Abstract
Background
Severe acute malnutrition (SAM) in infants may present as one of two distinct syndromic forms: non-edematous (marasmus), with severe wasting and no nutritional edema; or edematous (kwashiorkor) with moderately severe wasting. These differences may be related to developmental changes prior to the exposure to SAM and phenotypic changes appear to persist into adulthood with differences between the two groups. We examined whether the different response to SAM and subsequent trajectories may be explained by developmentally-induced epigenetic differences.
Methods
We extracted genomic DNA from muscle biopsies obtained from adult survivors of kwashiorkor (n = 21) or marasmus (n = 23) and compared epigenetic profiles (CpG methylation) between the two groups using the Infinium® 450 K BeadChip array.
Findings
We found significant differences in methylation of CpG sites from 63 genes in skeletal muscle DNA. Gene ontology studies showed significant differential methylation of genes in immune, body composition, metabolic, musculoskeletal growth, neuronal function and cardiovascular pathways, pathways compatible with the differences in the pathophysiology of adult survivors of SAM.
Interpretation
These findings suggest persistent developmental influences on adult physiology in survivors of SAM. Since children who develop marasmus have lower birth weights and after rehabilitation have different intermediary metabolism, these studies provide further support for persistent developmentally-induced phenomena mediated by epigenetic processes affecting both the infant response to acute malnutrition and later life consequences.
Funding
Supported by a Grant from the Bill and Melinda Gates Foundation (Global Health OPP1066846), Grand Challenge “Discover New Ways to Achieve Healthy Growth.”
Evidence Before This Study
Previous research has shown that infants who develop either kwashiorkor or marasmus in response to SAM differ in birth weight and subsequently have different metabolic patterns in both infancy and adulthood.
Added Value of This Study
This study demonstrates epigenetic differences in the skeletal muscle of adult survivors of marasmus versus kwashiorkor and these differences are in genes that may underlie the longer-term consequences.
Implications of All the Available Evidence
These data are compatible with the different clinical responses to SAM arising from developmentally-induced epigenetic changes laid down largely before birth and provide evidence for the predictive adaptive response model operating in human development.
Keywords: Severe acute malnutrition, Epigenetic, Birthweight, Muscle, Jamaica
Highlights
-
•
Infants who develop either kwashiorkor or marasmus differ in birth weight and subsequently have different metabolic patterns in later life.
-
•
We found epigenetic differences in the skeletal muscle of adult survivors of marasmus versus kwashiorkor which may underlie the longer-term consequences.
-
•
Different clinical responses to malnutrition may arise from developmentally-induced epigenetic changes resulting in predictive adaptive responses.
1. Introduction
Severe acute malnutrition (SAM) in infants may present in one of two distinct syndromic forms: non-edematous (marasmus), characterized by severe wasting (weight-for-age < 60%) with no nutritional edema; or edematous (kwashiorkor) characterized by moderately severe wasting (weight-for-age of 60–80%) with edema (Bender, 2005). Globally about 17 million children each year are affected by SAM of which about 10% will die. Sustained dietary inadequacy and superimposed infections during childhood are the main precipitants of SAM. These syndromes arise during periods when the child should be growing rapidly, but falters because s/he is underfed, receiving little or no breastfeeding, inadequate or inappropriate weaning foods or, in the older child, energy- and nutrient-deficient mixed diets. Children who initially are successfully treated for kwashiorkor or marasmus maintain persistent phenotypic and pathophysiological differences into adulthood. As adults survivors of marasmus are shorter, have lower lean body mass, and are more glucose intolerant than their kwashiorkor counterparts (Forrester et al., 2012, Francis-Emmanuel et al., 2014). Both syndromes show persistent behavioral and cognitive deficits throughout childhood and adolescence, which carry through into adulthood when economic underperformance and psychiatric morbidities are added to the disease burden (Galler et al., 2012, Galler et al., 2013).
It is unclear why some severely malnourished children develop the edematous forms of SAM and others the non-edematous. Our previous work has shown that these children have persistent differences in protein, amino acid, lipid and energy metabolism when acutely ill, as well as after nutritional rehabilitation, such that kwashiorkor survivors appear to be nutritionally profligate whereas marasmic survivors appear to be better able to conserve and utilize nutrients (Forrester et al., 2012). These persistent physiological differences previously led us to consider whether there may be factors that pre-date the episode of SAM and influence the response to the severe undernutrition stress. In a study of over 1100 infants who developed either syndrome, we found that infants who develop marasmus had much lower birth weights (Forrester et al., 2012). We proposed that marasmic infants were preconditioned to better respond to severe postnatal malnutrition by the deficient intrauterine environment that had impaired their growth and led them to predict a nutritionally poor postnatal environment. This is in accord with the predictive adaptive response hypothesis (Bateson et al., 2014), particularly given the higher survival of marasmic infants compared to kwashiorkor infants.
