Abstract
Nuclear factor of activated T cells 5 (NFAT5) has been implicated in the pathogenesis of various human diseases, including cancer and arthritis. However, therapeutic agents inhibiting NFAT5 activity are currently unavailable. To discover NFAT5 inhibitors, a library of > 40,000 chemicals was screened for the suppression of nitric oxide, a direct target regulated by NFAT5 activity, through high-throughput screening. We validated the anti-NFAT5 activity of 198 primary hit compounds using an NFAT5-dependent reporter assay and identified the novel NFAT5 suppressor KRN2, 13-(2-fluoro)-benzylberberine, and its derivative KRN5. KRN2 inhibited NFAT5 upregulation in macrophages stimulated with lipopolysaccharide and repressed the formation of NF-κB p65-DNA complexes in the NFAT5 promoter region. Interestingly, KRN2 selectively suppressed the expression of pro-inflammatory genes, including Nos2 and Il6, without hampering high-salt-induced NFAT5 and its target gene expressions. Moreover, KRN2 and KRN5, the latter of which exhibits high oral bioavailability and metabolic stability, ameliorated experimentally induced arthritis in mice without serious adverse effects, decreasing pro-inflammatory cytokine production. Particularly, orally administered KRN5 was stronger in suppressing arthritis than methotrexate, a commonly used anti-rheumatic drug, displaying better potency and safety than its original compound, berberine. Therefore, KRN2 and KRN5 can be potential therapeutic agents in the treatment of chronic arthritis.
Keywords: NFAT5 suppressor, κB inhibitor, Small molecules, High-throughput drug screening, Chronic arthritis
Highlights
-
•
We identify a novel NFAT5 suppressor KRN2, 13-(2-fluoro)-benzylberberine, and its derivative KRN5 to inhibit NFAT5 activity.
-
•
KRN2 inhibits the transcriptional activation of NFAT5 and the pro-inflammatory responses.
-
•
KRN2 and KRN5 suppress experimentally induced arthritis in mice.
NFAT5 has been implicated in the pathogenesis of arthritis. However, therapeutic agents specifically inhibiting NFAT5 activity are currently unavailable. To discover NFAT5 inhibitors, a library of > 40,000 chemicals was screened, leading to the discovery of novel berberine-based NFAT5 suppressors, KRN2 and its oral derivative KRN5. KRN2 inhibited the transcriptional activation of NFAT5 by blocking NF-κB binding to the NFAT5 promoter region, thereby reducing the expression of pro-inflammatory genes. Moreover, KRN2 and KRN5 ameliorated experimentally induced arthritis in mice without serious adverse effects. Therefore, we propose that KRN2 and KRN5 may be potential therapeutic agents in the treatment of chronic arthritis.
1. Introduction
Rheumatoid arthritis (RA) is a chronic autoimmune inflammatory disease characterized by synovial hyperplasia, causing cartilage and bone destruction (Firestein, 1996). The synovial tissues of RA patients contain diverse innate and adaptive immune cells activated by self or non-self antigens (Firestein, 2003). In particular, synovial macrophages are activated by the stimulation of a variety of inflammatory mediators secreted from surrounding inflammatory cells or via cell-to-cell contact. Activated macrophages, in turn, release matrix metalloproteinases and pro-inflammatory cytokines and chemokines, such as interleukin-1 (IL-1), interleukin-6 (IL-6), tumor necrosis factor-α (TNF-α), granulocyte/macrophage colony-stimulating factor (GM-CSF), and monocyte chemoattractant protein (MCP-1), thereby contributing to chronic inflammation (Firestein, 2003, Kinne et al., 2000). Moreover, the number of synovial macrophages, but not the number of lymphocytes, correlates with the progression of RA (Mulherin et al., 1996). In sum, previous studies suggest that macrophages are the major cell type responsible for RA pathology.
Nuclear factor of activated T cells 5 (NFAT5), also known as tonicity-responsive enhancer-binding protein (TonEBP), is a transcription factor whose DNA binding domain shares structural homology with NF-κB and other members of the NFAT family (Lopez-Rodriguez et al., 1999). In response to osmotic stress, NFAT5 is activated via p38 mitogen-activated protein kinase (MAPK) signaling to protect cells from hypertonic stimulation (Ko et al., 2002). Therefore, NFAT5 has important roles in different tissues normally exposed to hypertonicity, such as kidney, skin, and eye (Miyakawa et al., 1999, Go et al., 2004, Neuhofer, 2010, Sawazaki et al., 2014). It has also been implicated in several physiologic and pathologic conditions, including cancer cell proliferation and invasion (Kuper et al., 2014, Jauliac et al., 2002). Recently, evidence has emerged that NFAT5 is activated by isotonic stimuli. For example, NFAT5 induces the expression of toll-like receptor (TLR)-mediated inflammatory genes in macrophages in a tonicity-independent manner (Buxadé et al., 2012, Kim et al., 2013). High salt and TLR ligation activate distinct sets of downstream target genes in a NFAT5-dependent manner (Kim et al., 2014). While ROS are essential for this, their source differs depending on the context: mitochondria for high salt and xanthine oxidase for TLR (Kim et al., 2014). Moreover, the two pathways are mutually suppressive (Kim et al., 2013). Therefore, to apply anti-NFAT5 therapies to chronic inflammatory diseases, it may be necessary to selectively inhibit its inflammatory effects without affecting its osmotic effects since the latter are involved in cellular homeostasis and cytoprotection (Miyakawa et al., 1999).
In a previous study, we firstly uncovered the crucial role of NFAT5 in the development of autoimmune disease by demonstrating that NFAT5-deficient mice showed a marked reduction of antibody-induced arthritis (Yoon et al., 2011). Moreover, we have demonstrated that NFAT5 is highly expressed in the synovia of RA patients and regulates synoviocyte proliferation and angiogenesis (the so-called pannus formation), the pathologic hallmark of RA (Yoon et al., 2011). We also identified a significant decrease in the incidence of TLR-induced chronic arthritis in NFAT5 haplo-insufficient mice as compared to their wild-type littermates (Kim et al., 2014). Taken together, our previous results strongly suggest that NFAT5 is crucial in RA pathogenesis and should be further studied for the development of RA treatment. Despite the pivotal role of NFAT5 in the pathogenesis of cancer and arthritis (Kuper et al., 2014, Jauliac et al., 2002, Yoon et al., 2011, Kim et al., 2014), specific drugs that inhibit NFAT5 activity, especially those selectively targeting the inflammatory effects of NFAT5, are currently unavailable.
Conventional disease-modifying anti-rheumatic drugs (DMARDs) must be discontinued within the second year in two-thirds of RA patients due to drug toxicity or therapy-independent relapse (Smolen and Aletaha, 2015, van der Kooij et al., 2007). Thus, new therapeutic agents with different modes of action are required for optimal treatment of RA. In the present study, we attempted to identify NFAT5 inhibitors with this in mind. Using high-throughput screening (HTS), > 40,000 compounds were screened with a cell-based inhibition assay for the induction of nitric oxide (NO), a target of NFAT5. We validated the anti-NFAT5 activity of the primary hit compounds using an NFAT5-specific reporter and identified novel NFAT5 suppressors, KRN2 and its derivative KRN5, the latter of which exhibits high oral bioavailability and metabolic stability. KRN2 reduced the expression of inflammatory NFAT5-target genes, including Nos2, Il6, Tnf, and Csf2, in TLR4-stimulated RAW 264.7 macrophages. We also found that the suppressive effect of KRN2 on NFAT5 expression was mediated through inhibition of NF-κB p65 binding to the promoter region of the Nfat5 gene. Interestingly, high salt-induced NFAT5 and its target genes, including Ar, Bgt and Smit, were unaffected by KRN2. Finally, using in vivo models of chronic arthritis, we showed that KRN2 and KRN5 ameliorated arthritis severity, decreasing proinflammatory cytokine production. Therefore, KRN2 and KRN5 demonstrate potential of NFAT5 inhibitors as possible therapeutic agents to treat chronic inflammatory arthritis, including RA.
2. Materials and Methods
2.1. Synthesis of KRN2 and KRN5 From Berberine
Detailed methods are described in the Supplemental methods section.
2.2. High-throughput Screening (HTS)
RAW 264.7 (murine macrophage/monocyte) cells were grown in modified DMEM, and nitrate was measured using Griess reagent as previously reported (Kim et al., 2013, Kim et al., 2014). Briefly, cells were seeded in 96-well plates at a density of 2.5 × 104 cells/well and left overnight. Compound (1 μM) and LPS (100 ng/ml) were added to the cells, which were then incubated at 37 °C, 5% CO2 and 95% humidity for 21 h. Equal volumes of supernatant and Griess reagent (Sigma) were mixed and incubated at room temperature for 15 min. Absorbance was measured at 540 nm with an Envision Multilabel Reader (PerkinElmer). Since there are no reported NFAT5 inhibitors, 1,4-PBIT (S,S′-1,4-phenylene-bis[1,2-ethanediyl]bis-isothiourea) (Cayman), an iNOS enzyme inhibitor, was used as a reference in HTS setup and performance using Biomek FX and ORCA robot systems (Beckman Coulter).
2.3. Isolation and Cell Culture of Murine Macrophages and Splenocytes
Murine peritoneal macrophages were obtained from C57BL/6 mice peritoneally injected with 3% thioglycollate for 3 days. The cells were incubated in RPMI 1640 medium with 10% fetal bovine serum (FBS, Gibco BRL) at 37 °C in a 5% CO2 atmosphere. To isolate splenocytes, lipopolysaccharide (LPS) was peritoneally injected into C57BL/6 mice for 24 h and their spleens were then dissected. After red blood cell (RBC) lysis, isolated splenocytes were incubated in RPMI 1640 supplemented with 10% FBS. RAW 264.7 macrophages were obtained from American Type Culture Collection (ATCC) and were maintained in RPMI 1640 medium supplemented with 10% FBS.
