Abstract
HPA axis genes implicated in glucocorticoid regulation play an important role in regulating the physiological impact of social and environmental stress, and have become a focal point for investigating the role of glucocorticoid regulation in the etiology of disease. We conducted a systematic review to critically assess the full range of clinical associations that have been reported in relation to DNA methylation of CRH, CRH-R1/2, CRH-BP, AVP, POMC, ACTH, ACTH-R, NR3C1, FKBP5, and HSD11β1/2 genes in adults. A total of 32 studies were identified. There is prospective evidence for an association between HSD11β2 methylation and hypertension, and functional evidence of an association between NR3C1 methylation and both small cell lung cancer (SCLC) and breast cancer. Strong associations have been reported between FKBP5 and NR3C1 methylation and PTSD, and biologically-plausible associations have been reported between FKBP5 methylation and Alzheimer's Disease. Mixed associations between NR3C1 methylation and mental health outcomes have been reported according to different social and environmental exposures, and according to varying gene regions investigated. We conclude by highlighting key challenges and future research directions that will need to be addressed in order to develop both clinically meaningful prognostic biomarkers and an evidence base that can inform public policy practice.
Keywords: Methylation, HPA axis, Glucocorticoids, NR3C1, HSD11β2, FKBP5, Adverse childhood experiences (ACE), Stress, Hypertension, Cancer, PTSD, Depression, Alzheimer's
Highlights
-
•
Systematic review assessing clinical associations reported in relation to DNA methylation of HPA axis genes in adults.
-
•
Strong associations have been reported for hypertension, SCLC, breast cancer, and Alzheimer's.
-
•
Strong associations have been reported for PTSD, however mixed associations are reported for other mental health outcomes.
1. Introduction
Genes implicated in glucocorticoid regulation play an important role in regulating the physiological impact of social and environmental stress, and have become a focal point for investigating the role of stress in the etiology of disease (Moisiadis and Matthews, 2014). As such, a growing number of studies have identified DNA methylation of genes within the hypothalamic-pituitary-adrenal (HPA) axis as an important mechanism through which exposure to stressful physical and social environments may alter glucocorticoid regulation (Palma-Gudiel et al., 2015, Daskalakis and Yehuda, 2014, Zannas et al., 2016, Needham et al., 2015). What remains unclear, however, is the extent to which such epigenetic regulation might be associated with risk of various clinical diseases. In this systematic review, we outline the full range of clinical associations that have been reported in relation to DNA methylation of HPA axis genes in order to better understand the underlying biological and epigenetic pathways of stress-related disease, and to explore new avenues for diagnosis and treatment.