To provide further support for early developmental differences that might explain the persistent differences in pathophysiology that are observed into adulthood we have now examined whether there may be epigenetic differences in adult survivors of these two SAM-associated syndromes. Accordingly, we extracted DNA from muscle biopsies obtained from adult survivors of kwashiorkor and marasmus and compared epigenetic profiles between the two groups. We found significant differences in DNA methylation in genes lying within functional pathways that appear relevant to several aspects of the different adult outcomes of these two syndromes.
2. Methods
2.1. Study Design and Participants
21 adult survivors of kwashiorkor and 23 survivors of marasmus consented (written) to muscle biopsies. The protocols were approved by The Faculty of Medical Sciences/University Hospital of the West Indies Ethics Committee and fulfilled the principles of the Declaration of Helsinki. At the time of admission in infancy for management of malnutrition, written consent was given from the next of kin, caretakers, or guardians on behalf of the minors/children participants, for their information to be stored in the hospital database and used for research.
2.2. Study Subjects
Adult subjects were recruited from those who had experienced SAM in childhood and who had been rehabilitated on the metabolic ward of the Tropical Metabolism Research Unit, University of the West Indies, Kingston, Jamaica (Forrester et al., 2012). We reviewed the admission records for 1417 patients admitted with SAM between 1963 and 1994 and abstracted clinical (age, gender, presence of edema), anthropometric (weight and height at admission), and survival data as well as recalled birth weight. The mortality rate was 4.1% during this period. Using the last known address and name of the parent, we traced 729 individuals in the community. Of those 729, 312 were included in the cohort, and a further 163 were unavailable to the study because of refusal (14), migration (53), illness (18), pregnancy (3) or death (75). The remaining 688 cohort members have not been traced. For this study, 44 subjects responded to invitations to participate in detailed protocols including muscle biopsy. Details of phenotypic measurements and methodologies are presented in Appendix S1.
2.3. Statistics for Epigenetic Analyses
Differentially methylated CpGs (dmCpGs) within chromosomal regions of interest were identified by computational analysis of Infinium 450K array data using a threshold significance of 0.05. Standard analysis workflows based on the Bayesian statistical tool known as ‘BATMAN chromosome walking’ predicated a Mann-Whitney comparison between phenotypes. The biological significance of dmCpGs was assessed by standard functional gene and network enrichment analysis. Details of the analysis approach are presented in Supplemental Materials and Methods.
We next defined nutritional insults acting in intrauterine life as ‘Hit 1’ and those in infancy and childhood around the episode of SAM as ‘Hit 2’. We explored the relationships between anthropometry and body composition, immunity and glucose metabolism in adult survivors of SAM with birth weight, which we designated as a measure of Hit 1 and during infancy by the severity of stunting (height/age) and wasting (weight/height) at admission with SAM as a measure of Hit 2. To determine the contribution of prenatal (Hit 1) and postnatal (Hit 2) factors to the methylation variance measured in the kwashiorkor and marasmus groups, multiple linear regression analysis of methylation on the dmCpGs of interest against clinical measures of interest were performed in Matlab 2013b software (Mathworks Inc., MA, USA) using our errors-in-variables approach (Pleasants et al., 2014). The following regression model was used: Yij = μ + αiGender + β1Hit1ij + β2Hit2ij + εij.
3. Results
In a cohort, such as this, there were challenges in achieving full data sets. After accounting for missing data in the various clinical measures, the sample size ranged from 9 to 19 per K or M group.
3.1. Clinical Features of Kwashiorkor or Marasmus Survivors in Adulthood and Infancy
Among the adult survivors of SAM, kwashiorkor survivors had higher body mass indices, and trended to higher bone mineral density (Table 1). Kwashiorkor infants were heavier both at birth and, when they presented with malnutrition, by definition, less wasted than their marasmic counterparts, and they were taller. Individual markers of infections at admission in infancy as well as composite scores were not significantly different between the two clinical syndromes, but kwashiorkor infants mounted a greater leucocyte response. The mean gestational age of survivors of kwashiorkor was longer than that of children who developed marasmus.
Table 1.