2.4. Cloning of NFAT5 Expression Reporter
To construct an NFAT5 expression reporter system, mouse genomic DNA, encompassing base-pair positions − 3000 to + 1 relative to the start codon of Nfat5, was cloned by PCR using primers (5′-catatgcatacaataaggca-3′ and 5′-tcgactcgcagctcgacccagcc-3′) containing restriction enzyme sites of NdeI and SalI. The 3 kb Nfat5 promoter region, which includes two NF-κB consensus sequences, was transferred into the pEGFP-N1 vector (Clontech). Then the cytomegalovirus promoter was removed using AseI and SalI restriction enzymes (BioLabs). The recombinant reporter gene construct was verified by sequencing (Cosmo Genetech). To analyze the transcriptional activity of NFAT5, the Nfat5 consensus sequence with tandem repeats (3 copies of TGGAAAATTACCG) was inserted into the pEGFP-N1 vector (Clontech) and the pDsRed-Express-N1 vector (Clontech) as described previously (Kim et al., 2013, Kim et al., 2014). To construct the cells that are highly expressed with NFAT5 reporter genes, RAW 264.7 macrophages were seeded to 40–50% confluence in 12-well plates and then transduced with a GFP-NFAT5 promoter reporter using Lipofectamine 2000 (Invitrogen). After 2–3 days, cells were reseeded and then selected with 50 μg/ml geneticin (Invitrogen) for 3 weeks, as previously described (Kim et al., 2013, Kim et al., 2014).
2.5. Flow Cytometry Analysis
Green fluorescence protein (GFP) expression levels were detected using a FACS Canto II system (BD Biosciences). GFP intensity was analyzed using FlowJo software (Tree Star). Data are shown as percentage change in mean fluorescence intensity (% ΔMFI), which was calculated by the following formula: (MFI of treated sample − MFI of untreated sample) × 100 / MFI of untreated sample.
2.6. Quantitative Real-time PCR
Total RNA was isolated with an RNeasy Mini kit according to the manufacturer's protocol (Qiagen). Isolated RNA was reverse-transcribed to cDNA using reverse transcriptase (Takara, Shiga, Japan). Real-time quantitative PCR was performed with a CFX96™ machine (Bio-Rad) using SYBR Green PCR Master Mix (Bio-Rad) and the following primers:
Nfat5 (forward: 5′-cagagctgcagtatgtg-3′ and reverse: 5′-cctctgctttggatttcg-3′),
Il6 (forward: 5′-ttccatccagttgccttcttg-3′ and reverse: 5′-aggtctgttgggagtggtatc-3′),
Csf2 (forward: 5′-cctgggcattgtggtct-3′ and reverse: 5′-gaaatccgcataggtggta-3′),
Tnf (forward: 5′-atagctcccagaaaagcaag-3′ and reverse: 5′-caccccgaagttcagtagac-3′),
Mcp1 (forward: 5′-tctctt cctccaccacctg-3′ and reverse: 5′-ggaaaaatggatccacacct-3′),
Smit (forward: 5′-ccgggcgctctatgacctggg-3′ and reverse: 5′-caaacagagaggcaccaatcg-3′),
Bgt1 (forward: 5′-ctgggagagacgggttttgggtattacatc-3′ and reverse: 5′-ggaccccaggtcgtggat-3′), and
Ar (forward: 5′-agtgcgcattgctgagaactt-3′ and reverse: 5′-gtagctgagtagagtggccatgtc-3′).
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. Gene expression levels were calculated using the comparative 2− ΔΔCt algorithm.
2.7. Fractionation and Western Blot Analysis
RAW 264.7 cells were lysed in RIPA lysis buffer for 15 min at 4 °C. Protein concentrations in the lysates were measured using the Bradford protein assay (Bio-Rad). Electrophoresis was performed using SDS-PAGE, and the blot was transferred to a nitrocellulose membrane (Bio-Rad). The membrane was incubated with the following antibodies: anti-iNOS (1:1000; Santa Cruz Biotechnology), anti-NFAT5 (1:1000; gifted from KHM in Ulsan National Institute of Science and Technology), and anti-β-actin (1:10,000; Abcam). Membranes were visualized with an enhanced chemi-luminescent technique (ECL, Amersham Biosciences). To detect nuclear translocation of NFAT5 and p65, cells were harvested and then incubated in cytoplasmic lysis buffer for 15 min on ice (Kim et al., 2013, Kim et al., 2014). After centrifugation, the supernatant was used as the cytoplasmic fraction. The residual pellet was resuspended in nuclear lysis buffer and centrifuged for 20 min at 12,000 rpm as previously described (Kim et al., 2013, Kim et al., 2014). Each fractionated lysate was analyzed by western blot using antibodies to NFAT5, p65 (Abcam), NMP p84 (Abcam), and α-tubulin (Sigma).
2.8. Enzyme-linked Immunosorbent Assay (ELISA)
Cytokine (IL-6, TNF-α, and GM-CSF) levels in the culture supernatants and in plasma obtained from mice were assessed using ELISA kits according to the manufacturer's instructions (R&D).
2.9. Electrophoretic Mobility Shift Assay (EMSA)
To simulate the interaction of NF-κB p65 to its binding sites in the upstream site (base pairs − 3000 to + 1) of Nfat5 exon 1 in a solid phase, double stranded oligonucleotides encompassing the NF-κB p65 binding site (5′-AGAAAGGGGATTTCCTATAC-3′ for Nfat5 promoter 1 and 5′-ATGAAGGGACTTCCCTTGGG-3′ for Nfat5 promoter 2) and their mutant DNA oligonucleotides (5′-AGAAATTTTATTTCCTATAC-3′ as the mutant DNA for Nfat5 promoter 1 and 5′-ATGAATTTACTTCCCTTGGG-3′ as the mutant DNA for Nfat5 promoter 2) were used as DNA probes. The DNA probes (40 fM) and recombinant p65 (400 ng) were added in 20 μl of 1 × binding buffer supplemented with 50 ng of poly dl/dC provided by Pierce Biotechnology (Rockford) and incubated at 25 °C for 20 min. The protein-DNA complex was separated by 10% polyacrylamide gel using 0.5XTBE running buffer for 2 h and electro-transferred to the PVDF membrane for detection using an EMSA kit (Pierce Biotechnology).
2.10. Chromatin Immunoprecipitation Assay (ChIP Assay)
ChIP assay was performed according to the manufacturer's protocol (Millipore). Briefly, RAW 264.7 macrophages were fixed with 1% formaldehyde for 10 min at 37 °C. Nuclear fractions were isolated using fractionation buffers, and then chromatin was sheared with a sonicator (Misonic 3000). A small aliquot was stored as input DNA. Chromatin-containing lysates were incubated with anti-p65 (p65/NF-κB) antibody (Abcam) and then DNA–protein immunocomplexes were precipitated. DNA samples were extracted with phenol/chloroform and precipitated with ethanol. To analyze the p65/NF-κB binding capacity of the Nfat5 promoter, immunoprecipitated DNA samples were amplified by PCR using primer pairs for the Nfat5 promoter (5′-tttggaggatccctcttcac-3′ and 5′-acaagtcaagaagggccaag-3′) as described previously. Primers for the Nfat5 exon region (5′-gcgagatgatgtcacttcag-3′ and 5′-gtggaagtttgactgtggac-3′) were used as negative controls (Kim et al., 2013, Kim et al., 2014).
2.11. Collagen Induced Arthritis (CIA) and Adjuvant Induced Arthritis (AIA) Mouse Models
To induce adjuvant induced arthritis (AIA) in mice, 2 mg of complete Freund's adjuvant (CFA: Chondrex) was injected intradermally to 8-week-old C57BL/6 mice. KRN2 (3 mg/kg) was given to AIA mice via daily peritoneal injection, and swelling in the ankle and footpad was measured for 2 weeks. To induce Collagen induced arthritis (CIA) in mice, 8-week-old DBA/1J mice were immunized with bovine type II collagen (CII, Chondrex) emulsified in CFA on day 0. On day 14, a booster injection of CII in incomplete Freund's adjuvant (IFA, Chondrex) was given in the left footpad. After one week, KRN2 (3 mg/kg) was injected daily into the CIA mice. Other CII-immunized mice received KRN5 orally (15 mg/kg and 60 mg/kg) on alternate days. During the development of arthritis, clinical severity was scored daily as previously described (Kim et al., 2002). Paws and ankles were harvested on day 40, and the degrees of inflammation, synovial proliferation, and bone destruction were evaluated using a standard scoring protocol as previously described (Kim et al., 2002, Kong et al., 2010).
2.12. Assay for IgG Antibodies to CII
Serum was harvested from each group of mice. Anti-CII IgG levels were measured using a commercially available ELISA kit (Chondrex).
2.13. Hematoxylin and Eosin Staining and Immunohistochemical Staining
Joint tissues of AIA and CIA mice were fixed in saline for 1 day and then decalcified in 5% formic acid for 2 weeks. Decalcified joints were embedded in a paraffin block. Paraffin-embedded tissue blocks were sectioned at 5 μm and then stained with hematoxylin and eosin according to the manufacturer's protocol (Sigma). For immunohistochemical analysis of macrophages, 5 μm tissue sections were deparaffinized in xylene and rehydrated in a graded series of ethanol. After blocking with 10% goat serum for 1 h at room temperature, sections were incubated with an anti-F4/80 macrophage marker antibody (1/50, Santa Cruz Biotechnology) overnight at 4 °C. The sections were then incubated with biotinylated goat anti-rabbit IgG (Santa Cruz Biotechnology). After washing, the sections were incubated with peroxidase-conjugated streptavidin (Vector Laboratories) for 30 min at room temperature, followed by incubation with 3′3-diaminobenzidine tetrahydrochloride (DAB, Vector Laboratories). The sections were counterstained with hematoxylin.
2.14. Statistical Analysis
Data are shown as mean ± standard deviation (SD). Comparisons between groups were performed by paired or unpaired Mann-Whitney U test. P values < 0.05 were considered to be statistically significant.
2.15. Study Approval
This study was performed with the approval of the institutional review board (CUMC09U034).
3. Results
3.1. Discovery of Novel Small Molecules Inhibiting NFAT5 Activity Using HTS
Other investigators, like ourselves, have demonstrated that NFAT5 is essential for the expression of NO and its synthetic enzyme, the inducible form of nitric oxide synthase (iNOS), in TLR-stimulated macrophages (Buxadé et al., 2012, Kim et al., 2013, Kim et al., 2014). Therefore, using HTS, we screened > 40,000 compounds by assaying for suppression of NO production induced by lipopolysaccharides (LPS) in RAW 264.7 cells. From a number of NFAT5 targets, NO was particularly selected for HTS since NO cannot only be easily determined using a Griess reaction, and thus adequate for drug screening, but also is one of the most over-susceptible targets to NFAT5 activity (Buxadé et al., 2012, Kim et al., 2013, Kim et al., 2014). As a result, we identified 204 small molecules that showed > 30% inhibition of NO at 1 μM concentrations (Fig. 1a). We next tested NFAT5 inhibitory activity on the selected molecules in the presence of LPS using a cell-based reporter system where NFAT5 consensus sequences (3 copies of TGGAAAATTACCG) fused to a green fluorescent protein (GFP) reporter were stably transfected into RAW 264.7 macrophages (Fig. 1b) (Kim et al., 2013). Five small molecules (as indicated by the red bar in Fig. 1b) out of 198 primary hits (6 hits were excluded because of out of stock) were identified as showing marked suppression of NFAT5-dependent reporter activity (Fig. 1b). Finally, we selected the molecule, KRN2 that showed the strongest inhibition of NFAT5 activity with an acceptable toxicity profile (Supplementary Table 1).
