Abstract
The with-no-lysine kinase (WNK) 4 gene is a causative gene in pseudohypoaldosteronism type II. Although WNKs are widely expressed in the body, neither their metabolic functions nor their extrarenal role is clear. In this study, we found that WNK4 was expressed in mouse adipose tissue and 3T3-L1 adipocytes. In mouse primary preadipocytes and in 3T3-L1 adipocytes, WNK4 was markedly induced in the early phase of adipocyte differentiation. WNK4 expression preceded the expression of key transcriptional factors PPARγ and C/EBPα. WNK4-siRNA-transfected 3T3-L1 cells and human mesenchymal stem cells showed reduced expression of PPARγ and C/EBPα and lipid accumulation. WNK4 protein affected the DNA-binding ability of C/EBPβ and thereby reduced PPARγ expression. In the WNK4−/− mice, PPARγ and C/EBPα expression were decreased in adipose tissues, and the mice exhibited partial resistance to high-fat diet-induced adiposity. These data suggest that WNK4 may be a proadipogenic factor, and offer insights into the relationship between WNKs and energy metabolism.
Keywords: WNK4, Hypertension, Metabolic syndrome
Highlights
-
•
WNK4 regulates adipocyte differentiation in mouse and human preadipocytes.
-
•
WNK4−/− mice exhibit reduced adiposity and increased insulin sensitivity.
-
•
WNK4 may be a drug target for diet-induced obesity and salt-sensitive hypertension.
The with-no-lysine kinase (WNK) 4 gene is a causative gene in pseudohypoaldosteronism type II, a hereditary hypertensive disease. Although WNKs are widely expressed in the body and are involved in the pathogenesis of hypertension, neither their metabolic functions nor their extrarenal role is clear. This study demonstrated a contribution of WNK4 to the regulation of core transcriptional factors for adipogenesis and that its depletion indicates some beneficial effects for obesity by a high-fat diet. This study suggests a role of hypertension-causing WNK4 as a proadipogenic factor and offers insights into the relationship between WNKs and energy metabolism.
1. Introduction
Metabolic syndrome (MetS), characterized by insulin resistance, dyslipidemia, hypertension, and central obesity, is a significant clinical problem associated with cardiovascular disease and diabetes (Alberti et al., 2009). Because white adipose tissue (WAT) is the main energy storage organ and secretes several humoral factors (Cristancho and Lazar, 2011), it has important roles in MetS. Core transcriptional factors regulating adipocyte differentiation include the master regulators, peroxisome proliferator-activated receptor gamma (PPARγ), and CCAAT/enhancer-binding protein alpha (C/EBPα) (Lefterova et al., 2008), and moderate reduction of PPARγ activity was shown to prevent obesity and improve insulin sensitivity (Kubota et al., 1999, Yamauchi et al., 2001, Jones et al., 2005).
The with-no-lysine kinases (WNKs) are a family of serine/threonine kinases composed of four human genes, WNK1, WNK2, WNK3, and WNK4. WNK1 and WNK4 were identified as being responsible for pseudohypoaldosteronism type II (PHAII; (Wilson et al., 2001), a hereditary hypertensive disease characterized by hyperkalemia, metabolic acidosis, and thiazide sensitivity (Gordon, 1986). We and others have clarified that WNK regulates the Na—Cl cotransporter (NCC) in the distal convoluted tubules of the kidney through phosphorylation of oxidative stress-responsive 1 (OSR1) and Ste20-like proline/alanine-rich kinase (SPAK) (Yang et al., 2007, Chiga et al., 2011). Recent studies have added further information that insulin phosphorylates WNK1 through protein kinase B (PKB)/Akt (Jiang et al., 2005), and that insulin is a powerful activator of the WNK4–OSR1/SPAK–NCC signaling cascade in vitro and in vivo (Sohara et al., 2011, Nishida et al., 2012, Takahashi et al., 2014). These data suggest that activation of the WNK–OSR1/SPAK–NCC signaling cascade caused by hyperinsulinemia may underlie the pathogenesis of salt-sensitive hypertension in MetS.
In extrarenal organs, recent studies have suggested that WNKs regulate cell growth, differentiation, and development. For example, WNK1 is required for mitosis in cultured cells (Tu et al., 2011). In addition, WNK2 is known to suppress cell growth (Hong et al., 2007), while WNK3 increases cell survival (Veríssimo et al., 2006), and WNK4 is required for the anterior formation of Xenopus embryos (Shimizu et al., 2013). However, the functions of WNKs in energy metabolism remain unknown.
In the present study, we reported a role of WNK4 as a regulator of adipocyte development. We found that WNK4 was expressed in the mouse adipose tissue. In primary preadipocytes and 3T3-L1 adipocytes, WNK4 was increased dramatically in the early phase of differentiation. The suppression of endogenous WNK4 decreased expression of PPARγ and C/EBPα, resulting in the impaired formation of mature adipocytes in 3T3-L1 cells and human mesenchymal stem cells. We also found that WNK4 was involved in cell cycle progression during mitotic clonal expansion (MCE), which might be the cause of decreased expression of PPARγ by C/EBPβ by WNK4 suppression. Consistent with the reduced expression of PPARγ, adult WNK4−/− (WNK4-KO) mice exhibited reduced adiposity and the decreased expression of adipogenic genes on high-fat diets (HFDs), suggesting the involvement of WNK4 in the development of obesity. These results suggested that the hypertension-causing gene WNK4 not only is involved in the regulation of salt-sensitive hypertension, but also in energy metabolism.
2. Materials and Methods
2.1. Animals
The generation of the WNK4-KO mice, WNK4D561A/+ knock-in mice, and WNK4 transgenic (WNK4-Tg) mice and their genotyping strategies were described previously (Takahashi et al., 2014, Wakabayashi et al., 2013, Yang et al., 2007). Studies were performed on each strain using littermates. The mice were raised under a 12-h day and night cycle, and were fed a normal rodent diet [6% kcal% fat] (Oriental Yeast, Tokyo, Japan) or high-fat diet [60% kcal% fat] (Research Diets, New Brunswick, Canada) and plain drinking water. This experiment was approved by the Animal Care and Use Committee of the Tokyo Medical and Dental University, Tokyo, Japan.
2.2. Cell Culture
3T3-L1 cells (ATCC; CL-173) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS), 2 mM l-glutamine, 100 U per ml penicillin, and 0.1 mg/ml streptomycin at 37 °C in a humidified 5% CO2 incubator. The cells were induced to differentiate with Adipoinducer Reagent (Takara, Shiga, Japan) containing 10 μg/ml insulin, 2.5 μM dexamethasone, and 0.5 mM 3-isobutyl-1-methylxantine (IBMX) (MDI). After 48 h of induction, 3T3-L1 cells were maintained in the medium containing DMEM supplemented with 10% FBS, penicillin/streptomycin, and 10 μg/ml insulin. For bumetanide stimulation experiments, the 3T3-L1 cells were exposed to MDI supplemented with 10 μM bumetanide/DMSO or DMSO as a negative control. The mpkDCT cells were maintained as described previously (Sohara et al., 2011). The human mesenchymal stem cells from adipose tissue (hMSC-AT) cells were purchased from PromoCell GmbH (Heidelberg, Germany). The hMSC-AT cells were cultured in Mesenchymal Stem Cell Growth Medium 2 (PromoCell), and were induced to differentiate with Mesenchymal Stem Cell Adipogenic Differentiation Medium 2 (PromoCell), according to the manufacturer's instructions. The preparation of stromal vascular fraction (SVF) from mice adipose tissue, culture of SVF cells, and adipogenic induction of SVF cells were performed as described previously (Suganami et al., 2005, Tanaka et al., 2014).
2.3. Transfection
3T3-L1 cells (3 × 105 cells/6-cm dish or 1 × 105 cells/3.5-cm dish) were transfected with the indicated amount of plasmid DNA or siRNA with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, Canada). For the knockdown experiments in 3T3-L1 cells, we used 20 nM cocktails of three duplexes of siRNAs. The oligonucleotide sequences of mouse si-WNK1, si-WNK4, si-SPAK and si-Nega were described previously (Sohara et al., 2011, Zeniya et al., 2013). The mouse si-OSR1 and si-C/EBPβ was obtained from Origene (MD, USA) and Santa Cruz Biotechnology (TX, USA), respectively. The hMSC-AT cells were transfected with the indicated amount of siRNA with Xfect RNA Transfection Reagent (Takara), as the manufacturer's instructions. For the knockdown experiments in hMSC-AT cells, we used 50 nM cocktails of two duplexes of siRNAs. The oligonucleotide sequences of human si-WNK4 were as follows: human si-WNK4-A ggaggacgacggcgagaagTT, human si-WNK4-B ugacagagugguugagugcTT. The differentiation of 3T3-L1 cells and hMSC-AT cells to adipocytes was induced 3 days after transfection.
