant YycF (r-YycF) was able to bind promoters of all eight selected genes: the r-YycF resulted in a mobility shift of the 188, 180, 167, 103, 159, 249, 232, and 150-bp fragments upstream of murE, qoxB, pstS, sceD, arlR, pitR, ssaA1, and rpsA, respectively, in a concentration-dependent manner. The negative control, a 125-bp DNA fragment of the yycF coding sequence, did not form a complex with r-YycF under the same conditions (Figure 7). The r-YycF protected region in the promoters of arlR, qoxB, and pitR was detected using the DNase I footprinting assay. A 60-nt protected region located upstream of the translational start site of arlR (−107 to −48 bp) was identified (Figure 8A). The 52-nt protected region in the promoter region of qoxB was located at −103 to −52 bp (Supplementary Figure 4A). Two separate protected regions (−282 to −196 bp, 87 nt; −152 to −114 bp, 39 nt) were identified in the pitR promoter region (Supplementary Figure 4B). The sequences of the protected region of the arlR, qoxB, and pitR promoters fit the consensus motif (Table 3).
Figure 7.
Binding of r-YycF to the YycF regulon and biofilm-related genes. Electrophoretic mobility shift assay using purified r-YycF with promoter regions of YycF target genes. DNA segments were amplified from the promoters of predicted YycF target genes and biofilm-related genes. For ica, all 164 nt between the coding sequence of icaA and icaR were used. For sarX, the promoter of serp3220, which is located upstream of sarX in the same operon, was amplified. A segment of the yycF coding sequence was used as a negative control.
Figure 8.
Identification of the YycF-protected cis-elements in the promoter regions of (A) arlR and (B) ica using DNase I footprinting assays. The regions protected by r-YycF are marked with frames of dashed lines. The DNA sequences of the protected regions are provided, and the sequences that are consistent with the previously reported YycF binding pattern are underlined. The arrows on these lines indicate the direction of the corresponding genes of the promoters.
To investigate whether the YycG/F TCS could bind the promoters of other genes in a motif-independent way, we further assessed the binding of r-YycF to promoters of the biofilm-related genes listed in Table 2. The r-YycF led to a mobility shift of the 83, 271, 322, and 361-bp fragments upstream of ica, aap, sarA, and sarX, respectively, in a concentration-dependent manner (Figure 7). The DNase I footprinting assay was used to identify an 83-bp r-YycF protected area in the region between icaR and icaA (Figure 8B).
Based on the high similarity of the YycF helix-turn-helix domain between B. subtilis and S. epidermidis, we gathered together previously reported atypical promoter sequences that YycF binds to in B. subtilis as well as those discovered in S. epidermidis in this study (Supplementary Table 1). A motif-based sequence analysis was performed to generate a new pattern with relaxed restrictions (Figure 9). By performing an in silico search of the S. epidermidis genome for the new pattern, more than 300 potential YycF binding sites were identified (Supplementary Table 2). The corresponding genes included genes that are involved in metabolism (fmtC, tdk, gpmA, and glmU), translation (rbfA, rpsF, rrsD, and prfB), and biofilm formation (atlE, rsbU, ebh, and sarR).
Figure 9.

A graphic depiction of an extended motif for YycF to recognize target genes in S. epidermidis. The conservative property of each base is indicated with heights of each letter.
Discussion
In the current study, we investigated the regulatory role of YycFG TCS in S. epidermidis biofilm formation by means of in vitro experiments and in silico techniques. With the results combined, we showed that YycFG TCS is a key regulator for S. epidermidis viability and negatively regulates S. epidermidis biofilm formation in an ica-dependent way.
Since YycFG TCS is essential for bacteria survival, we could not create knockout mutants of the S. epidermis genes. To study the regulatory functions of YycFG TCS in S. aureus, several methods have been used including site mutations (Fabret and Hoch, 1998), truncation of YycF (Gutu et al., 2010), and replacement of the promoter in the genome with an inducible promoter (Fukuchi et al., 2000; Dubrac and Msadek, 2004). In the present study, an asRNA silencing technique was applied. Our results showed that asRNA was highly effective for silencing genes, so it was able to reduce the yycF mRNA level by more than 99% at 6 h (Figure 2B). However, the efficacy decreases over time, as was reported by other researches (Ji et al., 2004; Stary et al., 2010). By detection of asRNA and mRNA of yycF or yycG, we showed that while the asRNA levels decreased from 6 to 12 h, the levels of target mRNAs increased (Figure 2). The result was consistent with the change of inhibitory effects of asRNAyycF or asRNAyycG on bacterial growth and biofilm formation (Figures 3A, 4A,B). However, the reason for the decrease of asRNA expression with time remains to be investigated in further study. In addition, the asRNAs acted specifically against the target mRNAs. The asRNAyycF targeting the 5′ end of yycF (yycF-5) led to a decrease of over 99% in the yycF-5 mRNA level (Figure 1), but it barely affected yycF-3 or the three following genes (yycG, yycH, and yycI) (Supplementary Figure 5). The base pairing characteristic confers the asRNA technology the advantage to specifically silence the target gene without affecting the other genes in the same operon, and thus prevents the polar effects to the other genes in the same operon brought by promoter replacement (Fan et al., 2001).