Fig. 1.
Discovery of novel small molecules to inhibit NFAT5 activity using HTS. (a) Distribution map indicates small molecules with the inhibitory activity on NO production in LPS-stimulated RAW 264.7 cells using HTS and 204 small molecules with > 30% of inhibition rate were identified as primary hits out of 40,000 compounds. (b) RAW 264.7 cells were transduced stably with NFAT5 consensus sequences fused to GFP reporter construct. The NFAT5-dependent GFP expression was measured in transduced cells stimulated with LPS (1 μg/ml) for 20 h by flow cytometry analysis (left panel). Through this analysis, 5 compounds (as indicated in red bar) with marked suppression of NFAT-dependent reporter activity were identified out of 198 hits compounds (right panel). (c) Chemical structure of KRN2. (d) KRN2 inhibition of NFAT5-dependent reporter activity. RAW 264.7 cells transfected with NFAT5-GFP reporter system were stimulated with LPS (1 μg/ml) for 20 h in the presence of KRN2 or BBR in a dose-dependent manner. The NFAT5-dependent GFP expression was measured by flow cytometry analysis. A representative histogram is shown on the upper panel. The bar graph shows the mean ± SD of three independent experiments. *P < 0.01, **P < 0.001 and versus LPS-stimulated cells. (e) KRN2 suppression of NFAT5 protein expression. NFAT5 protein expression was determined by western blot analysis in the same condition as described in (d). The bar graph shows NFAT5 protein band intensity normalized by β-actin as a loading control using ImageJ software (upper panel). Data are the representative of three independent experiments with similar results. (f) KRN2 suppression of NFAT5 mRNA expression. RAW 264.7 cells were pre-incubated with KRN2 (0.8 μM) or SB253580 (30 μM) for 1 h and then stimulated with LPS (1 μg/ml) for 12 h (upper panel) or 24 h (lower panel). The levels of NFAT5 mRNA (upper) and protein (lower) were determined by real-time PCR and western blot analysis. Data are the mean ± SD of three independent experiments. *P < 0.05 and **P < 0.01 versus LPS-stimulated cells. (g) KRN2 inhibition of NFAT5 translocation to nucleus. RAW 264.7 cells were stimulated with LPS (1 μg/ml) for 12 h in the absence or presence of KRN2 (0.8 μM) and then fractionated into cytosolic (C) and nuclear extracts (N). Translocation of NFAT5 protein was determined by western blot analysis in each fraction. The bar graph represents the nuclear NFAT5 levels normalized by the cytosolic NFAT5 levels using ImageJ software. Data are the representative of three independent experiments with similar results. (h) No effect of KRN2 on high salt-induced NFAT5 expression. RAW 264.7 cells were pre-incubated with KRN2 (1 μM) or SB203580 (5 μM) for 1 h and then stimulated with NaCl (45 mM) for 12 h. The expression levels of NFAT5 mRNA (upper) and protein (lower) were measured by real-time PCR and western blot assay. Data are expressed as the mean ± SD of three independent experiments. *P < 0.001 versus NaCl-stimulated cells.
As shown in Fig. 1c, KRN2 is 13-(2-fluorobenzyl)-berberine, a non-naturally-occurring quaternary iminium protoberberine. Protoberberines are isoquinoline alkaloids found in many plant families (Grycova et al., 2007). Some of them have been used as traditional phytomedicines because they have a variety of biological and pharmacological activities including inhibition of DNA synthesis, protein synthesis, oxidative phosphorylation and membrane permeability (Grycova et al., 2007, Tillhon et al., 2012). Berberine (BBR), a traditional medicine used in the treatment of gastroenteritis and secretory diarrhea, is known to have potent anti-inflammatory activity as it reduces intracellular superoxide levels in macrophages triggered by TLR (Yan et al., 2012, Sarna et al., 2010, Cheng et al., 2015). The 13-fluorobenzyl derivative KRN2 showed much stronger inhibition of NFAT5-dependent reporter activity in RAW 264.7 macrophages than did BBR; the half maximal inhibitory concentration (IC50) value was 0.1 μM for KRN2 and 4 μM for BBR (Fig. 1d). In parallel, 0.5 μM of KRN2 significantly suppressed the LPS-stimulated increase in NFAT5 protein expression in RAW 264.7 macrophages (Fig. 1e). To summarize, through HTS of > 40,000 compounds, we discovered a novel synthetic protoberberine, KRN2, which shows significant inhibition of NFAT5.
NFAT5 can be activated by isotonic inflammatory stimuli, such as TLR activation, as well as by hypertonic stress (Buxadé et al., 2012, Kim et al., 2013). Cells discriminate between TLR ligation and osmotic stimuli via ROS, directing NFAT5 activity toward proinflammatory or hypertonic responses in a context-dependent manner (Kim et al., 2013, Kim et al., 2014). Thus, we explored whether KRN2 inhibits high salt-induced NFAT5 expression. As seen in Fig. 1f, we confirmed that LPS-induced NFAT5 mRNA and protein expression was nearly completely blocked by KRN2. Similarly, KRN2 inhibited the translocation of NFAT5 into the nucleus of RAW 264.7 cells stimulated with LPS (Fig. 1g). However, KRN2 did not repress the high salt-induced increase in NFAT5 mRNA and protein in RAW 264.7 macrophage, while SB253580, a selective p38 MAP kinase (MAPK) inhibitor (Cuenda et al., 1995), completely blocked it (Fig. 1h). These results indicate that KRN2 selectively inhibits LPS-induced (‘inflammatory’) NFAT5 expression but not high-salt induced (‘osmotic’) NFAT5 expression, suggesting that KRN2 may be effective under LPS-stimulated conditions without altering cellular homeostasis in response to hypertonicity.
The discovery that KRN2 down-regulates LPS-induced NFAT5 mRNA levels suggests that a decrease in NFAT5 reporter activity is mediated by transcriptional inhibition of KRN2. We recently demonstrated that TLR-induced NFAT5 activation required XO-derived ROS and p38 kinase in RAW 264.7 macrophages (Kim et al., 2013, Kim et al., 2014). To further characterize how KRN2 inhibits LPS-induced NFAT5 mRNA transcription, we examined if KRN2 suppression of NFAT5 activity is mediated secondarily via the ROS-p38 MAPK pathway. The result showed that neither ROS nor p38 MAPK activity was affected by KRN2 (Supplementary Fig. 1a, b and c), indicating that ROS and p38 MAPK, upstream regulators of NFAT5, are not involved in the KRN2 inhibition of NFAT5 mRNA transcription. Moreover, KRN2 at a concentration of 1 μM only slightly inhibited NFAT1-dependent reporter activity in HEK293 cells stimulated with PMA (1 μg/ml) (Supplementary Fig. 2a). CREB- and ELK-dependent reporter (luciferase) activities also were not suppressed by KRN2 (Supplementary Fig. 2b, c and d). Collectively, these results suggest that KRN2 selectively regulates LPS-induced NFAT5 mRNA transcription without greatly influencing the aforementioned transcription factors involved in NFAT5 transcription upon TLR4 ligation.
3.2. KRN2 Inhibits Transcriptional Activation of the Inflammatory Nfat5 Gene by Blocking NF-κB Binding to the Nfat5 Promoter
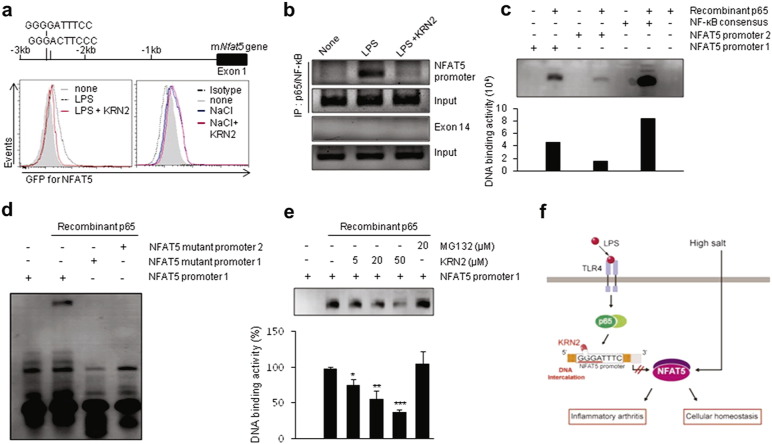
NFAT5 has two NF-κB consensus binding sites upstream of Nfat5 exon 1, and TLR4-induced NFAT5 expression is modulated by the NF-κB pathway in LPS-stimulated macrophages (Buxadé et al., 2012). Since BBR directly interacts with specific DNA sequences (Xu et al., 2012, Mazzini et al., 2003), it is possible that KRN2, a BBR analogue, may interrupt the interaction of the Nfat5 promoter with NF-κB. To address this issue, we monitored the effect of KRN2 on NF-κB binding activity upstream of Nfat5 exon 1. We first constructed a GFP reporter vector that includes the upstream site (base pairs − 3000 to + 1) of Nfat5 exon 1, which contains the NF-κB consensus sequences (Fig. 2a, upper panel). Using flow cytometry, we demonstrated that KRN2 specifically repressed LPS-induced NFAT5 promoter activity, whereas it failed to reduce high salt-induced NFAT5 activity in the same cells (Fig. 2a, lower panel), which is consistent with the selective inhibition of TLR4-activated NFAT5, but not hypertonicity-induced NFAT5, by KRN2 (Fig. 1f and h). During the activation of the NF-κB pathway, IκB is phosphorylated and degraded, and then the p65 subunit of NF-κB is phosphorylated and translocated from cytosol to nucleus (Lawrence, 2009). We found that KRN2 neither influenced the phosphorylation and translocation of p65 nor blocked the degradation of IκB (Supplementary Fig. 1d), which suggests that KRN2 directly suppresses the NF-κB binding to its putative binding sites in the Nfat5 promoter, rather than indirectly affecting IκB phosphorylation or NF-κB translocation. To clarify this, the interaction between NF-κB and NF-κB consensus binding sites in the promoter region of Nfat5 was analyzed by chromatin immunoprecipitation (ChIP) assay using an anti-NF-κB p65 antibody. As reported previously (Buxadé et al., 2012), LPS treatment markedly increased NF-κB p65 binding to the Nfat5 promoter in RAW 264.7 macrophages (Fig. 2b). However, treatment with KRN2 nearly completely eliminated the recruitment of NF-κB p65 to the Nfat5 promoter region following LPS stimulation (Fig. 2b), demonstrating the direct inhibitory effect of KRN2 on the interaction between NF-κB p65 and its DNA binding sites.