Current anthropometry and body composition of adult survivors of marasmus and kwashiorkor, and their clinical features when they presented as infants. Data are mean ± SD.
| Kwashiorkor |
Marasmus |
p | |
|---|---|---|---|
| N = 18 | N = 19 | ||
| Men/women | 8/10 | 9/10 | 1.00 |
| Age, yrs | 29.82 ± 9.03 | 25.02 ± 5.69 | 0.06 |
| BMI, kg/m2 | 25.21 ± 5.04 | 21.95 ± 4.73 | 0.05 |
| Fat mass, kg | 16.65 ± 11.13 | 13.31 ± 11.26 | 0.37 |
| Lean mass, kg | 45.52 ± 11.73 | 42.16 ± 10.78 | 0.37 |
| BMD, g/cm2 | 1.27 ± 0.10 | 1.21 ± 0.10 | 0.06 |
| BMC, g | 2944 ± 547 | 2682 ± 633 | 0.19 |
| Clinical features on admission as infantsa | |||
| Gestational age, wks | 40.0 ± 0.0 | 36·6 ± 5.2 | 0·02 |
| Birth weight, kg | 3.17 ± 0.18 | 2.45 ± 0.20 | 0.02 |
| Age, months | 11.23 ± 1.46 | 11.04 ± 1.42 | 0.92 |
| Weight, kg | 6.61 ± 0.28 | 4.40 ± 0.27 | < 0.001 |
| Height, cm | 65.44 ± 1.73 | 61.31 ± 1.68 | 0.10 |
| Weight-for-height, %b | 85.62 ± 0.54 | 72.10 ± 0.54 | < 0.001c |
| Weight-for-age, %b | 64.07 ± 0.51 | 48.42 ± 0.51 | < 0.001c |
| Height-for-age, %b | 88.59 ± 1.14 | 84.30 ± 1.14 | 0.01 |
| Presence of any clinician-diagnosed infection, % | 94 | 88 | 0.52 |
| Infection severity scored | 1.5 ± 0.9 | 1.8 ± 1.3 | 0.46 |
| White blood cell count, × 109/mL | 11.2 ± 4.7 | 10.7 ± 4.3 | 0.79 |
| Neutrophils, % | 40 ± 11 | 23 ± 12 | 0.004 |
| Lymphocytes, % | 58 ± 13 | 72 ± 13 | 0.01 |
| Hemoglobin, g/dL | 9.1 ± 1.4 | 9.1 ± 1.9 | 0.95 |
Data were missing for 2 marasmus infants and 3 kwashiorkor infants.
These anthropometric measures were calculated by comparison to National Center for Health Statistics (NCHS) standard growth curves.
after log transformation to a normal distribution.
Infections severity score was calculated based on the clinical diagnosis of lower or upper respiratory tract infection, urinary tract infection, otitis media, gastroenteritis (diarrhea), skin infection, sepsis, meningitis or any other infection. One point was given for each infection and the score is the sum of all infections.
3.2. Distinct CpG Methylation Profiles Between Muscles of Adult Survivors of Marasmus or Kwashiorkor
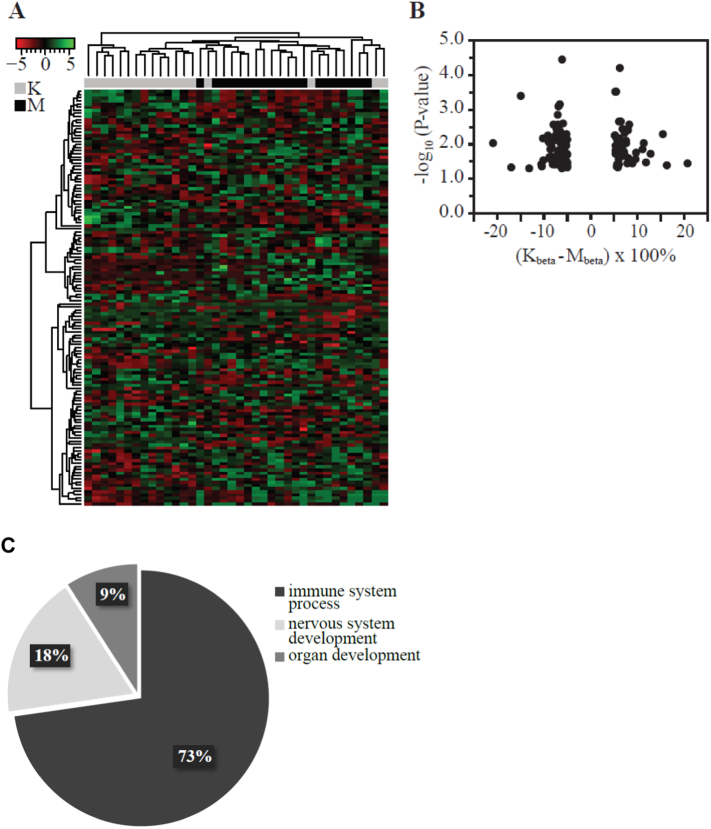
As illustrated in the heatmap (Fig. 1), kwashiorkor subjects form a distinct group compared to marasmus based on their CpG methylation profiles. We found strong representation of the dmCpGs on several genes which were relevant to systems and processes that are known to be affected in SAM and/or to be affected to differing degrees in kwashiorkor compared to marasmus. Such systems included, immunity (including the major histocompatibility complex, class II HLA-DQB1), glucose metabolism (p21 protein (Cdc42/Rac)-activated kinase PAK1, C-reactive protein CRP1 and hexokinase HK2), body size and composition (paired-like homeodomain 2 (PITX2), tenascin TNXB), and central nervous system (tubulin folding cofactor TBCD) (Appendix S1). The distribution of the dmCpGs in genomic regions of interest was assessed by determining the odds ratio (OR) for the occurrence (overrepresentation) of dmCpGs in each gene feature (Appendix S4B). In brief, our data indicate that the most likely region to see variable methylation (OR: 1.45, p = 0·0036) is the gene body (Appendix S4B). Interestingly, emerging data suggest that methylation in gene bodies is a feature of transcribed genes (Jones, 2012). We also found that the dmCpGs were predominantly located on chromosome 6 (16% of total; OR: 2·4; p = 0·0004); 10·5% each was found on chromosomes 7 and 17 (OR: 1·8; p = 0·036 and OR: 1·9; p = 0·021 respectively) (Appendix S4). Full details of dmCpG distribution across gene features are detailed in Appendix S4.