As cell density affects biofilm formation, we explored the effects of asRNAyycF when the OD600 of each bacterial culture reached 0.6. Our results suggested that YycF upregulates cell aggregation (Figure 4C), PIA formation (Figure 5A), bacterial autolysis (Figure 5D), and the expression of biofilm-related genes (arlRS, icaA, sbp, sarA, and sarX, as shown in Figure 6). No significant change in Aap production was observed. However, Aap-mediated cell aggregation may be enhanced by an elevated expression of Sbp, which increases the bridging of Aap B domains between bacterial cells (Decker et al., 2015). Although asRNAyycF repressed autolysis, it did not affect eDNA release. The abnormal morphology of bacterial cells after asRNAyycF silencing may be attributable to the repression of SsaA, and ssaA silenced by asRNA had a similar impact on the bacteria (Figure 3C).
After prediction and verification of the YycF regulon with the conserved motif, we assessed the effects of asRNAyycF on transcriptional levels of all YycF regulon genes by qRT-PCR, The expression of most of the regulon genes was not affected, except that rpsA was up regulated for about 4-folds and ssaA down regulated for about 5-folds (Supplementary Figure 6). Meanwhile, based on the detection of asRNA effects on biofilm formation, we found that many other biofilm genes that did not appear in the predicted YycF regulon showed significant transcriptional change. The expression of icaA, sbp, sarA, and sarX (especially icaA) increased considerably (by more than 20-fold) after asRNAyycF induction, while these genes were not identified from the YycF regulon (Figure 6). Furthermore, YycF is able to bind to the promoter regions of ica, aap, sarA, and sarX (Figures 8, 9B), proving that the recognition and regulation of target genes by YycF in S. epidermidis is not limited to the previously reported conserved pattern.
It has previously been reported that ArlRS positively regulates S. epidermidis biofilm formation in an ica-dependent manner (Wu et al., 2012). The mRNA level of icaA in ΔarlS was lower than that in the wild type strain (SE1457). When asRNAyycF was introduced into the arlS knockout strain, induction of asRNAyycF significantly increased the expression of icaA (Supplementary Figure 7), indicating that the YycFG TCS modulates biofilm formation mainly via the ica-dependent pathway, by regulation of icaA with other transcriptional regulators including ArlRS.
YycFG has been reported to regulate target genes in a conserved-motif-independent way in multiple bacterial species. In B. subtilis, several cell wall metabolism-associated genes (yvcE1, yoeB2, etc.) without the consensus recognition sequence in their promoter regions have been found to be directly controlled by YycF (Bisicchia et al., 2007). In a more recent study of B. subtilis, YycF-bound DNA was obtained using chromatin immunoprecipitation (ChIP), including many sites (ggaA, lytE, dacA, etc.) that do not fit the pattern either (Salzberg et al., 2012). More exceptions have been reported in S. mutans (gtfB, smaA1, lysM, atlA, etc., Senadheera et al., 2005; Stipp et al., 2013). To overcome the limitation of the consensus pattern, it can be modified based on in vitro experiments (Salzberg et al., 2012; Ayala et al., 2014). We performed the similar strategy to generate a new motif for prediction of more YycF regulon genes. The extended pattern provides more insights into YycF regulation in S. epidermidis. However, while the relaxation of certain sites allows so many more genes to be putative regulon genes, the binding ability of YycF to promoters of these genes requires further verification with gel shift assay.
S. epidermidis YycG histidine kinase inhibitors as well as S. aureus WalK/WalR inhibitors have potent antibacterial activities. In the present study, silencing yycF had more effect on the biological phenotype than silencing yycG, which indicates that the effect of asRNA that targets yycF may have some differences compared to the effect of asRNA that targets YycG. This indicates that YycG may function in other pathways through crosstalk with other TCSs, which requires further study.