2.4. Plasmids
Mouse WNK4 cDNA was isolated by PCR using mouse WNK4 vector (Ohno et al., 2011) and cloned into 3XFLAG-CMV10 vector (Sigma-Aldrich, MO, USA). Mouse C/EBPβ cDNA was isolated by PCR using mouse adipose tissue and cloned into T7-pRK5 vector. Sequences of the amplification primers employed were as follows: WNK4 sense, 5′-CTTGCGGCCGCGATGCTAGCACCTCGAAATAC-3′ and WNK4 antisense, 5′-ACCGATATCTCACATCCTGCCAATATC-3′; C/EBPβ sense, 5′-AGCAAATGGGTCGCGAGATGGAAGTGGCCAACTTCTACTACG-3′ and C/EBPβ antisense, 5′-CCAAGCTTCTGCAGGCTAGCAGTGGCCCGCCGA-3′. The mouse WNK4 cDNA was cloned into 3xFLAG-CMV10 vector (Sigma-Aldrich, MO, USA). The kinase-dead mutation (D318A) was introduced using a QuickChange Site-directed Mutagenesis kit (Stratagene, La Jolla, Canada) with the following primers: D318A sense, 5′-CTGTCAAAATCGGAGCCCTCGGACTGGCCA-3′, and D318A antisense, 5′-TGGCCAGTCCGAGGGCTCCGATTTTGACAG-3′ (Mutagenesis sites are underlined).
2.5. RT-PCR
Total RNA from the mouse adipose tissue or cultured cells was extracted using TRIzol reagent (Invitrogen). The total RNA was reverse-transcribed using Omniscript reverse transcriptase (Qiagen, Hilden, Germany). Sequences of RT-PCR primers employed were as follows: mouse C/EBPβ sense, 5′-GTTTCGGGAGTTGATGCAATC-3′, C/EBPβ antisense, 5′-AACAACCCCGCAGGAACAT-3′; mouse CUL3 sense, 5′-GGATGAGTTCAGGCAACATC-3′, mouse CUL3 antisense, 5′-GCATGTCTTGGTGCTGGTGG-3′; human perilipin sense, 5′- ACCCCCCTGAAAAGATTGCTT-3′, antisense, 5′- GATGGGAACGCTGATGCTGTT-3′; human PPARγ2 sense, 5′-GCAGGAGATCTACAAGGACTTG-3′, antisense, 5′- CCCTCAGAATAGTGCAACTGG-3′; human β-actin sense, 5′-CCGTCTTCCCCTCCATCG-3′; antisense, 5′- GTCCCAGTTGGTGACGATGC-3′; the mouse C/EBPα, mouse PPARγ, mouse β-actin, mouse GAPDH, mouse FAS, mouse LPL, mouse FABP4, mouse WNK2, mouse WNK3, mouse WNK4, mouse KLHL2, mouse KLHL3, mouse F4/80, and mouse adiponectin primers were described previously (Zeniya et al., 2013, Madsen et al., 2003, O'reilly et al., 2006, Dutruel et al., 2014, Suganami et al., 2005). Relative mRNA levels were normalized by endogenous reference genes (β-actin and GAPDH).
2.6. Indirect Calorimetry
Male mice at 9 weeks-old on a normal diet were monitored individually in a metabolism measurement system (Muromachi Kikai, Tokyo, Japan) for 48 h. Each cage was monitored for oxygen consumption and activity at 5 min intervals throughout the period. Total oxygen consumption was calculated as accumulation of oxygen uptake for each mouse divided by its body weight.
2.7. Immunoblotting
Extraction of protein samples and semiquantitative immunoblotting were performed as previously described (Yang et al., 2007). For immunoblotting, we used entire samples without the nuclear fraction (600 × g) or the crude membrane fraction (17,000 × g). To separate nuclear extracts of the cells, Nuclear and Cytoplasmic Extraction Reagents (PIERCE, AZ, USA) were used, following the manufacturer's instructions. The relative intensities of immunoblot bands were analyzed and quantified using ImageJ software (National Institutes of Health (NIH), MD, USA). Rabbit anti-FLAG (Sigma-Aldrich), rabbit anti-WNK1 (A301-516A; BETHYL), sheep anti-WNK3 (S346C), rabbit anti-mouse WNK4, rabbit anti-phosphorylated SPAK, rabbit anti-SPAK, rabbit anti-phosphorylated OSR1/SPAK, mouse anti-OSR1, anti-NKCC1, anti-phosphorylated NKCC1 and anti-actin were used as described previously (Takahashi et al., 2014). Rabbit anti-PPARγ, rabbit anti-C/EBPα, rabbit anti-C/EBPβ, rabbit anti-phosphorylated C/EBPβ, rabbit anti-ERK1/2, rabbit anti-phosphorylated ERK1/2, anti-Cyclin D1, anti-Cyclin D3, anti-Cyclin B1, anti-Histon H3, anti-human WNK4 antibody were purchased from Cell Signaling (MA, USA). Anti-Cyclin A2, anti-FABP4, anti-Rb, and anti-phosphorylated Rb antibody (p780) was purchased from Abcam (MA, USA). Alkaline-phosphatase-conjugated anti-IgG antibodies (Promega, WI, USA) were used as secondary antibodies for immunoblotting. Western Blue (Promega) was used for the development of immunoblots. The relative intensities of the immunoblot bands were determined by densitometry with ImageJ software.
2.8. Blood Samples
Glucose tolerance tests were performed after a 16 h overnight fast. Mice were injected glucose (1.0 g per kg of body weight) intraperitoneally. Blood glucose concentrations were determined with a glucose sensor (MEDISAFE MINI, Terumo, Tokyo, Japan). Blood samples were collected from the tail vein at 0, 15, 30, 60, 90, and 120 min after the injection. Insulin tolerance tests were performed after a 6 h fast. Mice were injected insulin (0.75 units/kg; Humulin R, Ely Lilly, IN, USA) intraperitoneally. Blood samples were collected from the tail vein at 0, 15, 30, 60, and 90 min after the injection. Plasma insulin, leptin, and adiponectin levels were measured by using ELISA kits (Morinaga, Kanagawa, Japan), respectively. Quantification of plasma triglycerides and total cholesterol were performed by using Triglycerides E-test kit and Cholesterol E-test kit (Wako, Osaka, Japan), respectively. Blood samples for leptin, adiponectin, triglyceride and cholesterol were obtained from the inferior vena cava under anesthesia with pentobarbital. For chronic hydrochlorothiazide (HCTZ) treatment, osmotic minipumps (Alzet, CA, USA) were implanted subcutaneously to deliver 30 mg/kg/day HCTZ dissolved in a 1.7% ethanolamine solution or vehicles (ethanolamine). Eight-week-old male C57BL6/J mice were randomly allocated into two groups (HCTZ-treated group and non-treated group). Blood electrolyte levels were determined using an i-STAT Portable Clinical Analyzer (Fuso Pharmaceutical Industries Ltd., Japan).
2.9. Histology
Sections were subjected to standard hematoxylin/eosin staining. For adipocyte size measurement, stained sections at 100 × magnification were subjected to ImageJ software (NIH). The adipocytes were measured by circling cells manually and the sizes were calculated automatically.
2.10. Oil-red O Stain and Adipogenesis Assay
For Oil-red O stain image, differentiated 3T3-L1 cells and hMSC-AT cells were washed twice with PBS and fixed in 10% formalin in PBS for 1 h. Cells were washed twice in PBS and incubated in freshly diluted Oil-red O solution (Sigma-Aldrich, MO, USA) for 15 min at room temperature. To quantity triglyceride content of day 8 or day 10 in the differentiated 3T3-L1 cells, adipogenesis assay kit (Chemicon, Temecula, Canada) was used as the manufacturer's instructions. Briefly, cells were washed twice with PBS and are incubated in the Oil-red O solution for 15 min. The stained cells were washed with wash solution, and dye extraction solution was added. Extracted dye was quantified by measuring OD495, using a microplate reader (Molecular Devices, CA, USA).
2.11. Cell Proliferation Assay
3T3-L1 preadipocytes (5 × 103 cells) were transfected with si-WNK4 or si-Nega and seeded in a 96-well flat-bottom tissue culture plate. After indicated times, 10 μl of Cell Count Reagent SF (Nacalai tesque, Kyoto, Japan) was added to each well and incubated at 37 °C for 2 h. Then, cell growth was measured at OD450 on a microplate reader (Molecular Devices).