Fig. 2.
KRN2 selectively represses transcriptional activation of inflammatory NFAT5 by blocking the NF-κB binding to Nfat5 promoter. (a) RAW 264.7 cells transfected with GFP reporter system containing two NF-κB binding sites in the upstream of Nfat5 gene were pre-incubated with KRN2 (1 μM) for 1 h and then stimulated with LPS (1 μg/ml) or NaCl (45 mM) for 20 h. GFP expression level was measured by flow cytometry. Data are the representative of three independent experiments with similar results. (b) RAW 264.7 cells were pre-treated with KRN2 (1 μM) for 1 h and then stimulated with LPS (1 μg/ml) for 12 h. ChIP assay for the NF-κB binding site within the putative Nfat5 promoter region was performed with anti-p65/NF-κB antibody. Exon14 was used as a negative control. Data are the representative of three independent experiments with similar results. (c) Electrophoretic mobility shift assay (EMSA) using recombinant NF-κB p65 and biotinylated DNA probes corresponding to NF-κB binding site in Nfat5 promoter region. Biotinylated 20 mer-DNA oligonucleotides (40 fM of 5′-AGAAAGGGGATTTCCTATAC-biotin-3′ for NF-κB binding site 1 [Nfat5 promoter 1] and 40 fM of 5′-ATGAAGGGACTTCCCTT-GGG-biotin-3′ for NF-κB binding site 2 [Nfat5 promoter 2]) were tested as DNA probes whether they recognize recombinant NF-κB p65 (400 ng). NF-κB consensus sequence (5′-AGTTGAGGGGACTTTCCCAGG-3′) was used as a positive control. The bar graph in the lower panel shows levels of DNA binding activity determined by ImageJ software. Data are representative of three independent experiments with similar results. (d) Sequence specificity in interaction of NF-κB with its DNA binding sites. Four GGGG or three GGG in DNA probes encompassing NF-κB binding site 1 were mutated to TTTT or TTT, respectively, and EMSA was done with the original DNA probe (40 fM) and two mutant probes (40 fM, 5′-AGAAATTTTATTTCCTATAC-3′ for NF-κB binding site 1 [Nfat5 mutant promoter 1] and ATGAATTTACTTCCCTTGGG for NF-κB binding site 2 [Nfat5 mutant promoter 2]). A representative of three independent experiments is shown. (e) KRN2 inhibition of the NF-κB p65 binding to its DNA binding site. Nfat5 promoter 1 probe (40 fM) was pre-incubated with KRN2 (5, 20, 50 μM) or MG132 (20 μM) for 20 min before addition of recombinant p65 (400 ng). The bar graph in the lower panel represents levels of DNA binding activity determined by ImageJ software. Data are mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 versus p65 only without KRN2. (f) Hypothetical scheme illustrating action mechanism of KRN2 for the suppression of pro-inflammatory response and chronic arthritis.
We next sought to investigate whether KRN2 directly interferes with NF-κB binding sites in the Nfat5 promoter. To this end, we assessed the binding activity of NF-κB p65 to 20-base pair DNA probes encompassing the putative NF-κB binding sites versus their mutant DNA (20-mer) using an electrophoretic mobility shift assay (EMSA). As expected, the NF-κB p65 could bind to the DNA probes harboring putative NF-κB binding sites (GGGGATTTCC and GGGACTTCCC), and its binding activity was much stronger with the probes for the upstream NF-κB binding site (Nfat5 promoter 1) than those for the downstream NF-κB binding site (Nfat5 promoter 2) (Fig. 2c). By contrast, substitution of Nfat5 promoter 1 with TTTTATTTCC and TTTACTTCCC resulted in the complete absence of the NF-κB binding activity (Fig. 2d), indicating that GGGG and GGG sequences are required for the interaction of NF-κB p65 with Nfat5 promoter 1 and promoter 2, respectively. We next tested whether KRN2 competes with recombinant p65 in binding with the Nfat5 promoter 1. As seen in Fig. 2e, KRN2 dose-dependently inhibited the NF-κB p65 binding to Nfat5 promoter 1, whereas MG132, which shows inhibitory action on NF-κB by preventing degradation of IκB (Ortiz-Lazareno et al., 2008), did not affect it. Together, these observations indicate that KRN2 directly blocks the interaction between NF-κB p65 and its DNA binding sequence in the upstream site (base pairs − 3000 to + 1) of Nfat5 exon 1.
Collectively, our data indicate that KRN2 selectively inhibits LPS-, but not high salt-, induced NFAT5 mRNA and protein expression at least partially through blocking the interaction of NF-κB with its binding site in the regulatory region of the Nfat5 gene (Fig. 2f).
3.3. KRN2 Suppresses the Expression of Pro-inflammatory Genes Governed by NFAT5
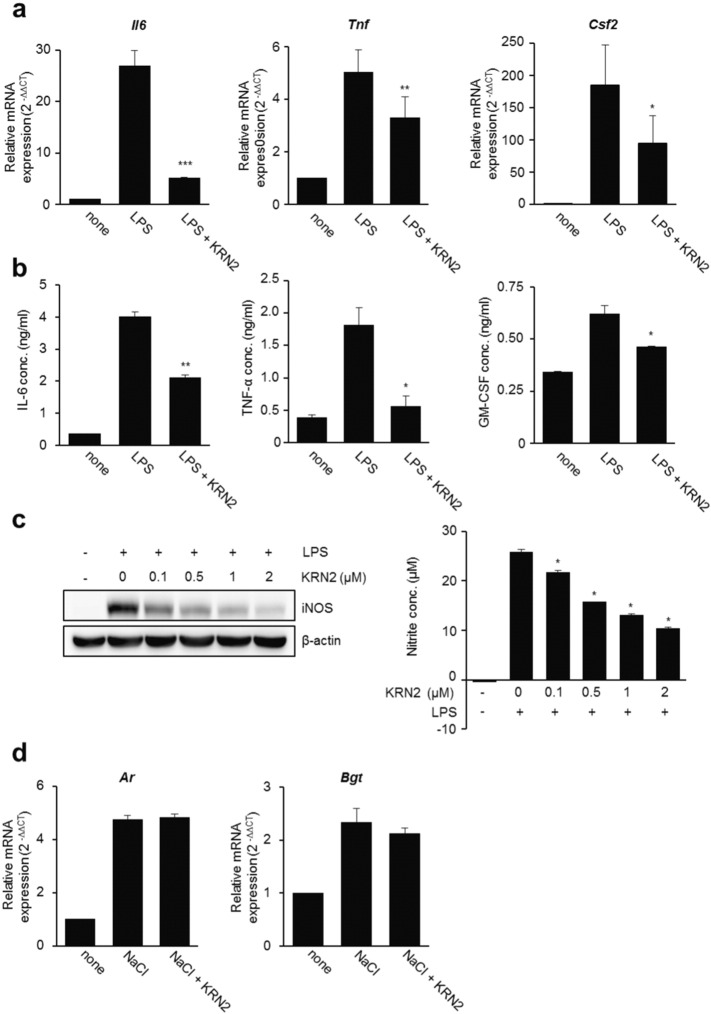
Previous reports have demonstrated that genes induced by TLR4 stimulation, including Nos2, Tnf, Csf2 and Mcp1, are NFAT5-dependent (Buxadé et al., 2012, Kim et al., 2013, Kim et al., 2014). We also have reported that TLR4-stimualted IL-6 production is dependent on NFAT5 activation in RAW 264.7 macrophages (Kim et al., 2013, Kim et al., 2014). Accordingly, we examined the ability of KRN2 to prevent the production of pro-inflammatory cytokines governed by NFAT5. As shown in Fig. 3a and b, the mRNA and protein expression levels of NFAT5 target genes in RAW 264.7 macrophages, including IL-6, TNF-α, and GM-CSF, significantly decreased in the presence of KRN2. Furthermore, KRN2 dose-dependently lowered the expression level of iNOS, another NFAT5-target gene, and NO production by LPS-stimulated RAW 264.7 cells (Fig. 3c). As described above, it is well known that NFAT5 is strongly induced under hypertonic conditions and regulates the tonicity-inducible genes Ar, Bgt, and Smit Miyakawa et al., 1999. Thus, we tested whether the expression levels of ‘osmotic’ (high-salt induced) NFAT5-target genes are suppressed by KRN2. As expected, KRN2 had no effect on the mRNA expression levels of Ar and Bgt in RAW 264.7 macrophages stimulated with high salt (Fig. 3d), which is in agreement with our data that showed no change in high salt-induced NFAT5 expression and reporter activity in the presence of KRN2 (Fig. 2c and d).
Fig. 3.
KRN2 suppresses the expression of pro-inflammatory genes governed by inflammatory NFAT5. RAW 264.7 cells were pre-incubated with KRN2 (0.8 μM) for 1 h and then stimulated with LPS (1 μg/ml) for 12 h for detecting mRNA expression levels or for 20 h for detecting protein expression levels. (a) The mRNA levels of Il6, Tnf, and Csf2 were measured by real-time PCR. Data are expressed as the mean ± SD. *P < 0.05, **P < 0.01 and ***P < 0.001 versus only LPS-stimulated cells. (b) The concentration of pro-inflammatory cytokines (IL-6, TNF-α, and GM-CSF) in the culture supernatants were measured by ELISA. Data are expressed as the mean ± SD. *P < 0.01 and **P < 0.001 versus only LPS-stimulated cells. (c) RAW 264.7 cells were pre-incubated with KRN2 for 1 h in a dose-dependent manner and then stimulated with LPS (1 μg/ml) for 24 h. The expression of iNOS protein was detected by western blot analysis (left panel). The nitrite concentration in the culture supernatants was measured by Griess assay (right panel). Values are the mean ± SD of three independent experiments in triplicate. *P < 0.001 versus only LPS-stimulated cells. (d) RAW 264.7 cells were stimulated with NaCl (45 mM) for 12 h in the absence or presence of KRN2 (0.8 μM). The mRNA levels of Ar and Bgt were measured by real-time PCR. Data are the mean ± SD of three independent experiments in duplicates.