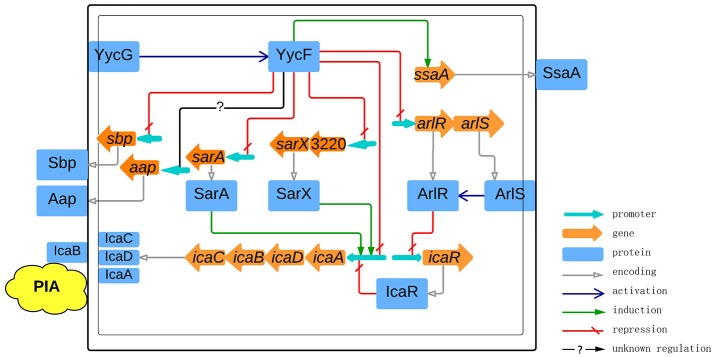
In summary, by using a conserved and a modified motif pattern to search for the S. epidermidis YycF regulon, we found several YycFG target genes involved in energy production, translation, and cell wall metabolism, as well as biofilm formation. Based on confirmation of the regulation of biofilm-related genes by YycF, a model was established for the role of YycFG TCS in S. epidermidis biofilm formation (Figure 10). In addition to previous discoveries, we showed that YycF not only regulates biofilm-associated regulators such as arlRS, sarA, and sarX, but that it also binds to the promoters of icaADBC to directly modulate PIA production. The interaction of YycFG with other TCSs in S. epidermidis (by inter-regulation and crosstalk) warrants further investigation.
Figure 10.
Schematic representation of the roles of YycFG in PIA-dependent biofilm formation in S. epidermidis. The cytoplasmic cell membrane is represented by a thin border, in which YycG, ArlR, IcaA, IcaD, and icaC are embedded. The cell wall is represented by bold border, in which IcaB and PIA are embedded. The phosphorylation of YycF and ArlR by YycG and ArlS is signified by blue arrows. The genes are shown as brown arrows with an associated promoter (light blue arrows). The gray arrows represent the translation into protein. The induction and repression of transcription are represented by green arrows and red broken lines, respectively, while the question mark indicates that the regulation of transcription is indirect or that it occurs via an unclear mechanism.
Author contributions
DQ, YZ, FG, RB, and TX designed the work and revised the manuscript; TX, YW, and ZL completed all the experiments; TX and YW performed the statistically analysis and made the figures; TX, YW, and DQ wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Timothy J. Foster, Trinity College, Dublin, Ireland, for providing the strain E. coli DC10B, and A. L. Cheung, Dartmouth Medical School of Hanover, for providing the plasmid pALC2073. We appreciate the help from Liam Good, The Royal Veterinary College, United Kingdom, who provided the plasmid pHN678 and suggestions about asRNA. This study was supported by the National Natural Science Foundation of China (81671982, 81271791, and 81571955) and the National Science and the Technology Major Project of China (2012ZX09301002-005 and 2012ZX10003008-010).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00724/full#supplementary-material
Representation of pMX6. The relevant features are indicated with boxes (rep, replicon in S. aureus; cole1, replicon in E. coli), genes with arrows (tetR, TET repressor; bla, ampicillin resistant gene; cat, chloramphenicol resistance gene), promoters with triangles, and paired termini with rectangles.
Detection of asRNAyycG and its effects on yycG mRNA. Total RNA of cultures grown for 6 or 12 h was extracted and expression levels of asRNA (A) and mRNAs (B) were examined by qRT-PCR.
Alignment of amino acid sequences of YycF from S. epidermidis RP62A, S. aureus RN4220, and B. subtilis str. 168. Shading indicates the conserved residues, and the two boxes indicate the DNA-binding winged helix-turn-helix domains. The amino acids at identity positions are marked with black backgrounds and the consensus positions are marked in gray.
DNase I footprinting assay of YycF binding property to promoters of qoxB (A) and pitR (B).
Impacts of asRNAyycF on relative expression of genes in the yyc operon. The yycF coding sequence were divided into two parts yycF-5 and yycF-3, in which specific RT primers were located. The endogenous gene gyrB was used as an internal control. Similar results were obtained from three independent experiments.
Effects of asRNAyycF on expression of regulon genes. The selected genes in the list of YycF target genes were detected by qRT-PCR, using gyrB as an internal control. Genes showing significant changes (cutoff = 2-fold) on expression level were shown. Similar results were obtained from three independent experiments.