2.12. Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assay (ChIP) was performed using a SimpleChIP Plus Enzymatic Chromatin IP Kit (Cell Signaling) according to the manufacturer's instructions. Briefly, 3T3-L1 adipocytes were exposed 1% formaldehyde for 10 min at room temperature followed by SDS lysis buffer. After sonication, DNA samples were immunoprecipitated with anti-C/EBPβ antibody (E299, Abcam, MA, USA) or normal rabbit IgG (Cell Signaling) as a negative control. Immunoprecipitated samples were analyzed by PCR using primers flanking the mouse PPARγ2 promoter region. Primers employed for RT-PCR were described previously (Salma et al., 2004).
2.13. BrdU Assay
BrdU assay was performed using FLOUS in situ detection kit (Roche, Basel, Switzerland) on 5 replicates. BrdU was added after 24 h after the onset of adipocyte differentiation for 30 min. Immunodetection of BrdU was performed following the manufacturer's instructions. Cells were also stained with Hoechist to detect nucleus. Analysis of percentage of BrdU-positive cells was shown by dividing the total number (Hoechist-positive) of cells.
2.14. Statistics
Statistical significance was evaluated using an un-paired t-test when only two value sets were compared. For multiplex comparisons, one-way ANOVA test with Fischer's post-hoc test was used. For growth curve, two-way ANOVA test with Tukey's test was used. The results with P values < 0.05 were considered statistically significant and is indicated by *. Data were presented as mean ± 95% confidence interval (95%CI). The tests were performed using the Prism 7 software (GraphPad, CA, USA).
3. Results
3.1. WNK4 Expression in the Mouse White Adipose Tissue and in 3T3-L1 Adipocytes
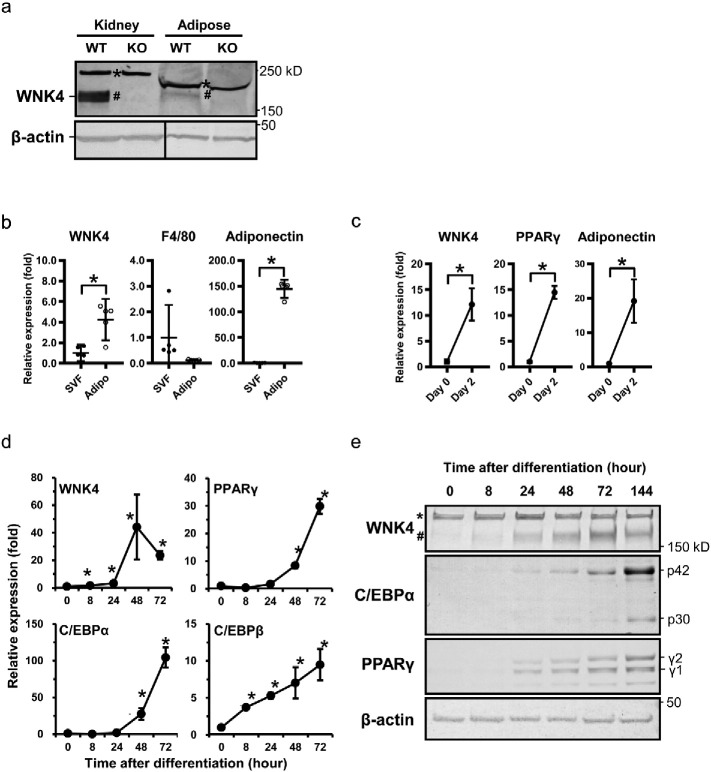
We investigated expression of WNK4 in the extrarenal organs by immunoblotting, and identified that WNK4 was expressed in WAT (Fig. 1a). The adipose tissue was composed of various cell types: mature adipocytes and the stromal vascular fraction (SVF), which includes blood cells, endothelial cells, and macrophages (Suganami et al., 2005). To analyze which cell type in the adipose tissue contained WNK4, we isolated mouse SVF from mature adipose tissue. The quality of the separation was assessed by quantitative (q) RT-PCR of macrophage marker F4/80 and adipose tissue marker adiponectin. We found that WNK4 was expressed predominantly in mature adipocytes (Fig. 1b). We next induced SVF into adipocyte differentiation and found that differentiated primary adipocytes expressed WNK4 (Fig. 1c).
Fig. 1.
WNK4 is expressed in mice adipose tissue and induced in the early phase of 3T3-L1 adipocyte differentiation. (a) Immunoblot analysis of WNK4 in the white adipose tissue of WT and WNK4−/− mice. The asterisks (*) and the sharps (#) show nonspecific bands and WNK4, respectively. (b) qRT-PCR analysis of mouse primary preadipocytes. WNK4 was expressed predominantly in mature adipocytes. (c) Primary preadipocyte differentiation into adipocytes and analysis of WNK4. The SVF cells treated with adipogenic stimuli showed increased WNK4 expression at day 2 of differentiation. (d) qRT-PCR analysis of differentiating 3T3-L1 adipocytes. The level of WNK4 mRNA was increased significantly by 8 h of differentiation. Representative results are shown for three experiments, and values are expressed as mean ± 95%CI. *P < 0.05. (e) Western blot analysis of WNK4 in differentiating 3T3-L1 adipocytes. The WNK4 protein was increased dramatically along with the process of differentiation.
To evaluate the potential function of WNK4 in WAT in more detail, we examined WNK4 expression in 3T3-L1 preadipocytes, a well-established model of adipocyte differentiation (Green and Kehinde, 1975). Confluent 3T3-L1 preadipocytes differentiate upon exposure to the adipogenic inducers, fetal bovine serum (FBS), dexamethasone (Dexa), isobutylmethylxanthine (IBMX), and insulin. This cocktail (MDI) activates an adipogenic program, which consists of two well-defined phases. The stimulated cells immediately reenter the cell cycle and progress through at least two cell-cycle divisions, called MCE. During this time, the cells express specific adipogenic transcription factors, including C/EBPβ as well as cell-cycle regulators that together facilitate expression of PPARγ and C/EBPα. Following this event, the cells undergo terminal differentiation manifested by production of lipid droplets as well as expression of multiple metabolic programs characteristic of mature fat cells. The validity of this 3T3-L1 system as an appropriate model of adipocyte formation in the animal has been demonstrated (Farmer, 2006). The expression level of WNK4 mRNA in the undifferentiated 3T3-L1 cells was low, but after stimulation with MDI, WNK4 was increased significantly by 1.8-fold at as early as 8 h, and dramatically increased to 44.2-fold at day 2 (Fig. 1d). An increase in WNK4 was followed by the expression of PPARγ and C/EBPα, which was supported by the immunoblot analysis (Fig. 1e). In contrast, WNK2 and WNK3 were not expressed in the mouse adipose tissue nor in the 3T3-L1 cells (Fig. S1a). WNK1 already was expressed in the undifferentiated 3T3-L1 cells, and neither the protein levels of WNK1 nor the phosphorylation status of OSR1/SPAK was changed throughout (Fig. S1b). Since WNK4 is a substrate for the KLHL2-Cullin3 and KLHL3–Cullin3 E3 ligase complex (Takahashi et al., 2013, Wakabayashi et al., 2013), we also examined qRT-PCR analysis and immunoblots of KLHL2, KLHL3, and CUL3 to examine whether WNK4 degradation also was regulated in differentiating 3T3-L1 cells. Since the mRNA levels of these proteins were not decreased during adipocyte differentiation (Fig. S1c), the increase of WNK4 protein during adipocyte differentiation would be mainly due to the increased WNK4 transcription.
We also examined which component of MDI was responsible for WNK4 expression, and found that Dexa was a major inducer of WNK4 (Fig. S2a). Since C/EBPβ also is one of a downstream protein induced by Dexa, we examined the relation between C/EBPβ and WNK4. Although we transfected T7-C/EBPβ to the undifferentiated 3T3-L1 cells, the expression of WNK4 was not induced (Fig. S2b). We also performed knockdown of C/EBPβ, and found that the expression of WNK4 protein after differentiation was not significantly affected (Fig. S2c). Therefore, C/EBPβ is not a major regulator for WNK4 expression during early adipocyte differentiation. It also is notable that increases in WNK4 protein levels were not observed in different cell types (mpkDCT cells) when using Dexa, suggesting that WNK4 induction by Dexa in 3T3-L1 cells was not a common effect across different cell types (Fig. S2d), and, in fact, may be involved in the complex adipocyte differentiation machinery. These findings indicated that WNK4 expression was induced in the early phase of adipocyte differentiation, preceding PPARγ and C/EBPα expression.