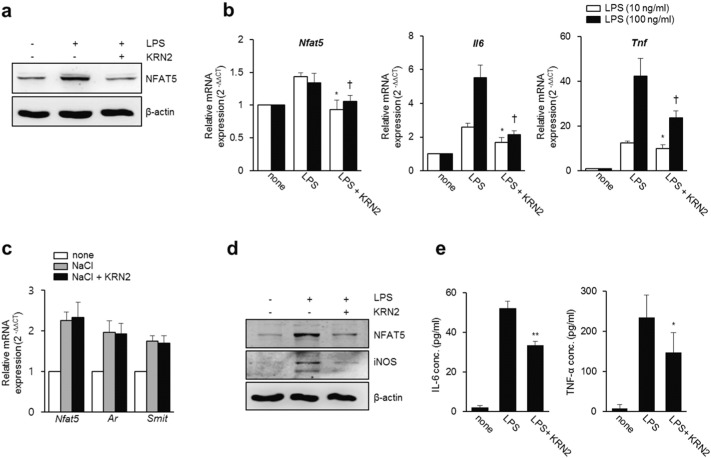
Based on the data in RAW 264.7 macrophages, we next investigated whether KRN2 is similarly effective in suppressing NFAT5 and its pro-inflammatory targets in primary macrophages. Peritoneal macrophages were isolated from mice challenged by injection with 3% thioglycollate and then stimulated with LPS in the presence or absence of KRN2. Consistent with the data in RAW 264.7 cells, KRN2 mitigated the LPS-stimulated increase in expression levels of NFAT5 protein and mRNA in murine peritoneal macrophages (Fig. 4a and b). Moreover, as shown in Fig. 4b, the expression levels of Il6 and Tnf, target genes of inflammatory NFAT5, were also decreased by KRN2 under the same conditions. In contrast, KRN2 neither mitigated the high salt-induced increase in osmotic Nfat5 mRNA expression nor blocked the mRNA expressions of Ar and Smit, NFAT5-dependent tonicity-inducible genes (Fig. 4c). Furthermore, KRN2-induced decreases in iNOS, IL-6, and TNF-α expressions were also reproducible in total splenocytes of mice stimulated with LPS (Fig. 4d and e). Collectively, these results show that KRN2 blocked the expressions of NFAT5 and its pro-inflammatory target genes in primary macrophages and thus may be effective in suppressing chronic inflammatory diseases in vivo.
Fig. 4.
KRN2 inhibits the expression of NFAT5 and its target genes in primary macrophages. (a) Murine peritoneal macrophages were incubated with LPS (1 μg/ml) for 24 h in the absence or presence of KRN2 (0.8 μM). NFAT5 expression levels were determined by western blot analysis. Data are the representative of three independent experiments. (b–c) Murine peritoneal macrophages were stimulated with LPS (10 and 100 ng/ml) (b) or NaCl (45 mM) (c) for 6 h in the absence or presence of KRN2 (0.8 μM). The mRNA expression levels of Nfat5, Il6, Tnf, Ar and Smit were assessed by real-time PCR. Data are the mean ± SD of three independent experiments in duplicates. *P < 0.05 and †P < 0.05 versus LPS alone. (d) Murine splenocytes were incubated with LPS (1 μg/ml) for 24 h in the absence or presence of KRN2 (0.8 μM). The expression levels of NFAT5 and iNOS were determined by western blot analysis. Data are the representative of three independent experiments with similar results. (e) Murine splenocytes were incubated in the same condition as described in (d). The concentration of IL-6 and TNF-α in the culture supernatants were measured by ELISA. Data shows the mean ± SD of three independent experiments. *P < 0.05, **P < 0.005 versus only LPS-stimulated cells.
3.4. KRN2 Inhibits Experimentally Induced Arthritis in Mice
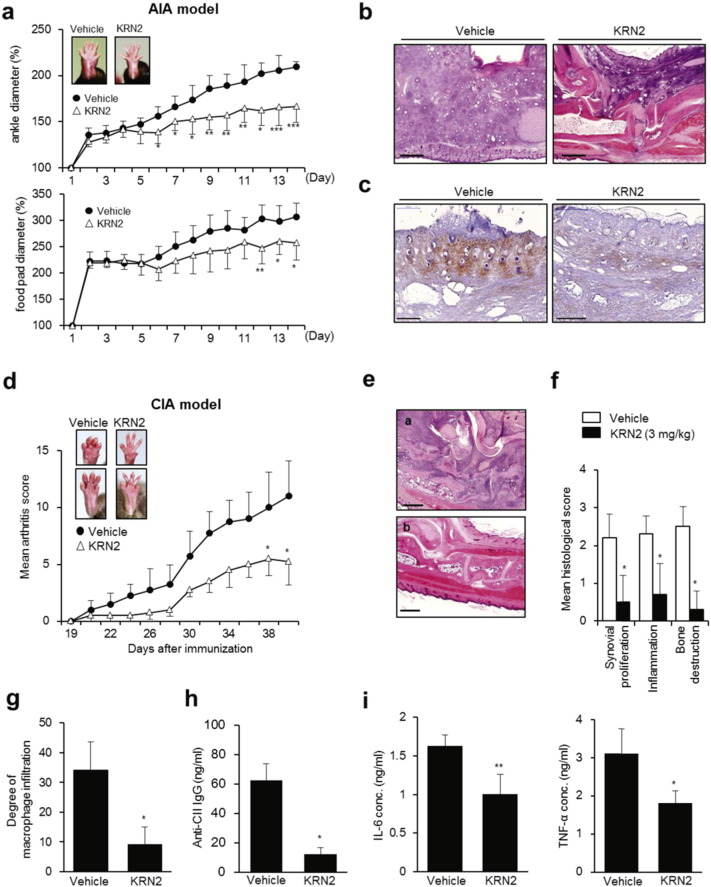
TLR-activated macrophages are crucial in the pathogenesis of chronic arthritis (Huang et al., 2007). To explore the possibility that KRN2 can be used as a therapeutic agent in chronic arthritis, we asked whether KRN2 suppresses the development of adjuvant-induced arthritis (AIA) in mice, a model of chronic arthritis dependent on TLR2 and TLR4 (Billiau and Matthys, 2001). To this end, KRN2 (3 mg/kg) was injected peritoneally into mice every day for 2 weeks. We found that paw swelling, which was assessed by measuring the diameter of arthritic ankles and footpads, was significantly lower in KRN2-injected mice than in vehicle-treated mice (Fig. 5a). Moreover, compared to vehicle-treated mice, KRN2-treated mice exhibited lesser degrees of inflammatory cell infiltration, synovial hyperplasia, and joint destruction, as assayed with hematoxylin and eosin (H&E) staining (Fig. 5b). The number of anti-F4/80 + cells infiltrated, indicating macrophages, also declined in the joints of mice treated with KRN2 (Fig. 5c). Together, these results indicate that KRN2 is effective in suppressing AIA in which innate immune cells play a predominant role (Billiau and Matthys, 2001).
Fig. 5.
KRN2 inhibits experimentally induced arthritis in mice. (a) KRN2 (3 mg/kg) was injected peritoneally into AIA-induced C57BL/6 mice every day for 2 weeks and then the disease severity was assessed by ankle and foot pad thickness index calculated at the indicated time points. Values are the mean ± SD of 6 mice per group. *P < 0.05, **P < 0.01 and ***P < 0.001 versus mice group with vehicle (PBS). (b) Hematoxylin and eosin staining of ankle joint sections obtained from mice treated with vehicle (left) or KRN2 (right). Bar indicates 500 μm. (c) Immunohistochemical staining of joint sections from mice treated with vehicle (left) or KRN2 (right) using anti-F4/80 antibody. (d) Disease severity of CIA-induced DBA/1J mice injected with KRN2 (3 mg/kg) or vehicle (PBS) every day for 3 weeks from day 21 (n = 6, respectively). Values are the mean ± SD. *P < 0.05 versus mice group with vehicle. (e) Hematoxylin and eosin staining of ankle joint sections obtained from mice injected with KRN2 or vehicle. Bar indicates 500 μm. (f) Mean histological scores of synovial proliferation, inflammatory cell infiltration, and joint destruction in mice injected KRN2 versus vehicle (n = 5, respectively). Values are the mean ± SD. *P < 0.0001 versus vehicle mice. (g) Degrees of macrophage infiltration obtained from ankle joint sections in mice injected KRN2 versus vehicle (n = 5, respectively), as determined by immunohistochemical staining using anti-F4/80 antibody. Values are the mean ± SD. *P < 0.001 versus vehicle mice. (h–i) Sera from CIA induced mice injected with KRN2 or vehicle (PBS) (n = 5, respectively) were collected on day 40. Anti-bovine CII IgG level (h) and the concentrations of IL-6 and TNF-α (i) were measured by ELISA. Values are the mean ± SD. *P < 0.005, **P < 0.001 versus mice group with vehicle.
We further analyzed the therapeutic efficacy of KRN2 in mice with collagen-induced arthritis (CIA), a classical animal model of RA in which adaptive immune cells, including T and B lymphocytes, play a dominant role (Brand et al., 2003). When KRN2 (3 mg/kg) was injected peritoneally every day for 3 weeks beginning on day 21, arthritis severity significantly decreased as assessed by visual inspection (Fig. 5d). Moreover, KRN2-injected mice developed a limited degree of inflammatory cell infiltration, synovial hyperplasia, and joint destruction as compared with mice treated with vehicle only (Fig. 5e and f). As assessed with immunohistochemical staining of the joints, the number of anti-F4/80 + macrophages was significantly lower in mice treated with KRN2 than in vehicle-treated mice (Fig. 5g). In parallel, serum levels of anti-type II collagen antibody, IL-6, and TNF-α significantly decreased in mice treated with KRN2, as expected (Fig. 5h and i). Together, these results indicate that KRN2 is effective in suppressing CIA as well as AIA in mice, decreasing the production of pro-inflammatory cytokines and autoantibodies as well as macrophage infiltration.