Impacts of asRNAyycF on relative expression of icaA in SE1457 and arlRS mutant strain. The endogenous gene gyrB was used as an internal control. Similar results were obtained from three independent experiments.
Genes directly regulated by YycF with atypical promoter sequences.
Predicted regulon genes of YycF based on the new motif in S. epidermidis.
References
- Agalarov S., Kalinichenko A. A., Kommer A. A., Spirin A. S. (2006). Ribosomal protein S1 induces a conformational change of the 30S ribosomal subunit. FEBS Lett. 580, 6797–6799. 10.1016/j.febslet.2006.11.036 [DOI] [PubMed] [Google Scholar]
- Al Laham N., Rohde H., Sander G., Fischer A., Hussain M., Heilmann C., et al. (2007). Augmented expression of polysaccharide intercellular adhesin in a defined Staphylococcus epidermidis mutant with the small-colony-variant phenotype. J. Bacteriol. 189, 4494–4501. 10.1128/JB.00160-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala E., Downey J. S., Mashburn-Warren L., Senadheera D. B., Cvitkovitch D. G., Goodman S. D. (2014). A biochemical characterization of the DNA binding activity of the response regulator VicR from Streptococcus mutans. PLoS ONE 9:e108027. 10.1371/journal.pone.0108027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H., Sang G., You Y., Xue X., Zhou Y., Hou Z., et al. (2012). Targeting RNA polymerase primary sigma as a therapeutic strategy against methicillin-resistant Staphylococcus aureus by antisense peptide nucleic acid. PLoS ONE 7:e29886. 10.1371/journal.pone.0029886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman B. T., Donegan N. P., Jarry T. M., Palma M., Cheung A. L. (2001). Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 69, 7851–7857. 10.1128/IAI.69.12.7851-7857.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera A., Herbert S., Jakob A., Vollmer W., Gotz F. (2005). Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55, 778–787. 10.1111/j.1365-2958.2004.04446.x [DOI] [PubMed] [Google Scholar]
- Bisicchia P., Noone D., Lioliou E., Howell A., Quigley S., Jensen T., et al. (2007). The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol. 65, 180–200. 10.1111/j.1365-2958.2007.05782.x [DOI] [PubMed] [Google Scholar]
- Botella E., Hubner S., Hokamp K., Hansen A., Bisicchia P., Noone D., et al. (2011). Cell envelope gene expression in phosphate-limited Bacillus subtilis cells. Microbiology 157, 2470–2484. 10.1099/mic.0.049205-0 [DOI] [PubMed] [Google Scholar]
- Cameron D. R., Jiang J. H., Kostoulias X., Foxwell D. J., Peleg A. Y. (2016). Vancomycin susceptibility in methicillin-resistant Staphylococcus aureus is mediated by YycHI activation of the WalRK essential two-component regulatory system. Sci. Rep. 6:30823. 10.1038/srep30823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F. Y., Torriani A. (1996). PstB protein of the phosphate-specific transport system of Escherichia coli is an ATPase. J. Bacteriol. 178, 3974–3977. 10.1128/jb.178.13.3974-3977.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christner M., Heinze C., Busch M., Franke G., Hentschke M., Bayard Duhring S., et al. (2012). SarA negatively regulates Staphylococcus epidermidis biofilm formation by modulating expression of 1 MDa extracellular matrix binding protein and autolysis-dependent release of eDNA. Mol. Microbiol. 86, 394–410. 10.1111/j.1365-2958.2012.08203.x [DOI] [PubMed] [Google Scholar]
- Conrady D. G., Brescia C. C., Horii K., Weiss A. A., Hassett D. J., Herr A. B. (2008). A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc. Natl. Acad. Sci. U.S.A. 105, 19456–19461. 10.1073/pnas.0807717105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker R., Burdelski C., Zobiak M., Büttner H., Franke G., Christner M., et al. (2015). An 18 kDa scaffold protein is critical for Staphylococcus epidermidis biofilm formation. PLoS Pathog. 11:e1004735. 10.1371/journal.ppat.1004735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaune A., Dubrac S., Blanchet C., Poupel O., Mader U., Hiron A., et al. (2012). The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect. Immun. 80, 3438–3453. 10.1128/IAI.00195-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaune A., Poupel O., Mallet A., Coic Y.-M., Msadek T., Dubrac S. (2011). Peptidoglycan crosslinking relaxation plays an important role in Staphylococcus aureus WalKR-dependent cell viability. PLoS ONE 6:e17054. 10.1371/journal.pone.0017054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman A., Bhatnagar S., Bhatnagar R. (2014). Functional characterization of WalRK: a two-component signal transduction system from Bacillus anthracis. FEBS Open Bio. 4, 65–76. 10.1016/j.fob.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrac S., Msadek T. (2004). Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 186, 1175–1181. 10.1128/JB.186.4.1175-1181.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrac S., Msadek T. (2008). Tearing down the wall: peptidoglycan metabolism and the WalK/WalR (YycG/YycF) essential two-component system. Adv. Exp. Med. Biol. 631, 214–228. 10.1007/978-0-387-78885-2_15 [DOI] [PubMed] [Google Scholar]