3.2. WNK4 is Required for the Expression of Core Transcriptional Factors in 3T3-L1 Adipocytes and hMSC-AT Cells
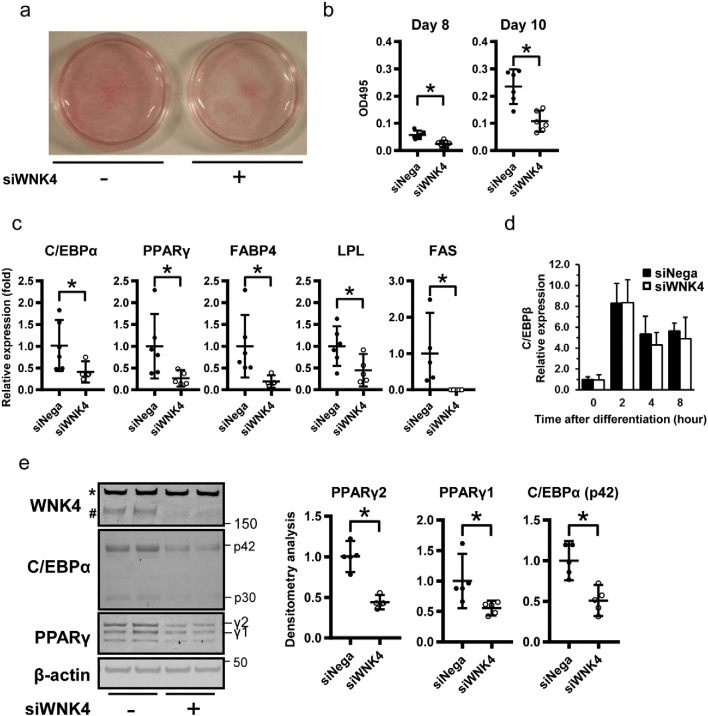
The potential function of WNK4 in the adipocyte was examined by siRNA specific to WNK4 (si-WNK4; (Sohara et al., 2011). Oil-Red O staining and adipogenesis assays showed reduced accumulation of triglyceride as lipid droplets in the si-WNK4 group (Fig. 2a and b). Given WNK4 levels increase in the very early phase of differentiation, we investigated whether si-WNK4 affects transcriptional factors and adipocyte markers during adipocyte differentiation. The mRNA expression levels were measured by RT-PCR on day 4 after differentiation. On day 4, si-WNK4 transfected cells showed marked reductions in mRNA levels of PPARγ, C/EBPα, and PPARγ targeting genes FABP4, lipoprotein lipase (LPL), and fatty acid synthase (FAS; Fig. 2c). This indicated that WNK4 inhibition resulted in the inhibition of terminal differentiation of 3T3-L1 adipocytes. In contrast, C/EBPβ mRNA levels, which are known to increase within 4 h of differentiation (Tang and Lane, 1999), were not significantly changed (Fig. 2d). Consistent with these results, the protein levels of PPARγ and C/EBPα were significantly suppressed in the WNK4-siRNA group (Fig. 2e). We next analyzed human mesenchymal stem cells from adipose tissue (hMSC-AT cells), to examine whether inhibition of WNK4 affected expression of transcriptional factors in human preadipocytes. Consistent with the results in Fig. 2, hMSC-AT cells also showed similar phenotypes as 3T3-L1 cells (Fig. S4). We also treated 3T3-L1 cells with either WNK1-siRNA (si-WNK1), SPAK-siRNA (si-SPAK), OSR1-siRNA (si-OSR1), or the NKCC1 inhibitor bumetanide, but no inhibition of PPARγ or C/EBPα expression was observed (Fig. S5). These results indicated that WNK4 was involved in adipocyte differentiation in mouse 3T3-L1 and in hMSC-AT cells, independent of other WNKs or the known downstream effectors of the OSR1/SPAK-NKCC1 signaling cascade.
Fig. 2.
Knockdown of WNK4 impairs lipid accumulation and expression of adipogenic genes in 3T3-L1 cells. (a) Oil-Red O staining of 3T3-L1 cells expressing si-WNK4 showed decreased lipid accumulation at day 6 of differentiation. (b) Lipids extracted from Oil-Red O stained cells were quantified (n = 6 per group). Cells expressing si-WNK4 showed impaired lipid accumulation. (c) Real-time PCR analysis of 3T3-L1 adipocytes expressing si-Nega or si-WNK4 at day 4 of differentiation. The mRNA levels of PPARγ, C/EBPα, and PPARγ targeting genes were reduced in si-WNK4-transfected 3T3-L1 cells (n = 6 per group). (d) Real-time PCR analysis of 3T3-L1 adipocytes expressing si-Nega or si-WNK4 within 8 h of differentiation (n = 5 per group). The level of C/EBPβ mRNA in the early phase of differentiation was not affected by si-WNK4. (e) WNK4 knockdown in 3T3-L1 cells prevented PPARγ and C/EBPα protein expression at day 2 of differentiation (n = 6 per group). Whole course of C/EBPβ, PPARγ, and C/EBPα proteins during adipogenesis under si-WNK4 are shown in Fig. S3.
3.3. The Role of WNK4 in Expression of Adipogenic Genes in 3T3-L1 Cells
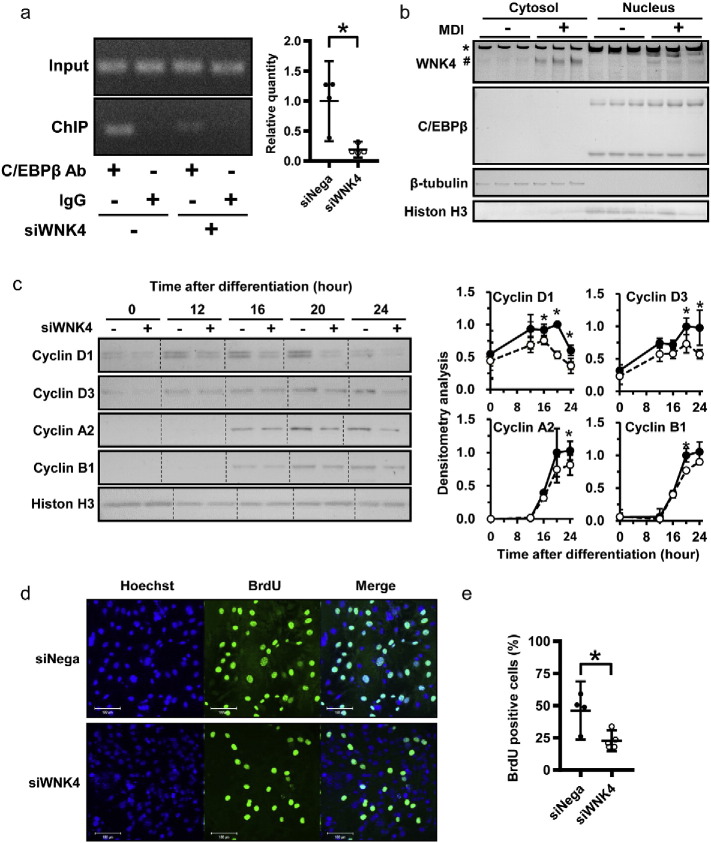
Our data showed C/EBPβ expression was not affected by si-WNK4 (Fig. 2d). Because C/EBPβ is an established activator of PPARγ during adipocyte differentiation (Guo et al., 2015), we hypothesized, therefore, that WNK4 might affect the C/EBPβ action to its downstream genes. To test this, we performed chromatin immunoprecipitation (ChIP) to examine whether the binding ability of endogenous C/EBPβ to the PPARγ2 promoter in 3T3-L1 cells was influenced by si-WNK4. DNA fragments were immunoprecipitated with the C/EBPβ antibody at day 2 of differentiation and the immunoprecipitated DNA fragments were amplified by PCR. The binding ability of C/EBPβ to the PPARγ2 promoter was impaired by si-WNK4 (Fig. 3a).
Fig. 3.