3.5. KRN5, an Oxo Derivative of KRN2, as an Anti-NFAT5 Drug Candidate
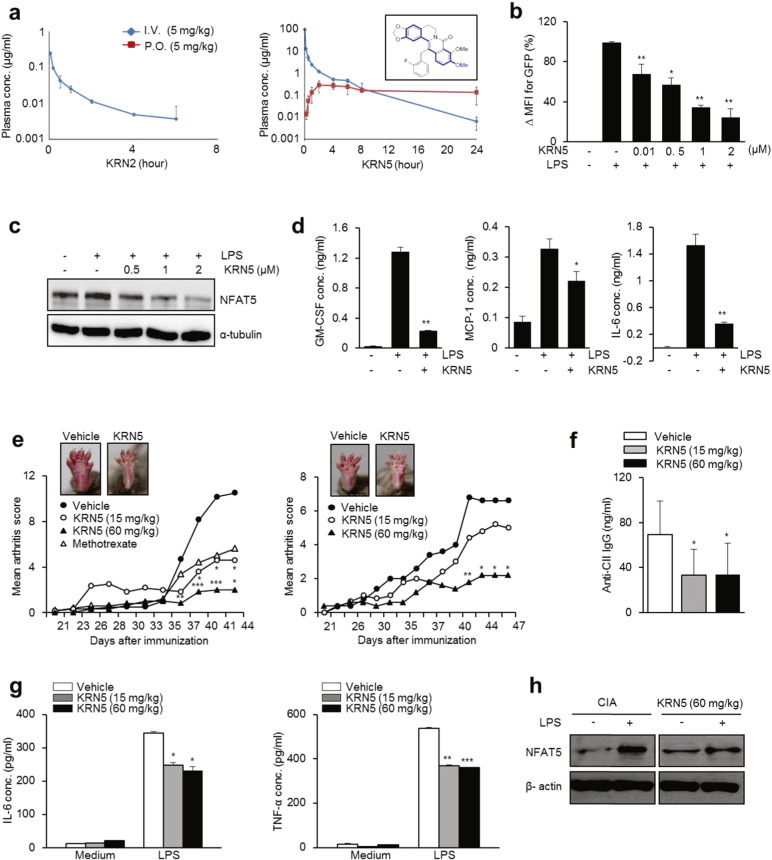
To be easily used in a clinical setting, drugs need to be orally bioavailable. In contrast to intraperitoneally injected KRN2, oral administration of KRN2 (3 mg/kg) every other day for 3 weeks beginning on day 21 failed to suppress CIA development in mice (data not shown). This was primarily due to the lack of absorption of orally administered KRN2 or low microsomal stability (Fig. 6a, left panel and Supplementary Table 2b). In fact, KRN2 was not detectable in plasma after oral administration (Fig. 6a, left panel). To overcome this limitation, we tried to modify the chemical structure of KRN2 and found that oxidation of KRN2 to KRN5, which resulted in removal of iminium positive charges, enhanced its oral bioavailability. As seen in Fig. 6a (right panel), plasma concentration following oral administration remarkably increased with the introduction of the 8-oxo group, and microsomal stability also improved (Supplementary Fig. 3). The plasma half-life of KRN5 was estimated at > 8 h when KRN5 was administered orally (Fig. 6a, right panel), and the bioavailability (F%) after oral and intravenous administration was 15% in rats (Supplementary Table 2b). The IC50 value of KRN5 was 0.75 μM as determined by NFAT5-dependent reporter assay in LPS-stimulated RAW 264.7 cells (Fig. 6b), suggesting that the in vitro NFAT5 inhibitory capacity can be maintained after chemical modification of KRN2 to KRN5. Moreover, KRN5 is less toxic than BBR as determined by a cytotoxicity assay, hERG K + channel assay, cytochrome inhibition assay, and liver microsomal metabolic stability test (Supplementary Table 1), which makes it a potential drug candidate.
Fig. 6.
Pharmacokinetic profiles of KRN5, an oxo derivative of KRN2, and its effects as an anti-NFAT5 drug candidate. (a) Comparison of pharmacokinetic (PK) profiles of KRN2 (left) and KRN5 (right) in rat according to Intravenous (I.V.) or Per os (P.O.) administration. Figure in the box shows the structure of KRN5 (right panel). (b) KRN5 was dose-dependently added to RAW 264.7 cells transfected with NFAT5-GFP reporter system for 1 h. GFP expression was measured by flow cytometry. Values are the mean ± SD for % ΔMFI. *P < 0.005, **P < 0.001. (c) RAW 264.7 cells were stimulated with LPS (1 μg/ml) for 24 h in the presence of KRN5 as indicated doses. NFAT5 protein was detected by western blot analysis. (d) RAW 264.7 cells were stimulated with LPS (1 μg/ml) for 24 h in the presence of KRN5 (1 μM). Level of cytokines (GM-CSF, MCP-1, and IL-6) in the culture supernatants were measured by ELISA. (e) Disease severity of CIA-induced mice (n = 5, respectively) orally administrated with KRN5 (15 and 60 mg/kg) or vehicle for 44 and 47 days. Methotrexate (15 mg/kg) was used as a positive control. Values are the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.005 versus mice group with vehicle. Each graph shows two independent experiments. (f) Sera from CIA-induced mice injected with KRN5 or vehicle were collected on day 44. Anti-bovine CII IgG level was measured by ELISA. Values are the mean ± SD. *P < 0.005 versus mice group with vehicle. (g–h) Mouse splenocytes isolated from CIA-induced mice treated with vehicle or KRN5 (15 mg/kg and 60 mg/kg) were incubated with LPS (1 μg/ml) for 24 h and then the culture supernatants were harvested. (g) IL-6 and TNF-α level in the supernatant were measured by ELISA. Data show the mean ± SD of three independent experiments. *P < 0.05, **P < 0.005 and ***P < 0.001 versus unstimulated cells. (h) NFAT5 protein level was determined by western blot analysis. Data are the representative of three independent experiments with similar results.
KRN5 at a concentration of 1 μM inhibited the expressions of NFAT5, IL-6, MCP-1, and GM-CSF, which are NFAT5 target molecules (Buxadé et al., 2012, Kim et al., 2013, Kim et al., 2014), in RAW 264.7 macrophages stimulated with LPS (Fig. 6c and d), which suggests that the therapeutic efficacy of KRN5 is comparable to that of its starting compound KRN2. To confirm this in vivo, we tested the effect of oral KRN5 on arthritis severity in mice with CIA. As shown in Fig. 6e, oral feeding of KRN5 every other day for 3 weeks from day 21 dose-dependently mitigated arthritis severity. When compared to methotrexate, a commonly used DMARD, the efficacy of oral KRN5 at 60 mg/kg was more potent in suppressing arthritis; a supra-therapeutic dose (5 mg/kg) of methotrexate was administered orally twice a week for the same experimental period. Of special interest, no side effects were noted throughout the course of our experiments (data not shown). The concentration of serum anti-type II collagen IgG also significantly decreased in the sera of KRN5-treated mice (Fig. 6f). In parallel, TNF-α and IL-6 production by LPS-stimulated splenocytes were significantly lower in KRN5-treated CIA mice than in vehicle-treated mice (Fig. 6g). NFAT5 expression in spleen cells stimulated by LPS was also reduced by KRN5 (Fig. 6h). Taken together, these results show that oral administration of KRN5, a KRN2 derivative with enhanced bioavailability and metabolic stability, significantly ameliorated arthritis in mice, suppressing the production of pro-inflammatory cytokines and autoantibodies.
4. Discussion
NFAT5 has been implicated in autoimmune responses and disease states. A recently growing body of research indicates that NFAT5 activity is regulated by either osmotic or pro-inflammatory stress in a context-dependent manner (Kim et al., 2013), suggesting a link between adaptation to hyperosmolarity and immune response. Lymphoid tissues are hyperosmolar compared with blood (Go et al., 2004). Indeed, NFAT5 has a critical role in regulating proliferation and inflammatory responses of macrophages and T lymphocytes, key cellular components of innate and adaptive immunity, respectively (Go et al., 2004, Buxadé et al., 2012, Kim et al., 2013, Lopez-Rodriguez et al., 2001). Elevation of sodium chloride to 40–80 mM, which is similar to the levels found in the interstitium of animals on a high salt diet, promotes the differentiation of CD4+ T cells into Th17 cells, important actors in autoimmune diseases, and enhances production of pathogenic Th17 cytokines and chemokines (Kleinewietfeld et al., 2013, Wu et al., 2013). Furthermore, genetic haplo-insufficiency of NFAT5 expression results in surprisingly dramatic suppression of arthritis severity (Yoon et al., 2011), which makes NFAT5 an attractive therapeutic target of RA.
In the present study, we identified novel NFAT5 suppressors for use in RA therapies. We screened > 40,000 small molecules which suppress NO induction and then validated anti-NFAT5 activity of the candidate molecules using an NFAT5-specific reporter. As a result, we discovered the novel NFAT5 inhibitor KRN2 and its derivative KRN5. KRN2 exhibited 40 times more anti-NFAT5 activity than BBR deprived of its 13-fluorobenzyl group. KRN2 and KRN5 inhibited the expressions of TLR4-stimulated (‘inflammatory’) NFAT5 and its target genes, including Il6, Nos2, and Csf2, in RAW 264.7 macrophages. Interestingly, high salt-stimulated (‘osmotic’) NFAT5 and tonicity-associated genes, including Ar, Bgt and Smit, were unaffected by KRN2, suggesting that KRN2 selectively works under LPS-stimulated conditions, but will not disturb cellular homeostasis and cytoprotection under hypertonic conditions.