- Dubrac S., Bisicchia P., Devine K. M., Msadek T. (2008). A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70, 1307–1322. 10.1111/j.1365-2958.2008.06483.x [DOI] [PubMed] [Google Scholar]
- Dubrac S., Boneca I. G., Poupel O., Msadek T. (2007). New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 189, 8257–8269. 10.1128/JB.00645-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabret C., Hoch J. A. (1998). A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J. Bacteriol. 180, 6375–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F., Lunsford R. D., Sylvester D., Fan J., Celesnik H., Iordanescu S., et al. (2001). Regulated ectopic expression and allelic-replacement mutagenesis as a method for gene essentiality testing in Staphylococcus aureus. Plasmid 46, 71–75. 10.1006/plas.2001.1526 [DOI] [PubMed] [Google Scholar]
- Fey P. D., Olson M. E. (2010). Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 5, 917–933. 10.2217/fmb.10.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., Kjelleberg S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. 10.1038/nrmicro.2016.94 [DOI] [PubMed] [Google Scholar]
- Fontecave M., Ollagnier-de-Choudens S. (2008). Iron-sulfur cluster biosynthesis in bacteria: mechanisms of cluster assembly and transfer. Arch. Biochem. Biophys. 474, 226–237. 10.1016/j.abb.2007.12.014 [DOI] [PubMed] [Google Scholar]
- Fukuchi K., Kasahara Y., Asai K., Kobayashi K., Moriya S., Ogasawara N. (2000). The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146(Pt 7), 1573–1583. 10.1099/00221287-146-7-1573 [DOI] [PubMed] [Google Scholar]
- Fukushima T., Furihata I., Emmins R., Daniel R. A., Hoch J. A., Szurmant H. (2011). A role for the essential YycG sensor histidine kinase in sensing cell division. Mol. Microbiol. 79, 503–522. 10.1111/j.1365-2958.2010.07464.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardete S., Ludovice A. M., Sobral R. G., Filipe S. R., de Lencastre H., Tomasz A. (2004). Role of murE in the expression of -lactam antibiotic resistance in Staphylococcus aureus. J. Bacteriol. 186, 1705–1713. 10.1128/JB.186.6.1705-1713.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard S., Cook G. M. (2008). Differential regulation of high-affinity phosphate transport systems of Mycobacterium smegmatis: identification of PhnF, a repressor of the phnDCE operon. J. Bacteriol. 190, 1335–1343. 10.1128/JB.01764-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon R. J., Quagliarello B., Lowy F. D. (2006). Ventricular assist device-related infections. Lancet Infect. Dis. 6, 426–437. 10.1016/S1473-3099(06)70522-9 [DOI] [PubMed] [Google Scholar]
- Grundling A., Schneewind O. (2007a). Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J. Bacteriol. 189, 2521–2530. 10.1128/JB.01683-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundling A., Schneewind O. (2007b). Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 104, 8478–8483. 10.1073/pnas.0701821104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutu A. D., Wayne K. J., Sham L. T., Winkler M. E. (2010). Kinetic characterization of the WalRKSpn (VicRK) two-component system of Streptococcus pneumoniae: dependence of WalKSpn (VicK) phosphatase activity on its PAS domain. J. Bacteriol. 192, 2346–2358. 10.1128/JB.01690-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartford O., O'Brien L., Schofield K., Wells J., Foster T. J. (2001). The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology 147, 2545–2552. 10.1099/00221287-147-9-2545 [DOI] [PubMed] [Google Scholar]
- Heilmann C., Gerke C., Perdreau-Remington F., Gotz F. (1996). Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64, 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman L. M., Fried M. G. (2007). Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2, 1849–1861. 10.1038/nprot.2007.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A., Dubrac S., Andersen K. K., Noone D., Fert J., Msadek T., et al. (2003). Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49, 1639–1655. 10.1046/j.1365-2958.2003.03661.x [DOI] [PubMed] [Google Scholar]