Fig. 1.
Methylation variance of dmCpGs from muscle biopsies of adult survivors of kwashiorkor or marasmus.
(A) Cluster analysis illustrating subject grouping into K (kwashiorkor) or M (marasmus) groups based on the methylation profile of 133 dmCpGs. Distance measure was calculated using Euclidean method. Clustering was done using complete agglomeration method. Red-black-green: Low to high methylation (row z-score). (B) Absolute methylation change between K and M subjects for the 133 dmCpGs (p < 0.05). (C) Biological processes associated with the differentially methylated genes in adult survivors of severe acute malnutrition.
3.3. Molecular Differences Between Kwashiorkor and Marasmus Survivors
To understand the gene patterns attributable to kwashiorkor and marasmus groups, we conducted functional enrichment analysis (Appendix S5–S7). In brief, our strategy was to test independently by using 6 different enrichment tools, and have arbitrarily determined that success in 4 out of 6 represents a potential lead candidate (detailed in Appendix S1: Supporting materials and methods). Significantly overrepresented ‘Gene Ontology (GO)’ terms associated with the differentially methylated genes are detailed in Appendices S5 and S6. Within the ‘Biological Process’ category, we broadly classified 73% of the overrepresented GO terms as processes of the immune system, and the remaining 27% attributed to nervous system and organ developmental processes (Fig. 1C). Results of the significantly enriched pathways in the ‘Kyoto Encyclopedia of Genes and Genomes (KEGG)’ database indicated predominantly immunity related pathways (Appendix S7), and are consistent with results of GO enrichment analysis. More importantly, identification of the following metabolic KEGG pathways - insulin signaling pathway, TGF-beta signaling pathway and Type I diabetes mellitus - implies differences in metabolic functions between kwashiorkor and marasmus subjects. From our list of differentially methylated genes, we identified critical gene nodes associated with immunity (e.g., HLA-DRB1), overall growth (e.g., PITX2) and glucose metabolism (e.g., p21 protein (Cdc42/Rac)-activated kinase PAK1, C-reactive protein CRP1 and hexokinase HK2) as critical gene nodes (Appendix S8).
We hypothesized that methylation differences might reflect developmental effects evoked by malnutrition experienced during developmentally plastic periods both during gestation and postnatally during the first year of life. We characterized epigenotype/phenotype associations that originated during gestation as Hit 1, and those arising from developmental influences in early months after birth encompassing the ages when SAM presented in this cohort (6–18 months) as Hit 2. We used birth weight as a proxy for exposures in utero that may contribute to the differential methylation in kwashiorkor and marasmus survivors (Hit 1). We used anthropometry of children when admitted for SAM as proxy measures for Hit 2 (weight for age, height for age, weight for height) (Fig. 2).
Fig. 2.