To understand the mode of action of KRN2, we investigated the transcriptional regulatory mechanism of KRN2 on NFAT5. We found that KRN2 nearly completely ameliorated the upregulation of NFAT5 mRNA and protein induced by LPS, demonstrating that KRN2-induced decreases in NFAT5 protein expression and nuclear translocation of NFAT5 are caused by its down-regulatory effect on NFAT5 mRNA transcription. To further understand the inhibitory mechanism of KRN2, we checked the activation of representative signaling molecules affecting NFAT5 mRNA transcription, p38 MAPK and NF-κB. p38 MAPK is phosphorylated in response to LPS stimulation (Han et al., 1993), and it then activates various transcription factors (Zarubin and Han, 2005). Especially, p38α has been identified as an important upstream molecule of NFAT5 (Ko et al., 2002, Kim et al., 2014). NF-κB has long been implicated in a pro-inflammatory signaling pathway (Ghosh and Hayden, 2008) and is recently identified as a critical regulator of NFAT5 in macrophages subjected to LPS stimulation (Buxadé et al., 2012, Kim et al., 2014). In this study, we found that p38 MAPK and its isoforms were unaffected by treatment with KRN2. The translocation and phosphorylation of NF-κB p65 upon LPS stimulation were not inhibited by KRN2 either. Furthermore, KRN2 had no effects on ROS expression and Elk-1- and CREB-dependent reporter activity, all of which are associated with NF-κB and p38 MAPK (Wen et al., 2010, Tian and Karin, 1999). Thus, we exclude the possibility that KRN2's actions on NFAT5 are indirect.
BBR is a traditional Chinese medicine used for the treatment of gastroenteritis without serious adverse effects and is known to have potent anti-inflammatory activity (Grycova et al., 2007, Tillhon et al., 2012, Yan et al., 2012, Sarna et al., 2010, Cheng et al., 2015). In this study, we discovered novel BBR-based NFAT5 suppressors to inhibit NFAT5-dependent reporter activity, NFAT5 mRNA and protein expressions, and NFAT5 translocation to nucleus. There are many physicochemical studies on BBR binding to double-stranded DNA (Xu et al., 2012, Mazzini et al., 2003). For example, evidence has shown that BBR binds the minor groove of the AT-rich region [d(AAGAATTCTT)]2 and that this binding is related to its effect on Topoisomerase-I and -II (Mazzini et al., 2003, Kim et al., 1998). Thus, KRN2, a BBR derivative, might interact with NFAT5-binding consensus sequence (TGGAAAATTACCG) through its BBR moiety, decreasing expression of NFAT5 target genes. It has also been demonstrated that BBR directly binds to specific DNA sequences and forms a complex with DNA triplexes or G-quadruplexes [d(TGGGGT)]4 (Xu et al., 2012).
In this study, we postulated that KRN2 could target two NF-κB consensus binding sites, GGGGATTTCC and GGGACTTCCC, located in the Nfat5 promoter site. We discovered BBR-based novel κB inhibitors to suppress NFAT5 expression, which have completely different mode of action from conventional NF-κB inhibitors, such as MG132 (Ortiz-Lazareno et al., 2008), that suppress I-κB expression or activity. Using a GFP reporter vector encompassing the upstream site (base pairs − 3000 to + 1) of Nfat5 exon 1, we demonstrated that KRN2 selectively blocked Nfat5 promoter activity induced by LPS, but not by high salt. Moreover, a ChIP assay using an anti-NF-κB antibody demonstrated a marked reduction of p65 binding to NF-κB consensus binding sites in the Nfat5 promoter. EMSA also revealed that KRN2 decreased the formation of NF-κB p65-DNA complexes in a dose-dependent manner. Given that NFAT5 expression is nearly completely dependent on NF-κB binding to its promoter (Buxadé et al., 2012), our data suggest that KRN2 inhibits transcriptional activation of NFAT5 at least partially through blocking NF-κB binding to the Nfat5 promoter. This notion is bolstered by our transcriptome data showing that NF-κB target genes significantly overlapped with the genes expressed differentially in NFAT5-deficient RAW 264.7 macrophages, particularly in the presence of LPS (Supplementary Fig. 4).
Interestingly, we found that high salt also increased the translocation of p65 as well the degradation of IκB in RAW 264.7 macrophages (Supplementary Fig. 5), which is in parallel with a previous report (Roth et al., 2010). These data indicates that the NF-κB pathway is activated by hypertonicity in these cells. Given the requirement of NF-κB activity for NFAT5 transcription, it is unclear why KRN2 failed to hamper high salt-induced NFAT5 upregulation and its target gene expression, including Ar and Smit. A possible explanation would be the differential activation of co-regulators depending on the kinds of stimuli. We have demonstrated that ROS are essential for NFAT5 transcription, but their source differs depending on the context: mitochondria for high salt and xanthine oxidase for TLR (Kim et al., 2013, Kim et al., 2014). Moreover, the two pathways are mutually exclusive and suppressive (Kim et al., 2013), suggesting that a distinct set of signal regulators can be activated for NFAT5 transcription according to high salt versus LPS. In this regard, other transcriptional factors or co-regulators, in addition to NF-κB, might also be targets of KRN2, since they can bind to the Nfat5 promoter region (− 3000 to + 1) and interact with each other to result in NFAT5 transcription (Buxadé et al., 2012). Further studies are required to clarify this issue.
In arthritic joints, various inflammatory cells, including macrophages, T cells, synoviocytes, and endothelial cells interact with each other via an array of cytokines and/or cell-to-cell contact, leading to prolonged inflammation and destruction of cartilage and bone. As demonstrated in NFAT5 haplo-insufficient mice (Yoon et al., 2011), the NFAT5 blockade by KRN2 and KRN5 successfully repressed experimentally-induced arthritis in mice with AIA and CIA where innate and adaptive immune cells play major roles, respectively, which confirms that NFAT5 is crucial in RA pathogenesis. Given that KRN2 and KRN5 inhibited macrophage activation in both mouse and human systems through suppressing the expression of pro-inflammatory mediators, such inhibition of macrophage activation may explain the therapeutic efficacy of KRN2 and KRN5 in vivo. However, NFAT5 controls antigen-specific T cell proliferation, Th17 cell activation, and Treg cell function (Go et al., 2004, Kleinewietfeld et al., 2013, Wu et al., 2013, Li et al., 2014). NFAT5 is also involved in synoviocyte proliferation and angiogenesis (Yoon et al., 2011), both pathologic hallmarks of RA. We believe that the pharmacological effects of KRN2 and KRN5 are not limited to macrophages in mice with AIA or CIA. Ultimately, the therapeutic effects may be demonstrated to result from the overall action of these small molecules on multiple types of cells, including macrophages, Th17 cells, Treg cells, and synoviocytes.
In sum, 13-(2-fluoro)-benzylberberine, KRN2 inhibits NFAT5-dependent reporter activity, NFAT5 mRNA and protein expression, NFAT5 translocation to the nucleus, and NFAT5 promoter activity. KRN2 exhibits 40 times more anti-NFAT5 activity (IC50 = 0.1 μM) than BBR deprived of its 13-fluorobenzyl group. KRN2 inhibition of transcriptional activation of NFAT5 was at least partially due to the decrease in the formation of NF-κB p65-DNA complexes in the Nfat5 promoter region. This compound selectively suppresses the expression of pro-inflammatory genes regulated by NFAT5 in TLR4-stimulated macrophages without hampering ‘osmotic’ NFAT5 and its target gene expressions. Moreover, KRN2 and its oral derivative KRN5 show suppressive effects on the development of experimental arthritis in mice. Given their high efficacy in arthritic mice, KRN2 and KRN5 should be considered as potential therapeutic agents for treating chronic arthritis, including RA. Particularly, KRN5, as an oral agent, seems to be promising since it was stronger in suppressing arthritis than methotrexate, a commonly used anti-rheumatic drug, displaying better potency and safety than its original compound BBR. In addition, our strategy of discovering NFAT5 suppressors using HTS and an NFAT5-depdendent reporter system can be applied to other chemical libraries to discover the ideal anti-NFAT5 drugs with high specificity and less cytotoxicity than KRN2 and KRN5. We anticipate that the resultant anti-NFAT5 small molecules will provide novel candidates in the treatment of other chronic inflammatory diseases and certain types of cancer in which NFAT5 plays a key role.
Funding Sources
This work was supported by grants from the Korea Healthcare Technology R&D Project, Ministry of Health, Welfare and Family Affairs (No. HI14C3417), the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2015R1A3A2032927, 2014R1A6A3A04054066, and 2015R1C1A2A01055547), and the Korea Research Council for Industrial Science and Technology (KK1303-D0 and KK-1403-D0 to HC). The chemical library used in this study was kindly provided by Korea Chemical Bank (http://www.chembank.org) of Korea Research Institute of Chemical Technology.
Conflicts of Interest
The authors declare no competing financial interests.
Author Contributions
E.J.H. and H.Y.K. performed the experiments. W.U.K. and H.C. designed the experiments and analyzed the data. Y.J.P., T.Q.D., D.Y.J., H.J.L., W.K.P., and G.H.L. provided the compounds. N.H.K., H.M.K. and D.M.J. contributed to data analysis. E.J.H. and N.L. drafted the paper. W.U.K. edited the paper. All authors commented on the manuscript. W.U.K. coordinated the study design and implementation.
Acknowledgments
We would like to thank all members of Center for Integrative Rheumatoid Transcriptomics and Dynamics for their help and advices.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2017.03.039.
Contributor Information
Heeyeong Cho, Email: hycho@krict.re.kr.
Wan-Uk Kim, Email: wan725@catholic.ac.kr.