- Hu J., Xu T., Zhu T., Lou Q., Wang X., Wu Y., et al. (2011). Monoclonal antibodies against accumulation-associated protein affect EPS biosynthesis and enhance bacterial accumulation of Staphylococcus epidermidis. PLoS ONE 6:e20918. 10.1371/journal.pone.0020918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson K. K., Cramton S. E., Gotz F., Pier G. B. (2003). Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol. Microbiol. 48, 889–899. 10.1046/j.1365-2958.2003.03482.x [DOI] [PubMed] [Google Scholar]
- Jefferson K. K., Pier D. B., Goldmann D. A., Pier G. B. (2004). The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186, 2449–2456. 10.1128/JB.186.8.2449-2456.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Yin D., Fox B., Holmes D. J., Payne D., Rosenberg M. (2004). Validation of antibacterial mechanism of action using regulated antisense RNA expression in Staphylococcus aureus. FEMS Microbiol. Lett. 231, 177–184. 10.1016/S0378-1097(03)00931-5 [DOI] [PubMed] [Google Scholar]
- Kloosterman H., Hessels G. I., Vrijbloed J. W., Euverink G. J., Dijkhuizen L. (2003). (De)regulation of key enzyme steps in the shikimate pathway and phenylalanine-specific pathway of the actinomycete Amycolatopsis methanolica. Microbiology 149, 3321–3330. 10.1099/mic.0.26494-0 [DOI] [PubMed] [Google Scholar]
- Knobloch J. K., Bartscht K., Sabottke A., Rohde H., Feucht H. H., Mack D. (2001). Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183, 2624–2633. 10.1128/JB.183.8.2624-2633.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S., Livesley M. A., Lambert P. A., Littler W. A., Elliott T. S. (2000). Identification of a novel antigen from Staphylococcus epidermidis. FEMS Immunol. Med. Microbiol. 29, 213–220. 10.1111/j.1574-695X.2000.tb01525.x [DOI] [PubMed] [Google Scholar]
- Lasa I., Penades J. R. (2006). Bap: a family of surface proteins involved in biofilm formation. Res. Microbiol. 157, 99–107. 10.1016/j.resmic.2005.11.003 [DOI] [PubMed] [Google Scholar]
- Lauderdale K. J., Boles B. R., Cheung A. L., Horswill A. R. (2009). Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 77, 1623–1635. 10.1128/IAI.01036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lofblom J., Kronqvist N., Uhlen M., Stahl S., Wernerus H. (2007). Optimization of electroporation-mediated transformation: Staphylococcus carnosus as model organism. J. Appl. Microbiol. 102, 736–747. 10.1111/j.1365-2672.2006.03127.x [DOI] [PubMed] [Google Scholar]
- Mack D., Davies A. P., Harris L. G., Jeeves R., Pascoe B., Knobloch J. K.-M., et al. (2013). Staphylococcus epidermidis in biomaterial-associated infections, in Biomaterials Associated Infection, eds Moriarty T. F., Zaat S. A. J., Busscher H. J. (New York, NY: Springer; ), 25–56. [Google Scholar]
- Mack D., Siemssen N., Laufs R. (1992). Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60, 2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechler L., Herbig A., Paprotka K., Fraunholz M., Nieselt K., Bertram R. (2015). A novel point mutation promotes growth phase-dependent daptomycin tolerance in Staphylococcus aureus. Antimicrob. Agents Chemother. 59, 5366–5376. 10.1128/AAC.00643-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel A., Agerer F., Hauck C. R., Herrmann M., Ullrich J., Hacker J., et al. (2006). Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188, 5783–5796. 10.1128/JB.00074-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk I. R., Shah I. M., Xu M., Tan M. W., Foster T. J. (2012). Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277-11. 10.1128/mBio.00277-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N., Tamura T., Good L. (2006). Paired termini stabilize antisense RNAs and enhance conditional gene silencing in Escherichia coli. Nucleic Acids Res. 34:e138. 10.1093/nar/gkl697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukui M., Mello L. V., Littlejohn J. E., Setlow B., Setlow P., Kim K., et al. (2007). Structure and molecular mechanism of Bacillus anthracis cofactor-independent phosphoglycerate mutase: a crucial enzyme for spores and growing cells of Bacillus species. Biophys. J. 92, 977–988. 10.1529/biophysj.106.093872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gara J. P. (2007). Ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270, 179–188. 10.1111/j.1574-6968.2007.00688.x [DOI] [PubMed] [Google Scholar]