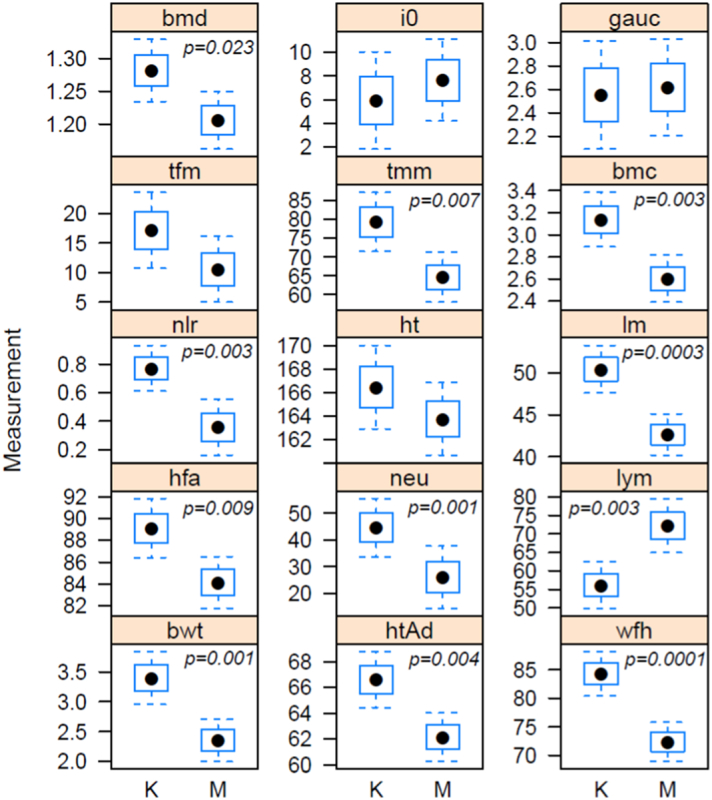
Clinical measures used as Hit 1 and Hit 2 proxies for the assessment of the prenatal and postnatal factors on the methylation variance measured in the kwashiorkor (K) and marasmus (M) groups.
Boxplots illustrate mean (dot), 25th and 75th percentile (box), and minimum and maximum (tails) values of each clinical measure after adjusting for gender (bwt); gender and age (hfa, wfh, ht., htAd, bmc, lm, tfm, tmm); gender and BMI (i0, gauc). Abbreviations (variable; y-axis unit of measurement): bwt: birth weight (kg); wfh: weight for height on admission (kg); hfa: height for age on admission (cm); htAd: height on admission (cm); ht.: adult height (attained height, cm); lm: total lean mass (kg); tfm: total fat mass (kg); tmm: thigh muscle mass (kg); bmc: bone mineral content (kg); bmd: bone mineral density (kg/m2); i0: insulin concentration at baseline (μIU/ml); gauc: glucose area under the curve; neu: neutrophil count (× 108/L) on admission; lymphocyte count (× 108/L) on admission; nlr: neutrophil to lymphocyte ratio on admission.
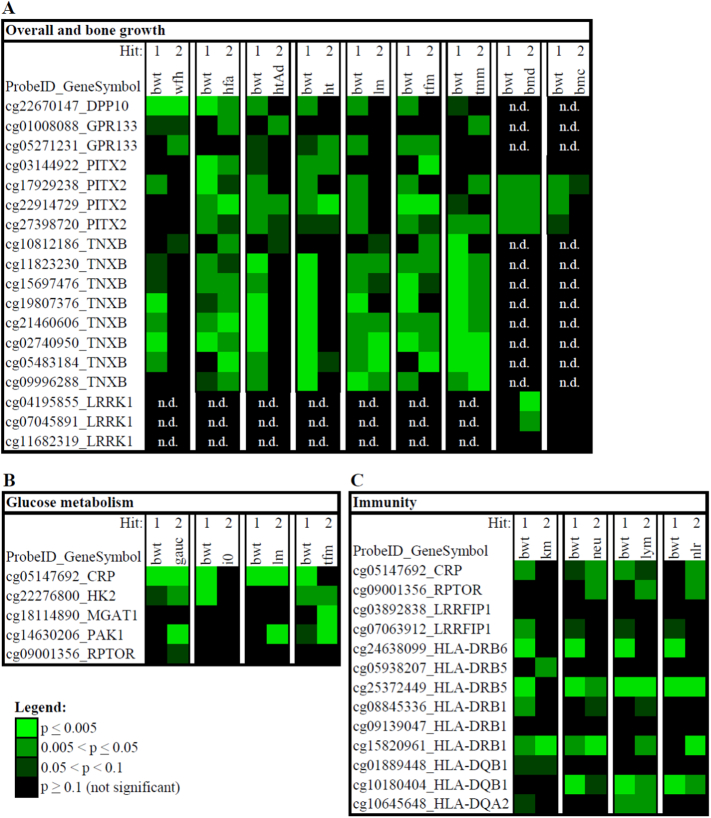
For the regression analysis, genes were broadly categorized by functional relevance, e.g., PITX2 relates to overall organism growth; LRRK1 relates to bone growth; CRP, PAK1 and HK2 are associated with glucose metabolism; CRP, LRRFIP1 and HLA-DRB1 relate to immunity. To assess differences in the severity of the malnutrition stimulus we used data from their hospital admission records when children were admitted with SAM. ‘Weight for height’ captured wasting, a measure of negative energy balance, and ‘height for age’ was an indicator of linear growth failure. We found that both Hit 1 and Hit 2 contribute significantly to different extents to the methylation variance of the various genes implicated in overall growth of kwashiorkor and marasmus subjects for each of the regression models assessed (Fig. 3). Overall, we found the methylation variance in kwashiorkor and marasmus subjects to predominantly associate with birth weight i.e., Hit 1. Similarly, we found the methylation variance of several genes which related to growth (Fig. 3A), glucose metabolism (Fig. 3B) and immunity (Fig. 3C) to be associated with Hit 1 and Hit 2 to different extents. Interestingly, we found that the methylation variance of genes which relate to glucose metabolism to be mostly associated with Hit 1 rather than Hit 2 (Fig. 3B). Complete results of the regression analysis for each model (regression coefficients, standard errors and p-values) are found in Appendices S9–S11. Notably, there were no differences by sex. In summary, our findings show that both Hit 1 and Hit 2 factors contribute to the differential methylation in kwashiorkor and marasmus adult survivors of malnutrition. In addition, our results suggest that prenatal factors predominantly contribute to the methylation variance observed in adulthood.
Fig. 3.
Multiple regression analysis of the dmCpGs associated with (A) overall and bone growth, (B) glucose metabolism and (C) immunity.