Appendix A. Supplementary Data
Supplementary material
References
- Billiau A., Matthys P. Modes of action of Freund's adjuvants in experimental models of autoimmune diseases. J. Leukoc. Biol. 2001;70:849–860. [PubMed] [Google Scholar]
- Brand D.D., Kang A.H., Rosloniec E.F. Immunopathogenesis of collagen arthritis. Springer Semin. Immunopathol. 2003;25:3–18. doi: 10.1007/s00281-003-0127-1. [DOI] [PubMed] [Google Scholar]
- Buxadé M., Lunazzi G., Minguillón J., Iborra S., Berga-Bolaños R., Del Val M., Aramburu J., López-Rodríguez C. Gene expression induced by Toll-like receptors in macrophages requires the transcription factor NFAT5. J. Exp. Med. 2012;209:379–393. doi: 10.1084/jem.20111569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W.E., Ying Chang M., Wei J.Y., Chen Y.J., Maa M.C., Leu T.H. Berberine reduces Toll-like receptor-mediated macrophage migration by suppression of Src enhancement. Eur. J. Pharmacol. 2015;757:1–10. doi: 10.1016/j.ejphar.2015.03.013. [DOI] [PubMed] [Google Scholar]
- Cuenda A., Rouse J., Doza Y.N., Meier R., Cohen P., Gallagher T.F., Young P.R., Lee J.C. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- Firestein G.S. Invasive fibroblast-like synoviocytes in rheumatoid arthritis. Passive responders or transformed aggressors? Arthritis Rheum. 1996;39:1781–1790. doi: 10.1002/art.1780391103. [DOI] [PubMed] [Google Scholar]
- Firestein G.S. Evolving concepts of rheumatoid arthritis. Nature. 2003;423:356–361. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Hayden M.S. New regulators of NF-kappaB in inflammation. Nat. Rev. Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- Go W.Y., Liu X., Roti M.A., Liu F., Ho S.N. NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10673–10678. doi: 10.1073/pnas.0403139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grycova L., Dostal J., Marek R. Quaternary protoberberine alkaloids. Phytochemistry. 2007;68:150–175. doi: 10.1016/j.phytochem.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Han J., Lee J.D., Tobias P.S., Ulevitch R.J. Endotoxin induces rapid protein tyrosine phosphorylation in 70Z/3 cells expressing CD14. J. Biol. Chem. 1993;268:25009–25014. [PubMed] [Google Scholar]
- Huang Q., Ma Y., Adebayo A., Pope R.M. Increased macrophage activation mediated through toll-like receptors in rheumatoid arthritis. Arthritis Rheum. 2007;56:2192–2201. doi: 10.1002/art.22707. [DOI] [PubMed] [Google Scholar]
- Jauliac S., López-Rodriguez C., Shaw L.M., Brown L.F., Rao A., Toker A. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat. Cell Biol. 2002;4:540–544. doi: 10.1038/ncb816. [DOI] [PubMed] [Google Scholar]
- Kim S.A., Kwon Y., Kim J.H., Muller M.T., Chung I.K. Induction of topoisomerase II-mediated DNA cleavage by a protoberberine alkaloid, berberrubine. Biochemistry. 1998;37:16316–16324. doi: 10.1021/bi9810961. [DOI] [PubMed] [Google Scholar]
- Kim W.U., Lee W.K., Ryoo J.W., Kim S.H., Kim J., Youn J., Min S.Y., Bae E.Y., Hwang S.Y., Park S.H. Suppression of collagen-induced arthritis by single administration of poly(lactic-co-glycolic acid) nanoparticles entrapping type II collagen: a novel treatment strategy for induction of oral tolerance. Arthritis Rheum. 2002;46:1109–1120. doi: 10.1002/art.10198. [DOI] [PubMed] [Google Scholar]
- Kim N.H., Hong B.K., Choi S.Y., Kwon H.M., Cho C.S., Yi E.C., Kim W.U. Reactive oxygen species regulate context-dependent inhibition of NFAT5 target genes. Exp. Mol. Med. 2013;45 doi: 10.1038/emm.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.H., Choi S., Han E.J., Hong B.K., Choi S.Y., Kwon H.M., Hwang S.Y., Cho C.S., Kim W.U. The xanthine oxidase-NFAT5 pathway regulates macrophage activation and TLR-induced inflammatory arthritis. Eur. J. Immunol. 2014;44:2721–2736. doi: 10.1002/eji.201343669. [DOI] [PubMed] [Google Scholar]
- Kinne R.W., Brauer R., Stuhlmuller B., Palombo-Kinne E., Burmester G.R. Macrophages in rheumatoid arthritis. Arthritis Res. 2000;2:189–202. doi: 10.1186/ar86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinewietfeld M., Manzel A., Titze J., Kvakan H., Yosef N., Linker R.A., Muller D.N., Hafler D.A. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature. 2013;496:518–522. doi: 10.1038/nature11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko B.C., Lam A.K., Kapus A., Fan L., Chung S.K., Chung S.S. Fyn and p38 signaling are both required for maximal hypertonic activation of the osmotic response element-binding protein/tonicity-responsive enhancer-binding protein (OREBP/TonEBP) J. Biol. Chem. 2002;277:46085–46092. doi: 10.1074/jbc.M208138200. [DOI] [PubMed] [Google Scholar]
- Kong J.S., Yoo S.A., Kim J.W., Yang S.P., Chae C.B., Tarallo V., De Falco S., Ryu S.H., Cho C.S., Kim W.U. Anti-neuropilin-1 peptide inhibition of synoviocyte survival, angiogenesis, and experimental arthritis. Arthritis Rheum. 2010;62:179–190. doi: 10.1002/art.27243. [DOI] [PubMed] [Google Scholar]
- van der Kooij S.M., de Vries-Bouwstra J.K., Goekoop-Ruiterman Y.P., van Zeben D., Kerstens P.J., Gerards A.H., van Groenendael J.H., Hazes J.M., Breedveld F.C., Allaart C.F. Limited efficacy of conventional DMARDs after initial methotrexate failure in patients with recent onset rheumatoid arthritis treated according to the disease activity score. Ann. Rheum. Dis. 2007;66:1356–1362. doi: 10.1136/ard.2006.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuper C., Beck F.X., Neuhofer W. NFAT5-mediated expression of S100A4 contributes to proliferation and migration of renal carcinoma cells. Front. Physiol. 2014;5:293. doi: 10.3389/fphys.2014.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Kong L.B., Li J.T., Guo Z.Y., Xue Q., Yang T., Meng Y.L., Jin B.Q., Wen W.H., Yang A.G. MiR-568 inhibits the activation and function of CD4(+) T cells and Treg cells by targeting NFAT5. Int. Immunol. 2014;26:269–281. doi: 10.1093/intimm/dxt065. [DOI] [PubMed] [Google Scholar]
- Lopez-Rodriguez C., Aramburu J., Rakeman A.S., Rao A. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7214–7219. doi: 10.1073/pnas.96.13.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rodriguez C., Aramburu J., Jin L., Rakeman A.S., Michino M., Rao A. Bridging the NFAT and NF-kappaB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress. Immunity. 2001;15:47–58. doi: 10.1016/s1074-7613(01)00165-0. [DOI] [PubMed] [Google Scholar]
- Mazzini S., Bellucci M.C., Mondelli R. Mode of binding of the cytotoxic alkaloid berberine with the double helix oligonucleotide d(AAGAATTCTT)(2) Bioorg. Med. Chem. 2003;11:505–514. doi: 10.1016/s0968-0896(02)00466-2. [DOI] [PubMed] [Google Scholar]
- Miyakawa H., Woo S.K., Dahl S.C., Handler J.S., Kwon H.M. Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2538–2542. doi: 10.1073/pnas.96.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulherin D., Fitzgerald O., Bresnihan B. Synovial tissue macrophage populations and articular damage in rheumatoid arthritis. Arthritis Rheum. 1996;39:115–124. doi: 10.1002/art.1780390116. [DOI] [PubMed] [Google Scholar]
- Neuhofer W. Role of NFAT5 in inflammatory disorders associated with osmotic stress. Curr. Genomics. 2010;11:584–590. doi: 10.2174/138920210793360961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Lazareno P.C., Hernandez-Flores G., Dominguez-Rodriguez J.R., Lerma-Diaz J.M., Jave-Suarez L.F., Aguilar-Lemarroy A., Gomez-Contreras P.C., Scott-Algara D., Bravo-Cuellar A. MG132 proteasome inhibitor modulates proinflammatory cytokines production and expression of their receptors in U937 cells: involvement of nuclear factor-kappaB and activator protein-1. Immunology. 2008;124:534–541. doi: 10.1111/j.1365-2567.2008.02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth I., Leroy V., Kwon H.M., Martin P.Y., Féraille E., Hasler U. Osmoprotective transcription factor NFAT5/TonEBP modulates nuclear factor-kappaB activity. Mol. Biol. Cell. 2010;21:3459–3474. doi: 10.1091/mbc.E10-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarna L.K., Wu N., Hwang S.Y., Siow Y.L., OK Berberine inhibits NADPH oxidase mediated superoxide anion production in macrophages. Can. J. Physiol. Pharmacol. 2010;88:369–378. doi: 10.1139/Y09-136. [DOI] [PubMed] [Google Scholar]
- Sawazaki R., Ishihara T., Usui S., Hayashi E., Tahara K., Hoshino T., Higuchi A., Nakamura S., Tsubota K., Mizushima T. Diclofenac protects cultured human corneal epithelial cells against hyperosmolarity and ameliorates corneal surface damage in a rat model of dry eye. Invest. Ophthalmol. Vis. Sci. 2014;55:2547–2556. doi: 10.1167/iovs.13-13850. [DOI] [PubMed] [Google Scholar]
- Smolen J.S., Aletaha D. Rheumatoid arthritis therapy reappraisal: strategies, opportunities and challenges. Nat. Rev. Rheumatol. 2015;11:276–289. doi: 10.1038/nrrheum.2015.8. [DOI] [PubMed] [Google Scholar]
- Tian J., Karin M. Stimulation of Elk1 transcriptional activity by mitogen-activated protein kinases is negatively regulated by protein phosphatase 2B (calcineurin) J. Biol. Chem. 1999;274:15173–15180. doi: 10.1074/jbc.274.21.15173. [DOI] [PubMed] [Google Scholar]
- Tillhon M., Guaman Ortiz L.M., Lombardi P., Scovassi A.I. Berberine: new perspectives for old remedies. Biochem. Pharmacol. 2012;84:1260–1267. doi: 10.1016/j.bcp.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Wen A.Y., Sakamoto K.M., Miller L.S. The role of the transcription factor CREB in immune function. J. Immunol. 2010;185:6413–6419. doi: 10.4049/jimmunol.1001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Yosef N., Thalhamer T., Zhu C., Xiao S., Kishi Y., Regev A., Kuchroo V.K. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature. 2013;496:513–517. doi: 10.1038/nature11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Yang H., Cui M., Song F., Liu Z., Liu S. Evaluation of alkaloids binding to the parallel quadruplex structure [d(TGGGGT)]4 by electrospray ionization mass spectrometry. J. Mass Spectrom. 2012;47:694–700. doi: 10.1002/jms.2997. [DOI] [PubMed] [Google Scholar]
- Yan F., Wang L., Shi Y., Cao H., Liu L., Washington M.K., Chaturvedi R., Israel D.A., Cao H., Wang B. Berberine promotes recovery of colitis and inhibits inflammatory responses in colonic macrophages and epithelial cells in DSS-treated mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;302:G504–G514. doi: 10.1152/ajpgi.00312.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H.J., You S., Yoo S.A., Kim N.H., Kwon H.M., Yoon C.H., Cho C.S., Hwang D., Kim W.U. NF-AT5 is a critical regulator of inflammatory arthritis. Arthritis Rheum. 2011;63:1843–1852. doi: 10.1002/art.30229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarubin T., Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material