- Otto M. (2009). Staphylococcus epidermidis–the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7, 555–567. 10.1038/nrmicro2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane-Farre J., Jonas B., Forstner K., Engelmann S., Hecker M. (2006). The sigma(B) regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 296, 237–258. 10.1016/j.ijmm.2005.11.011 [DOI] [PubMed] [Google Scholar]
- Qin Z., Ou Y., Yang L., Zhu Y., Tolker-Nielsen T., Molin S., et al. (2007). Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153, 2083–2092. 10.1099/mic.0.2007/006031-0 [DOI] [PubMed] [Google Scholar]
- Rohde H., Knobloch J. K., Horstkotte M. A., Mack D. (2001). Correlation of Staphylococcus aureus icaADBC genotype and biofilm expression phenotype. J. Clin. Microbiol. 39, 4595–4596. 10.1128/JCM.39.12.4595-4596.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe S. E., Mahon V., Smith S. G., O'Gara J. P. (2011). A novel role for SarX in Staphylococcus epidermidis biofilm regulation. Microbiology 157, 1042–1049. 10.1099/mic.0.046581-0 [DOI] [PubMed] [Google Scholar]
- Salzberg L. I., Powell L., Hokamp K., Botella E., Noone D., Devine K. M. (2012). The WalRK (YycFG) and sigma(I) RsgI regulators cooperate to control CwlO and LytE expression in exponentially growing and stressed Bacillus subtilis cells. Mol. Microbiol. 87, 180–195. 10.1111/mmi.12092 [DOI] [PubMed] [Google Scholar]
- Santelli E., Liddington R. C., Mohan M. A., Hoch J. A., Szurmant H. (2007). The crystal structure of Bacillus subtilis YycI reveals a common fold for two members of an unusual class of sensor histidine kinase regulatory proteins. J. Bacteriol. 189, 3290–3295. 10.1128/JB.01937-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller A., Slater A. W., Norambuena T., Cifuentes J. J., Almonacid L. I., Melo F. (2012). Computer-based annotation of putative AraC/XylS-family transcription factors of known structure but unknown function. J. Biomed. Biotechnol. 2012:103132. 10.1155/2012/103132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senadheera D. B., Cordova M., Ayala E. A., Chavez de Paz L. E., Singh K., Downey J. S., et al. (2012). Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J. Bacteriol. 194, 1307–1316. 10.1128/JB.06071-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senadheera M. D., Guggenheim B., Spatafora G. A., Huang Y. C., Choi J., Hung D. C., et al. (2005). A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J. Bacteriol. 187, 4064–4076. 10.1128/JB.187.12.4064-4076.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D., Fischer S., Grossman A., Downer C., Hota B., Heroux A., et al. (2005). Left ventricular assist device-related infection: treatment and outcome. Clin. Infect. Dis. 40, 1108–1115. 10.1086/428728 [DOI] [PubMed] [Google Scholar]
- Stapleton M. R., Horsburgh M. J., Hayhurst E. J., Wright L., Jonsson I. M., Tarkowski A., et al. (2007). Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J. Bacteriol. 189, 7316–7325. 10.1128/JB.00734-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stary E., Gaupp R., Lechner S., Leibig M., Tichy E., Kolb M., et al. (2010). New architectures for Tet-on and Tet-off regulation in Staphylococcus aureus. Appl. Environ. Microbiol. 76, 680–687. 10.1128/AEM.02416-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp R. N., Boisvert H., Smith D. J., Höfling J. F., Duncan M. J., Mattos-Graner R. O. (2013). CovR and VicRK regulate cell surface biogenesis genes required for biofilm formation in Streptococcus mutans. PLoS ONE 8:e58271. 10.1371/journal.pone.0058271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H., Bu L., Brooks C. L., III., Hoch J. A. (2008). An essential sensor histidine kinase controlled by transmembrane helix interactions with its auxiliary proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 5891–5896. 10.1073/pnas.0800247105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo M. A., Marti M., Valle J., Manna A. C., Cheung A. L., Lasa I., et al. (2005). SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J. Bacteriol. 187, 2348–2356. 10.1128/JB.187.7.2348-2356.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandecasteele S. J., Peetermans W. E., Merckx R., Rijnders B. J., Van Eldere J. (2003). Reliability of the ica, aap and atlE genes in the discrimination between invasive, colonizing and contaminant Staphylococcus epidermidis isolates in the diagnosis of catheter-related infections. Clin. Microbiol. Infect. 9, 114–119. 10.1046/j.1469-0691.2003.00544.x [DOI] [PubMed] [Google Scholar]
- Wagner E. G., Simons R. W. (1994). Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol. 48, 713–742. 10.1146/annurev.mi.48.100194.003433 [DOI] [PubMed] [Google Scholar]