WNK4 may be involved in the cell cycle regulation of 3T3-L1 adipocyte. (a) CHIP assay of C/EBPβ on PPARγ2 promotor. The 3T3-L1 cells expressing si-Nega or si-WNK4 were induced to differentiate for 2 days. Cells were subjected to chromatin immunoprecipitation (ChIP) with antibody specific to C/EBPβ, and the immunoprecipitated samples were normalized to the input internal control DNA (actin gene). The values are expressed as mean ± 95%CI (n = 3) relative to si-Nega-transfected cells. IgG immunoprecipitated samples only showed a signal < 10% that of the C/EBPβ-antibody immunoprecipitated samples. Similar results were obtained in three separate experiments. (b) Cellular localization of WNK4. In differentiated 3T3-L1 cells on day 2, WNK4 protein was detected in the cytosolic and nucleic fractions. The asterisks (*) and sharps (#) show nonspecific bands and WNK4, respectively. (c) Inhibition of cyclins during 3T3-L1 differentiation. In the si-WNK4 group, time-dependent increases of cyclin D1, D3, A2, and B1 after MDI stimulation were significantly inhibited. (d) BrdU assay during MCE. Representative image of a BrdU assay 24 h after induction of adipogenic differentiation in si-Nega- or si-WNK4-transfected 3T3-L1 cells; BrdU (green), Hoechst (blue). Scale bar is 100 μM. (e) Quantification analysis of a BrdU assay in 3T3-L1 cells. Values are expressed as mean ± 95%CI. *P < 0.05; **P < 0.01.
It is known that, when preadipocytes are stimulated with MDI, growth-arrested cells reenter 2 to 3 rounds of the cell cycle, called MCE. The MCE is a necessary step for terminal adipocyte differentiation, and C/EBPβ obtains its binding ability during this step (Guo et al., 2015). It has been reported that blocking the cell cycle at the G1-S stage can inhibit MCE and delay subsequent events in adipogenesis (Tang et al., 2003b). Among the WNKs, WNK1 has been reported to be expressed in the mitotic spindles where it regulates mitosis and cell division (Tu et al., 2011), while endogenous WNK3 has been shown to be able to translocate into the nucleus (Veríssimo et al., 2006). Thus, we examined whether WNK4 was expressed in the cell nucleus during adipocyte differentiation. Although WNK4 was expressed predominantly in the cytosolic extracts, we also observed the expression of WNK4 in the nuclear extracts of differentiated 3T3-L1 adipocytes (Fig. 3b). Because WNKs are known as cell cycle regulators (Moniz and Jordan, 2010), we also examined the effect of si-WNK4 on MCE. We found that the increase of cyclins and phosphorylation of retinoblastoma protein (p-pRB) during the MCE phase was significantly inhibited in the si-WNK4 group (Figs. 3c and S6a). Moreover, the 5-bromodeoxyuridine (BrdU) assay during MCE showed a smaller number of S-phase cells in the si-WNK4 group compared to the si-Nega group (Fig. 3d). Accordingly, the cell proliferation of 3T3-L1 cells in the early phase (day 1) was significantly inhibited in the si-WNK4 group (Fig. S6b). These results indicated that si-WNK4 inhibited cell cycle progression during MCE, which might have resulted in the reduced ability of C/EBPβ to bind PPARγ promoter.
3.4. WNK4 Promotes Expression of Adipogenic Genes in 3T3-L1 Adipocytes
We also examined the effect of overexpression of WNK4. FLAG-tagged WNK4-transfected 3T3-L1 cells maintained without MDI did not differentiate into adipocytes (Fig. S7a). However, when FLAG-tagged WNK4-transfected cells were cultured in MDI, we observed significant increases in the expression of PPARγ and C/EBPα on day 2 in mRNA and protein levels (Figs. S7b and S7c) and increased accumulation of lipids on day 8 (Figs. S7d and S7e). These findings indicated that exogenous overexpression of WNK4 could promote expression of adipogenic genes. Intriguingly, these results were not abolished when kinase-dead mutant-WNK4 (D318A; (Ahlstrom and Yu, 2009) was transfected (Fig. S7b–e). Thus, we speculated that the effect of WNK4 in adipocytes might be a kinase-independent function.
3.5. WNK4 Deficiency Prevents Diet-induced Obesity
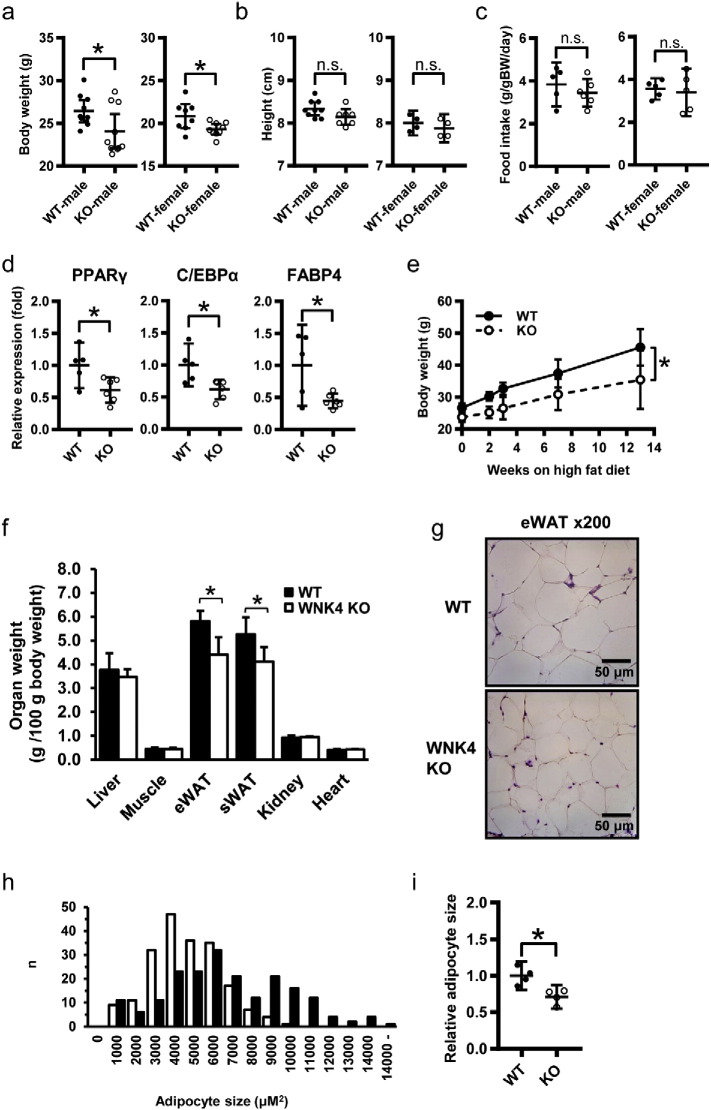
To assess the adipogenic properties of WNK4 in vivo, we analyzed WNK4 knockout mice. WNK4−/− mice were born at a normal birth rate and showed no obvious differences in body weight until 6 weeks of age (data not shown), but displayed moderately lower weights than the wild-type (WT) mice when 8 weeks old (Fig. 4a). This was not due to shorter body length or reduced food intake (Fig. 4b and c). As we reported previously, WNK4−/− mice had almost normal plasma electrolyte and normal blood pressure values when fed a normal diet (Takahashi et al., 2014). Indirect calorimetry at baseline showed normal energy expenditure and activity (Fig. S8), supporting our observation that the WNK4−/− mice were healthy.
Fig. 4.
Phenotypes of WNK4−/− mice. (a, b) The body weights (a) and body lengths (b) of 8-week-old WT and WNK4−/− mice on a normal diet (n = 8–9 per group). WNK4−/− mice exhibited lower average body weights but similar body lengths. (c) Total food intake was comparable between WT and WNK4−/− mice during a day (n = 5–6 per group). (d) Real-time PCR analyzes of adipogenic genes in the adipose tissue (n = 5–6 per group). (e) Body weight increases for WT and WNK4−/− male mice fed a HFD (n = 5–7). WNK4−/− mice exhibited lower body weights on a HFD. (f) Organ weights of the mice fed HFDs (n = 8–9 per group). The weights of epididymal (eWAT) and subcutaneous (sWAT) fat in WNK4−/− mice were significantly lower compared to those of WT mice. (g) Hematoxylin and eosin (HE) staining of adipose sections from WT and WNK4−/− mice. Scale bar, 50 μm. (h) The quantification of adipocyte size is shown (n = 200 cells per group). (i) The average adipocyte size of WNK4−/− mice was smaller than that of WT mice. Values are expressed as mean ± 95%CI. *P < 0.05.