The heatmap illustrates the level of significance (by p-values) of Hit 1 and Hit 2 variables for each regression model. Regression model: Methylation Yij = μ + αiGender + β1Hit1ij + β2Hit2ij + εij. Abbreviations: bwt: birth weight; wfh: weight for height; hfa: height for age; htAd: height on admission; ht.: adult height (i.e., attained height); lm: total lean mass; tfm: total fat mass; tmm: thigh muscle mass; bmd: bone mineral density; bmc: bone mineral content; gauc: glucose area under the curve; i0: insulin concentration at baseline; km: subject group (classified as kwashiorkor or marasmus); neu: neutrophil count; lym: lymphocyte count; nlr: neutrophil to lymphocyte ratio. ProbeID_GeneSymbol: Illumina probe identifier (ProbeID) corresponding to a CpG site on the gene of interest.
4. Discussion
Adult survivors of kwashiorkor or marasmus showed differential DNA methylation across 133 CpG loci associated with a number of gene pathways that could be related to persistent phenotypic differences between these two syndromes. Previously, in a larger group of adult survivors of SAM in this population, we described differences between marasmic and kwashiorkor survivors with marasmic survivors having worse glucose tolerance and beta cell function (Francis-Emmanuel et al., 2014). Also, both kwashiorkor and marasmus adult survivors had differences in cardiac structure and function, and blood pressure when compared with controls (Tennant et al., 2014). Others have documented substantial behavioral and cognitive impairment in all SAM survivors throughout childhood, adolescence and young adulthood, but the syndromic association is not clear. In preliminary data on a small group (unpublished) we found that marasmic survivors have more impaired electroencephalographic responses (N2, an index of response conflict, and P3 which indexes stimulus categorization) to repetitive and novel stimuli (Flanker and Oddball) than both kwashiorkor survivors and controls.
At clinical presentation in infancy, the two phenotypes of SAM are remarkably different. Both kwashiorkor and marasmus arise within the same families and coexist within populations. No differences in premorbid dietary intake are reliably found. Not only do they differ in their clinical signs and body composition, but also in their intermediary metabolism during the SAM event and after recovery. Children with kwashiorkor retain significant tissue reserves of protein and fat even at death, as these stores are mobilized inadequately during the wasting process, thus depriving the metabolic machinery of critical amino acids and non-esterified fatty acids to support gluconeogenesis and energy metabolism (Jahoor et al., 2008). On the other hand, children with marasmus are better able to maintain a supply of lipids and amino acids and maintain integrated metabolism by drawing down from protein and lipid stores (Forrester et al., 2012, Badaloo et al., 2006, Jahoor et al., 2005, Reid et al., 2002, Reid et al., 2000, Badaloo et al., 2002). Therefore, while they have greater tissue wasting on presentation, marasmic children have higher survival rates as they are able to sustain integrated metabolism.