- Wang C., Fan J., Niu C., Wang C., Villaruz A. E., Otto M., et al. (2010). Role of spx in biofilm formation of Staphylococcus epidermidis. FEMS Immunol. Med. Microbiol. 59, 152–160. 10.1111/j.1574-695X.2010.00673.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Li M., Dong D., Bach T. H., Sturdevant D. E., Vuong C., et al. (2008). SarZ is a key regulator of biofilm formation and virulence in Staphylococcus epidermidis. J. Infect. Dis. 197, 1254–1262. 10.1086/586714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Niu C., Sun G., Dong D., Villaruz A. E., Li M., et al. (2011). ygs is a novel gene that influences biofilm formation and the general stress response of Staphylococcus epidermidis. Infect. Immun. 79, 1007–1015. 10.1128/IAI.00916-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cen X. F., Zhao G. P., Wang J. (2012). Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J. Bacteriol. 194, 5237–5244. 10.1128/JB.00989-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M. E., Hoch J. A. (2008). Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J. Bacteriol. 190, 2645–2648. 10.1128/JB.01682-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstedt L., von Wachenfeldt C. (2000). Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa(3) or cytochrome bd, is required for aerobic growth. J. Bacteriol. 182, 6557–6564. 10.1128/JB.182.23.6557-6564.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Liu J., Jiang J., Hu J., Xu T., Wang J., et al. (2014). Role of the two-component regulatory system arlRS in ica operon and aap positive but non-biofilm-forming Staphylococcus epidermidis isolates from hospitalized patients. Microb. Pathog. 76, 89–98. 10.1016/j.micpath.2014.09.013 [DOI] [PubMed] [Google Scholar]
- Wu Y., Wang J., Xu T., Liu J., Yu W., Lou Q., et al. (2012). The two-component signal transduction system ArlRS regulates Staphylococcus epidermidis biofilm formation in an ica dependent manner. PLoS ONE 7:e40041. 10.1371/journal.pone.0040041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wu Y., Zhu T., Han H., Liu H., Xu T., et al. (2015). Staphylococcus epidermidis SrrAB regulates bacterial growth and biofilm formation differently under oxic and microaerobic conditions. J. Bacteriol. 197, 459–476. 10.1128/JB.02231-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher H. S., Green R. (2011). A primary role for release factor 3 in quality control during translation elongation in Escherichia coli. Cell 147, 396–408. 10.1016/j.cell.2011.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representation of pMX6. The relevant features are indicated with boxes (rep, replicon in S. aureus; cole1, replicon in E. coli), genes with arrows (tetR, TET repressor; bla, ampicillin resistant gene; cat, chloramphenicol resistance gene), promoters with triangles, and paired termini with rectangles.
Detection of asRNAyycG and its effects on yycG mRNA. Total RNA of cultures grown for 6 or 12 h was extracted and expression levels of asRNA (A) and mRNAs (B) were examined by qRT-PCR.
Alignment of amino acid sequences of YycF from S. epidermidis RP62A, S. aureus RN4220, and B. subtilis str. 168. Shading indicates the conserved residues, and the two boxes indicate the DNA-binding winged helix-turn-helix domains. The amino acids at identity positions are marked with black backgrounds and the consensus positions are marked in gray.
DNase I footprinting assay of YycF binding property to promoters of qoxB (A) and pitR (B).
Impacts of asRNAyycF on relative expression of genes in the yyc operon. The yycF coding sequence were divided into two parts yycF-5 and yycF-3, in which specific RT primers were located. The endogenous gene gyrB was used as an internal control. Similar results were obtained from three independent experiments.
Effects of asRNAyycF on expression of regulon genes. The selected genes in the list of YycF target genes were detected by qRT-PCR, using gyrB as an internal control. Genes showing significant changes (cutoff = 2-fold) on expression level were shown. Similar results were obtained from three independent experiments.
Impacts of asRNAyycF on relative expression of icaA in SE1457 and arlRS mutant strain. The endogenous gene gyrB was used as an internal control. Similar results were obtained from three independent experiments.
Genes directly regulated by YycF with atypical promoter sequences.
Predicted regulon genes of YycF based on the new motif in S. epidermidis.