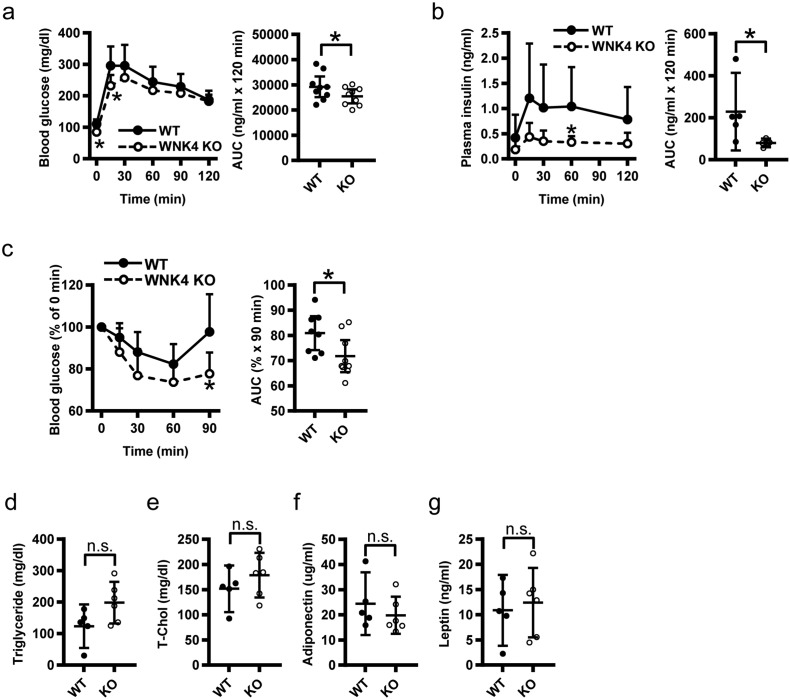
C57BL/6 male mice are prone to obesity, and are susceptible to insulin resistance and glucose intolerance in response to a HFD (Almind and Kahn, 2004). In the male WNK4−/− mice fed a HFD for 6 weeks, the expression of PPARγ, C/EBPα, and FABP4 in the adipose tissue was significantly lower in WNK4−/− mice compared to the WT mice (Fig. 4d). When WNK4−/− mice were fed a HFD, they ultimately weighed less than WT mice (Fig. 4e). This difference in body weight was not observed when wild-type mice on a HFD were treated with 30 mg/kg/day of hydrochlorothiazide (HCTZ) or vehicles for 6 weeks. In HCTZ-treated mice, K+ and HCO3– levels were tended to be lower and higher than those in non-treated mice, respectively, suggesting the effective administration of HCTZ to the mice (Table S1). However, body weight between the groups was not significantly different (Fig. S9). This indicating that decreased NCC activity in the kidney did not account for the phenotype of WNK4−/− mice. In WNK4−/− mice fed HFDs, the weights of the epididymal and subcutaneous fat pads were significantly decreased (Fig. 4f). In contrast, the weights of the liver, skeletal muscle (gastrocnemius), kidney, and heart were similar between the genotypes. HFD-fed WNK4−/− mice exhibited reduced and smaller adipocytes compared to WT mice (Figs. 4g–i). In addition, HFD-fed WNK4−/− mice had significantly lower fasting plasma glucose levels, lower blood glucose by intraperitoneal glucose and insulin tolerance tests (IPGTT and IPITT, respectively), lower plasma insulin levels, and a reduced hypoglycemic response to insulin (Figs. 5a–c). We also evaluated the metabolic parameters of fasted WNK4−/− mice, but plasma triglyceride (TG), total cholesterol (TC), adiponectin, and leptin levels were not significantly different (Figs. 5d–g). These results showed that WNK4−/− mice were protected from HFD-induced weight gain and insulin resistance despite normal adiponectin and leptin levels.
Fig. 5.
Phenotypes of HFD-fed WNK4−/− mice. (a–c) IPGTT (a), plasma insulin levels (b), IPITT (c), and their AUCs in HFD-fed WT and WNK4−/− mice. The HFD-fed WNK4−/− mice showed lower fasting glucose levels and presented lower plasma glucose levels 15 min after glucose injection (n = 5 per group; a). Plasma insulin level and AUC result during the glucose tolerance test (n = 5 per group; c). WNK4−/− mice exhibited significantly lower AUC values (n = 5–6 per group). (d–g) Fasting plasma triglyceride (d), total cholesterol (e), adiponectin (f), and leptin levels (g) were not affected in the WNK4−/− mice (n = 5–6). Values are expressed as mean ± 95%CI. *P < 0.05.
We explored whether the increased of WNK4 in adipocytes caused the opposite phenotypes. We examined WNK4 protein in adipose tissues of the WNK4 knock-in mice carrying PHAII mutation (WNK4D561A/+; Fig. S10a) and in the WNK4 transgenic (WNK4-Tg) mice. However, in contrast to the marked increase of WNK4 protein in the kidneys, they unexpectedly showed almost no or a minimal increase of WNK4 in their adipose tissues (Fig. S10b). As a result, WNK4-Tg mice showed similar body weights and insulin sensitivities, even after being fed a HFD (Fig. S10).
4. Discussion
We previously demonstrated the importance of the WNK4–OSR1/SPAK–NCC signaling cascade in the kidney for blood pressure regulation (Uchida, 2014). However, the extrarenal roles of WNK4 still required clarification. In this study, we found that WNK4 regulates expression of adipogenic genes and WNK4−/− mice had reduced adiposity.
Intriguingly, WNK4 in mouse adipose tissues was expressed predominantly in mouse mature adipocytes, rather than in primary preadipocytes (Fig. 1), and the expression was greatly induced with adipocyte differentiation. These data also were confirmed in the 3T3-L1 cells and hMSC-AT cells (Fig. 1 and Fig. S4). Then, WNK4 inhibition impaired the expression of PPARγ and C/EBPα, which are core transcriptional factors to form mature white adipocytes. These results indicated that WNK4 is an important factor for adipocyte differentiation.
Among WNKs, WNK1 and WNK4 were expressed in the adipose tissue (Fig. 1 and Fig. S1). In contrast to the dramatic change of WNK4 during adipocyte differentiation, the protein level of WNK1 was not changed through adipogenesis, and si-WNK1 did not affect the expression of adipogenic genes (Fig. 1 and Fig. S5). These results indicated that only WNK4 is involved in adipogenesis.
We indicated that the increase of WNK4 protein would be due mainly to the increase of WNK4 transcription, since WNK4 mRNA was, indeed, increased and KLHL2/3 or CUL3 mRNA levels were not decreased during 3T3-L1 differentiation. We also showed that WNK4 expression was induced by Dexa (Fig. S2) in 3T3-L1 cells. However, such induction by Dexa was not observed in different cell types (mpkDCT cells). Since microarray analysis revealed that hundreds of genes in 3T3-L1 cells are differently regulated by Dexa (Burton et al., 2004), the mechanism of WNK4 induction in Dexa could be involved in this complex machinery of adipogenesis, rather than a simple transcriptional activation by Dexa (Fig. S2d).
We sought to clarify the regulatory mechanism of adipogenic genes by WNK4. We showed the function of WNK4 in adipocytes that was independent of the WNK-OSR1/SPAK-NKCC1 signaling cascade (Fig. S5). Consistent with this finding, when kinase-dead mutant WNK4 (D318A) was exogenously expressed in 3T3-L1 cells, increased expression of PPARγ and C/EBPα was observed (Fig. S7). It was interesting that si-WNK4 did not affect the expression of C/EBPβ, but did affect the expression of C/EBPα and PPARγ. We also showed that WNK4 inhibition decreased the binding ability of C/EBPβ to the PPARγ2 promoter region, suggesting that WNK4 affected PPARγ transcription by C/EBPβ. Furthermore, we identified significant expression of WNK4 in the nuclei of differentiated 3T3-L1 adipocytes (Fig. 3b). Since WNK1 has been shown previously to be involved in cell division in Hela cells (Tu et al., 2011), we focused our attentions on MCE. Although MCE is considered an essential step in adipogenesis (Guo et al., 2015), its regulatory mechanism is poorly understood. Previously, it has been reported that blocking the cell cycle at the G1-S stage can inhibit MCE and thereby delay subsequent events in adipogenesis (Tang et al., 2003a). Similarly we revealed that si-WNK4 inhibited cell cycle progression during MCE, potentially indicating why the DNA-binding ability of C/EBPβ was affected by WNK4 (Fig. 3c and d), because C/EBPβ is known to obtain its binding ability during MCE (Guo et al., 2015). Thus, we determined that WNK4 in adipocytes had an important positive role in MCE. We believe that these results provide an insight into the regulatory mechanism of adipogenic gene expression by WNK4, and that they also shed light on the regulatory mechanisms of MCE.