These fundamental differences in intermediary metabolism led us to hypothesize that kwashiorkor and marasmus arise out of differential responses to essentially the same nutritional insult based on pre-existing developmentally induced differences in metabolic control (Forrester et al., 2012, Jahoor et al., 2006, Bateson et al., 2004, Gluckman and Hanson, 2004). Children who develop marasmus have much lower birth weights (Forrester et al., 2012) reflecting greater prenatal disadvantage and in particular a more restricted in utero nutritional environment. An evolutionary perspective on developmental plasticity argues that the fetal response to poor nutrition will induce epigenetic and physiological changes that manifest after weaning (Hanson & Gluckman, 2014). Where such responses have been induced, the individual will be better prepared to cope with acute and chronic nutritional deficiencies. This predictive adaptive response hypothesis (Bateson et al., 2014) would thus explain why there is an association between lower birth weight and a marasmic response to SAM. It would also explain why there are persistent abnormalities after rehabilitation and may explain why there are persistent abnormalities into adulthood. The current observations of persistent epigenetic differences in the skeletal muscle of adult survivors of the two syndromes are consistent with this. Pathway analysis showed that these epigenetic changes were associated with genes that may be related to the differential physiology of adult survivors.
Limitations
Our sample is modest in size, but the participants have the same characteristics as the larger population of survivors from which they were identified. It has not been possible to obtain matched controls based on birth weight and community to explore whether both groups are different from a control population. The small nature of the group reflects the circumstances of this population and their willingness to consent to biopsies. The Infinium® 450 K array represents only a small subset of potential CpG sites that may be methylated and where whole methylome technologies with sufficient reproducibility are available further studies may be indicated. Muscle tissue was chosen over other sources of DNA (blood, buccal smear) because of the clinical evidence suggesting that there are persistent growth and metabolic differences between survivors. As epigenetic profiles differ across tissue, studies in other tissues may reveal further differences. Unfortunately, we were unable to validate some of the identified loci using independent methods, such as pyrosequencing.
The present study reinforces the need to consider the longer-term consequences of SAM. With standardized clinical management using the principles of the WHO Treatment Manual, there has been significantly improved survival of infants with SAM. But there is increasing evidence for long-term sequelae including an increased risk of diabetes or raised blood pressure. Epigenetic differences may be playing a role in influencing both the acute presentation and long-term consequence of SAM. In principle, epigenetic changes are reversible and this may hold implications for further study to improve the health of those living in situations where SAM is unfortunately still present.
References cited in online Appendix A
Contributors
TF, MB and DT designed the clinical component of this study and were responsible for its conduct, sample and data collection management, and clinical data analysis. AS and SN designed the epigenetic component of this study. XL did the molecular work in the lab including sample quality checks and DNA extraction. SN and TP analyzed the epigenetic data. PDG is principle investigator on the grants that funded this study. All authors contributed to the writing of this manuscript.
Declaration of Interests
All authors declare no conflicts of interests.
Acknowledgments
This study was supported by a grant from the Bill and Melinda Gates Foundation (Global Health OPP1066846), Grand Challenge “Discover New Ways to Achieve Healthy Growth”. A previous grant from the Health Research Council of New Zealand (Grant 09/052), "Developmental Adaptation to an Obesogenic Environment" established this cohort. We thank Dr. Joanna Holbrook, Dr. Pan Haong and Dr. Pan Zengxiang for their assistance with microarray data preprocessing and Prof M Heymann for his input.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.03.001.