In WNK4−/− mice fed a HFD, the expression of PPARγ, C/EBPα, and FABP4 in the adipose tissue was significantly depressed. Furthermore, they had reduced insulin resistance and obesity induced by HFD. It has been reported previously that NCC knockout mice did not show lower body weight on a normal diet (Soleimani et al., 2012), and we have shown that administration of HCTZ did not inhibit the gain of body weight by HFD (Fig. S8). These results indicated that the decreased body weight of WNK4−/− mice was not due to low NCC activity or volume depletion. The association between reduced WNK4 expression and decreased expression of core transcriptional factors regulating adipocyte functions, both in 3T3-L1 adipocytes and in WNK4−/− mice, suggested that there may be an explanation for the observed phenotypes of the knockout mice. Although it is well known that PPARγ agonists improve insulin sensitivity, various studies have indicated that moderate reduction of PPARγ activity prevents insulin resistance and HFD-induced obesity (Kubota et al., 1999, Yamauchi et al., 2001, Jones et al., 2005). Intriguingly, adipose-specific PPARγ-knockout mice exhibited insulin sensitivity, reduced fat development, and resistance to HFD-induced obesity, despite their higher TG levels and lower plasma leptin and adiponectin levels (Jones et al., 2005). These phenotypes of the adipose-specific PPARγ-knockout mice were quite similar to those of our WNK4−/− mice. TG levels of WNK4-KO mice also tended to be higher than those of WT mice (Fig. 5). If we could increase the number of mice examined, the difference might reach statistical significance. Thus, most of the phenotypes observed in WNK4−/− mice could be explained by the reduced expression of PPARγ in adipocytes.
Since WNK4 overexpression accelerated expression of adipogenic genes (Fig. S7), we expected the overexpression of WNK4 in adipocytes would cause the phenotypes opposite to those in WNK4−/− mice. We first checked the WNK4 protein levels in the adipose tissues of WNK4D561A/+ and WNK4-Tg mice, which we have generated previously (Fig. S10). Unexpectedly, however, we found that the increase of WNK4 in adipose tissue of these mice was not so dramatic as we observed previously in the kidney (Figs. S10a and S10b). Indeed, the WNK4-Tg mice showed similar body weights to WT mice at 8 weeks of age and even after the introduction of a HFD (Fig. S10). There were no apparent differences in the metabolic phenotypes between the genotypes. The reason(s) for the unexpected minimal increase in the levels of WNK4 protein in the adipose tissue of WNK4-Tg mice is not clear, but some compensatory mechanisms may be working to prevent excessive increases in WNK4 in tissues other than kidney. Thus, it is not clear at present whether WNK4 overexpression in adipose tissue causes diet-induced obesity. WNK4 in adipose tissue of PHAII patients might not be increases as in the PHAII model mice, which may be consistent with the clinical observation that PHAII patients do not necessarily show metabolic disorders.
In conclusion, we revealed a role for WNK4 as a regulator of core adipogenic factors. Our results may offer insights into the complex process of obesity. Our data indicated that WNK4 inhibition would be beneficial in management of blood pressure and MetS. Since the recently reported pan-WNK inhibitor (Yamada et al., 2016) specifically targets WNK's kinase domain, it may not be effective for inhibiting WNK4’s adipogenic function. The strategy to inhibit WNK4 induction in the early adipogenesis and/or to stimulate WNK4 degradation by the recently identified KLHL3/Cul3 E3 ligase would be possible.
Acknowledgements and Funding Sources
We thank C. Iijima, K. Hashimoto, and A. Imono for help in the experiments. This study was supported in part by Grants-in-Aid for Scientific Research (S) from the Japan Society for the Promotion of Science (No. 25221306), a Health Labor Science Research Grant from the Ministry of Health Labour and Welfare (No. 1211001), the Salt Science Research Foundation (No. 1026, 1228), and the Takeda Science Foundation (No. X2365).
Conflicts of Interest
None.
Author Contributions
D.T. performed the studies, and S.U. designed and directed the project. D.T., T.R. and S.U. wrote the manuscript. M.T. and T.S. performed experiments on mouse primary preadipocytes. T.M., E.S., M.C., Y.I., M.Z., N.N., and T.R. helped with the general experimental procedures and contributed to the data discussion. H.O. and S.T. provided the equipment and supervised the indirect calorimetry experiments.
Footnotes
Supplementary data includes Supplemental Experimental Procedures and figures. Supplementary data associated with this article can be found in the online version, at 10.1016/j.ebiom.2017.03.011.
Appendix A. Supplementary Data
Supplementary material
References
- Ahlstrom R., Yu A.S. Characterization of the kinase activity of a WNK4 protein complex. Am. J. Physiol. Ren. Physiol. 2009;297:F685–F692. doi: 10.1152/ajprenal.00358.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti K.G., Eckel R.H., Grundy S.M., Zimmet P.Z., Cleeman J.I., Donato K.A., Fruchart J.C., James W.P., Loria C.M., Smith S.C., Prevention, I. D. F. T. F. O. E. A, Hational Heart, L. N., and Blood Institute, Association, A. H, Federation, W. H, Society, I. A, Obesity, I. A. F. T. S. O Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- Almind K., Kahn C.R. Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes. 2004;53:3274–3285. doi: 10.2337/diabetes.53.12.3274. [DOI] [PubMed] [Google Scholar]
- Burton G.R., Nagarajan R., Peterson C.A., Mcgehee R.E. Microarray analysis of differentiation-specific gene expression during 3T3-L1 adipogenesis. Gene. 2004;329:167–185. doi: 10.1016/j.gene.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Chiga M., Rafiqi F.H., Alessi D.R., Sohara E., Ohta A., Rai T., Sasaki S., Uchida S. Phenotypes of pseudohypoaldosteronism type II caused by the WNK4 D561A missense mutation are dependent on the WNK-OSR1/SPAK kinase cascade. J. Cell Sci. 2011;124:1391–1395. doi: 10.1242/jcs.084111. [DOI] [PubMed] [Google Scholar]
- Cristancho A.G., Lazar M.A. Forming functional fat: a growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutruel C., Bergmann F., Rooman I., Zucknick M., Weichenhan D., Geiselhart L., Kaffenberger T., Rachakonda P.S., Bauer A., Giese N., Hong C., Xie H., Costello J.F., Hoheisel J., Kumar R., Rehli M., Schirmacher P., Werner J., Plass C., Popanda O., Schmezer P. Early epigenetic downregulation of WNK2 kinase during pancreatic ductal adenocarcinoma development. Oncogene. 2014;33:3401–3410. doi: 10.1038/onc.2013.312. [DOI] [PubMed] [Google Scholar]
- Farmer S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon R.D. The syndrome of hypertension and hyperkalemia with normal glomerular filtration rate: Gordon's syndrome. Aust. NZ J. Med. 1986;16:183–184. doi: 10.1111/j.1445-5994.1986.tb01145.x. [DOI] [PubMed] [Google Scholar]
- Green H., Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- Guo L., Li X., Tang Q.Q. Transcriptional regulation of adipocyte differentiation: a central role for CCAAT/enhancer-binding protein (C/EBP) β. J. Biol. Chem. 2015;290:755–761. doi: 10.1074/jbc.R114.619957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C., Moorefield K.S., Jun P., Aldape K.D., Kharbanda S., Phillips H.S., Costello J.F. Epigenome scans and cancer genome sequencing converge on WNK2, a kinase-independent suppressor of cell growth. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10974–10979. doi: 10.1073/pnas.0700683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z.Y., Zhou Q.L., Holik J., Patel S., Leszyk J., Coleman K., Chouinard M., Czech M.P. Identification of WNK1 as a substrate of Akt/protein kinase B and a negative regulator of insulin-stimulated mitogenesis in 3T3-L1 cells. J. Biol. Chem. 2005;280:21622–21628. doi: 10.1074/jbc.M414464200. [DOI] [PubMed] [Google Scholar]
- Jones J.R., Barrick C., Kim K.A., Lindner J., Blondeau B., Fujimoto Y., Shiota M., Kesterson R.A., Kahn B.B., Magnuson M.A. Deletion of PPARgamma in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6207–6212. doi: 10.1073/pnas.0306743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota N., Terauchi Y., Miki H., Tamemoto H., Yamauchi T., Komeda K., Satoh S., Nakano R., Ishii C., Sugiyama T., Eto K., Tsubamoto Y., Okuno A., Murakami K., Sekihara H., Hasegawa G., Naito M., Toyoshima Y., Tanaka S., Shiota K., Kitamura T., Fujita T., Ezaki O., Aizawa S., Kadowaki T. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol. Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- Lefterova M.I., Zhang Y., Steger D.J., Schupp M., Schug J., Cristancho A., Feng D., Zhuo D., Stoeckert C.J., Liu X.S., Lazar M.A. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen L., Petersen R.K., Sørensen M.B., Jørgensen C., Hallenborg P., Pridal L., Fleckner J., Amri E.Z., Krieg P., Furstenberger G., Berge R.K., Kristiansen K. Adipocyte differentiation of 3T3-L1 preadipocytes is dependent on lipoxygenase activity during the initial stages of the differentiation process. Biochem. J. 2003;375:539–549. doi: 10.1042/bj20030503. [DOI] [PubMed] [Google Scholar]
- Moniz S., Jordan P. Emerging roles for WNK kinases in cancer. Cell. Mol. Life Sci. 2010;67:1265–1276. doi: 10.1007/s00018-010-0261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H., Sohara E., Nomura N., Chiga M., Alessi D.R., Rai T., Sasaki S., Uchida S. Phosphatidylinositol 3-kinase/Akt signaling pathway activates the WNK-OSR1/SPAK-NCC phosphorylation cascade in hyperinsulinemic db/db mice. Hypertension. 2012;60:981–990. doi: 10.1161/HYPERTENSIONAHA.112.201509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M., Uchida K., Ohashi T., Nitta K., Ohta A., Chiga M., Sasaki S., Uchida S. Immunolocalization of WNK4 in mouse kidney. Histochem. Cell Biol. 2011;136:25–35. doi: 10.1007/s00418-011-0827-x. [DOI] [PubMed] [Google Scholar]
- O'reilly M., Marshall E., Macgillivray T., Mittal M., Xue W., Kenyon C.J., Brown R.W. Dietary electrolyte-driven responses in the renal WNK kinase pathway in vivo. J. Am. Soc. Nephrol. 2006;17:2402–2413. doi: 10.1681/ASN.2005111197. [DOI] [PubMed] [Google Scholar]
- Salma N., Xiao H., Mueller E., Imbalzano A.N. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor gamma nuclear hormone receptor. Mol. Cell Biol. 2004;24:4651–4663. doi: 10.1128/MCB.24.11.4651-4663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M., Goto T., Sato A., Shibuya H. WNK4 is an essential effector of anterior formation in FGF signaling. Genes Cells. 2013;18:442–449. doi: 10.1111/gtc.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohara E., Rai T., Yang S.S., Ohta A., Naito S., Chiga M., Nomura N., Lin S.H., Vandewalle A., Ohta E., Sasaki S., Uchida S. Acute insulin stimulation induces phosphorylation of the Na-Cl cotransporter in cultured distal mpkDCT cells and mouse kidney. PLoS One. 2011;6:e24277. doi: 10.1371/journal.pone.0024277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani M., Barone S., Xu J., Shull G.E., Siddiqui F., Zahedi K., Amlal H. Double knockout of pendrin and Na-Cl cotransporter (NCC) causes severe salt wasting, volume depletion, and renal failure. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13368–13373. doi: 10.1073/pnas.1202671109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganami T., Nishida J., Ogawa Y. A paracrine loop between adipocytes and macrophages aggravates inflammatory changes: role of free fatty acids and tumor necrosis factor alpha. Arterioscler. Thromb. Vasc. Biol. 2005;25:2062–2068. doi: 10.1161/01.ATV.0000183883.72263.13. [DOI] [PubMed] [Google Scholar]
- Takahashi D., Mori T., Nomura N., Khan M.Z., Araki Y., Zeniya M., Sohara E., Rai T., Sasaki S., Uchida S. WNK4 is the major WNK positively regulating NCC in the mouse kidney. Biosci. Rep. 2014;34 doi: 10.1042/BSR20140047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi D., Mori T., Wakabayashi M., Mori Y., Susa K., Zeniya M., Sohara E., Rai T., Sasaki S., Uchida S. KLHL2 interacts with and ubiquitinates WNK kinases. Biochem. Biophys. Res. Commun. 2013;437:457–462. doi: 10.1016/j.bbrc.2013.06.104. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Ikeda K., Suganami T., Komiya C., Ochi K., Shirakawa I., Hamaguchi M., Nishimura S., Manabe I., Matsuda T., Kimura K., Inoue H., Inagaki Y., Aoe S., Yamasaki S., Ogawa Y. Macrophage-inducible C-type lectin underlies obesity-induced adipose tissue fibrosis. Nat. Commun. 2014;5:4982. doi: 10.1038/ncomms5982. [DOI] [PubMed] [Google Scholar]
- Tang Q.Q., Lane M.D. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 1999;13:2231–2241. doi: 10.1101/gad.13.17.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q.Q., Otto T.C., Lane M.D. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:850–855. doi: 10.1073/pnas.0337434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q.Q., Otto T.C., Lane M.D. Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:44–49. doi: 10.1073/pnas.0137044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S.W., Bugde A., Luby-Phelps K., Cobb M.H. WNK1 is required for mitosis and abscission. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1385–1390. doi: 10.1073/pnas.1018567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S. Regulation of blood pressure and renal electrolyte balance by Cullin-RING ligases. Curr. Opin. Nephrol. Hypertens. 2014;23:487–493. doi: 10.1097/MNH.0000000000000049. [DOI] [PubMed] [Google Scholar]
- Veríssimo F., Silva E., Morris J.D., Pepperkok R., Jordan P. Protein kinase WNK3 increases cell survival in a caspase-3-dependent pathway. Oncogene. 2006;25:4172–4182. doi: 10.1038/sj.onc.1209449. [DOI] [PubMed] [Google Scholar]
- Wakabayashi M., Mori T., Isobe K., Sohara E., Susa K., Araki Y., Chiga M., Kikuchi E., Nomura N., Mori Y., Matsuo H., Murata T., Nomura S., Asano T., Kawaguchi H., Nonoyama S., Rai T., Sasaki S., Uchida S. Impaired KLHL3-mediated ubiquitination of WNK4 causes human hypertension. Cell Rep. 2013;3:858–868. doi: 10.1016/j.celrep.2013.02.024. [DOI] [PubMed] [Google Scholar]
- Wilson F.H., Disse-Nicodème S., Choate K.A., Ishikawa K., Nelson-Williams C., Desitter I., Gunel M., Milford D.V., Lipkin G.W., Achard J.M., Feely M.P., Dussol B., Berland Y., Unwin R.J., Mayan H., Simon D.B., Farfel Z., Jeunemaitre X., Lifton R.P. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- Yamada K., Park H.M., Rigel D.F., Dipetrillo K., Whalen E.J., Anisowicz A., Beil M., Berstler J., Brocklehurst C.E., Burdick D.A., Caplan S.L., Capparelli M.P., Chen G., Chen W., Dale B., Deng L., Fu F., Hamamatsu N., Harasaki K., Herr T., Hoffmann P., Hu Q.Y., Huang W.J., Idamakanti N., Imase H., Iwaki Y., Jain M., Jeyaseelan J., Kato M., Kaushik V.K., Kohls D., Kunjathoor V., Lasala D., Lee J., Liu J., Luo Y., Ma F., Mo R., Mowbray S., Mogi M., Ossola F., Pandey P., Patel S.J., Raghavan S., Salem B., Shanado Y.H., Trakshel G.M., Turner G., Wakai H., Wang C., Weldon S., Wielicki J.B., Xie X., Xu L., Yagi Y.I., Yasoshima K., Yin J., Yowe D., Zhang J.H., Zheng G., Monovich L. Small-molecule WNK inhibition regulates cardiovascular and renal function. Nat. Chem. Biol. 2016;12:896–898. doi: 10.1038/nchembio.2168. [DOI] [PubMed] [Google Scholar]
- Yamauchi T., Waki H., Kamon J., Murakami K., Motojima K., Komeda K., Miki H., Kubota N., Terauchi Y., Tsuchida A., Tsuboyama-Kasaoka N., Yamauchi N., Ide T., Hori W., Kato S., Fukayama M., Akanuma Y., Ezaki O., Itai A., Nagai R., Kimura S., Tobe K., Kagechika H., Shudo K., Kadowaki T. Inhibition of RXR and PPARgamma ameliorates diet-induced obesity and type 2 diabetes. J. Clin. Invest. 2001;108:1001–1013. doi: 10.1172/JCI12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.S., Morimoto T., Rai T., Chiga M., Sohara E., Ohno M., Uchida K., Lin S.H., Moriguchi T., Shibuya H., Kondo Y., Sasaki S., Uchida S. Molecular pathogenesis of pseudohypoaldosteronism type II: generation and analysis of a Wnk4(D561A/+) knockin mouse model. Cell Metab. 2007;5:331–344. doi: 10.1016/j.cmet.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Zeniya M., Sohara E., Kita S., Iwamoto T., Susa K., Mori T., Oi K., Chiga M., Takahashi D., Yang S.S., Lin S.H., Rai T., Sasaki S., Uchida S. Dietary salt intake regulates WNK3-SPAK-NKCC1 phosphorylation cascade in mouse aorta through angiotensin II. Hypertension. 2013;62:872–878. doi: 10.1161/HYPERTENSIONAHA.113.01543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material