Appendix A. Supplementary data
Supplementary material
References
- Al-Shahrour F., Diaz-Uriarte R., Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20(4):578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- Badaloo A., Reid M., Forrester T., Heird W.C., Jahoor F. Cysteine supplementation improves the erythrocyte glutathione synthesis rate in children with severe edematous malnutrition. Am. J. Clin. Nutr. 2002;76(3):646–652. doi: 10.1093/ajcn/76.3.646. [DOI] [PubMed] [Google Scholar]
- Badaloo A.V., Forrester T., Reid M., Jahoor F. Lipid kinetic differences between children with kwashiorkor and those with marasmus. Am. J. Clin. Nutr. 2006;83(6):1283–1288. doi: 10.1093/ajcn/83.6.1283. [DOI] [PubMed] [Google Scholar]
- Bateson P., Barker D., Clutton-Brock T. Developmental plasticity and human health. Nature. 2004;430:419–421. doi: 10.1038/nature02725. [DOI] [PubMed] [Google Scholar]
- Bateson P., Gluckman P., Hanson M. The biology of developmental plasticity and the Predictive Adaptive Response hypothesis. J. Physiol. 2014;592(11):2357–2368. doi: 10.1113/jphysiol.2014.271460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender D.A. “Wellcome classification.” A Dictionary of Food and Nutrition. 2005. http://www.encyclopedia.com/doc/1O39-Wellcomeclassification.html (accessed August 15, 2016)
- Carmona-Saez P., Chagoyen M., Tirado F., Carazo J.M., Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome. Biol. 2007;8(1):R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.T., Nevins J.R. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22(23):2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- Dennis G., Jr., Sherman B.T., Hosack D.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- Forrester T.E., Badaloo A.V., Boyne M.S. Prenatal factors contribute to the emergence of kwashiorkor or marasmus in severe undernutrition: evidence for the predictive adaptation model. PLoS One. 2012;7(4):e35907. doi: 10.1371/journal.pone.0035907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis-Emmanuel P.M., Thompson D.S., Barnett A.T. Glucose metabolism in adult survivors of severe acute malnutrition. J. Clin. Endocrinol. Metab. 2014;99(6):2233–2240. doi: 10.1210/jc.2013-3511. [DOI] [PubMed] [Google Scholar]
- Galler J.R., Bryce C.P., Zichlin M.L., Fitzmaurice G., Eaglesfield G.D., Waber D.P. Infant malnutrition is associated with persisting attention deficits in middle adulthood. J. Nutr. 2012;142(4):788–794. doi: 10.3945/jn.111.145441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galler J.R., Bryce C.P., Zichlin M.L. Malnutrition in the first year of life and personality at age 40. J. Child Psychol. Psychiatry. 2013;54(8):911–919. doi: 10.1111/jcpp.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluckman P.D., Hanson M.A. Living with the past: evolution, development, and patterns of disease. Science. 2004;305:1733–1736. doi: 10.1126/science.1095292. [DOI] [PubMed] [Google Scholar]
- Hanson M.A., Gluckman P.D. Early developmental conditioning of later health and disease: physiology or pathophysiology? Physiol. Rev. 2014;94(4):1027–1076. doi: 10.1152/physrev.00029.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A.E., Murakami P., Lee H. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol. 2012;41(1):200–209. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahoor F., Badaloo A., Reid M., Forrester T. Protein kinetic differences between children with edematous and nonedematous severe childhood undernutrition in the fed and postabsorptive states. Am. J. Clin. Nutr. 2005;82(4):792–800. doi: 10.1093/ajcn/82.4.792. [DOI] [PubMed] [Google Scholar]
- Jahoor F., Badaloo A., Reid M., Forrester T. Unique metabolic characteristics of the major syndromes of severe childhood malnutrition. In: Forrester T., Picou D., Walker S., editors. The Tropical Metabolism Research Unit, The University of the West Indies, Jamaica 1956–2006: The House that John Built. Ian Randle Publishers; Kingston: 2006. pp. 23–60. [Google Scholar]
- Jahoor F., Badaloo A., Reid M., Forrester T. Protein metabolism in severe childhood malnutrition. Ann. Trop. Paediatr. 2008;28:87. doi: 10.1179/146532808X302107. [DOI] [PubMed] [Google Scholar]
- Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Pan H., Chen L., Dogra S. Measuring the methylome in clinical samples: improved processing of the Infinium Human Methylation450 BeadChip Array. Epigenetics. 2012;7(10):1173–1187. doi: 10.4161/epi.22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasants A.B., Wake G.C., Shorten P.R., Hassell-Sweatman C.Z., McLean C.A., Holbrook J.D., Gluckman P.D., Sheppard A.M. A new, improved and generalisable approach for the analysis of biological data generated by ‘-omic’ platforms. J. Dev. Orig. Health Dis. 6(1), 2015 doi: 10.1017/S2040174414000476. 17–26. [DOI] [PubMed] [Google Scholar]
- Reid M., Badaloo A., Forrester T. In vivo rates of erythrocyte glutathione synthesis in children with severe protein-energy malnutrition. Am. J. Physiol. Endocrinol. Metab. 2000;278(3):E405–E412. doi: 10.1152/ajpendo.2000.278.3.E405. [DOI] [PubMed] [Google Scholar]
- Reid M., Badaloo A., Forrester T., Morlese J.F., Heird W.C., Jahoor F. The acute-phase protein response to infection in edematous and nonedematous protein-energy malnutrition. Am. J. Clin. Nutr. 2002;76(6):1409–1415. doi: 10.1093/ajcn/76.6.1409. [DOI] [PubMed] [Google Scholar]
- Tennant I.A., Barnett A.T., Thompson D.S. Impaired cardiovascular structure and function in adult survivors of severe acute malnutrition. Hypertension. 2014;64(3):664–671. doi: 10.1161/HYPERTENSIONAHA.114.03230. [DOI] [PubMed] [Google Scholar]
- Wang J., Duncan D., Shi Z., Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41(Web Server issue):W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material