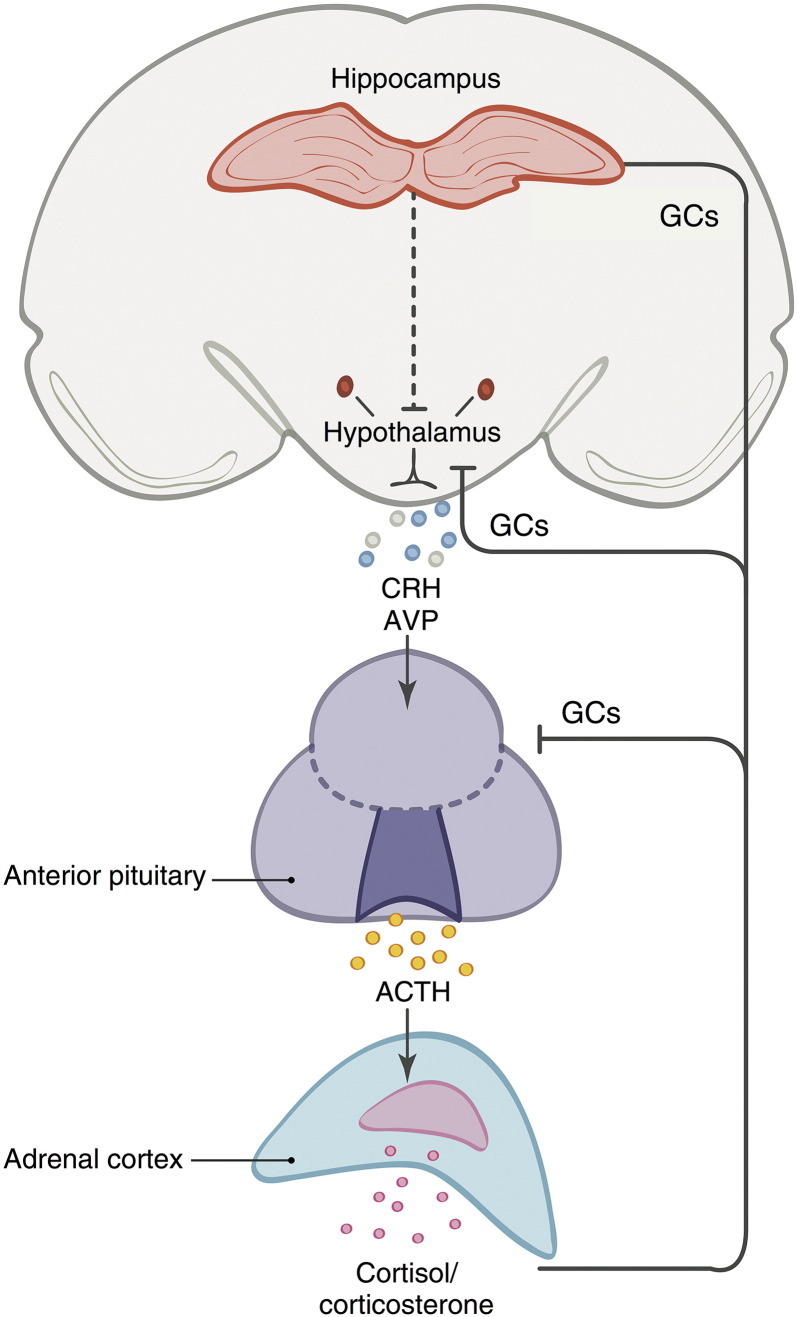
The HPA axis involves several biochemical feedback pathways between the hypothalamus, anterior pituitary gland, and adrenal glands (Fig. 1). It is the major neuroendocrine system that controls stress reactivity and regulates stress hormone (i.e., glucocorticoid) levels within the body. However, as the HPA axis also regulates many fundamental bodily processes such as the immune system, mood and cognition, and the metabolic system (Smith and Vale, 2006), disruption or dysregulation of this system can often increase risk of disease. The pathways and feedback mechanisms of the HPA axis involve interactions between a number of hormones, proteins, and receptors. First, acute stress triggers the release of corticotropin releasing hormone (CRH) and arginine vasopressin (AVP) from the hypothalamus, with circulating levels of CRH largely being regulated by CRH binding proteins (CRH-BP) (Westphal and Seasholtz, 2006). CRH is transported via the hypophyseal portal system of blood vessels to the anterior pituitary gland, where it binds to CRH-R1 and CRH-R2 receptors (Papadopoulou et al., 2005). This causes the pituitary gland to secrete adrenocorticotropic hormone (ACTH; whose precursor is POMC) into the bloodstream, which in turn binds to ACTH-R receptors in the adrenal gland and stimulates the production of glucocorticoids (e.g., cortisol) by the adrenal cortex (Khorram et al., 2011). When high concentrations of glucocorticoids are achieved in the bloodstream (i.e., during stressful experiences), glucocorticoids bind to glucocorticoid receptors (GR) in the hippocampus, hypothalamus, and anterior pituitary gland in order to suppress further release of CRH and ACTH in a negative feedback loop that stabilizes glucocorticoid levels within an appropriate physiological range (Tasker and Herman, 2011). Cortisol levels are further regulated by the HSD11β2 enzyme, which degrades cortisol to cortisone, whereas HSD11β1 catalyzes the conversion of the inactive cortisone to active cortisol (Tomlinson et al., 2004).
Fig. 1.
Overview of the hypothalamus-pituitary-adrenal (HPA) axis. Activation of the HPA axis leads to the production of corticotrophin-releasing hormone (CRH) and arginine vasopressin (AVP) in the hypothalamus. CRH is transported via the hypophyseal portal system of blood vessels to the anterior pituitary gland, which causes the pituitary gland to secrete adrenocorticotropic hormone (ACTH) into the bloodstream. ACTH then stimulates the production of glucocorticoids (e.g., cortisol) by the adrenal cortex. Glucocorticoids (GCs) produced by the adrenal cortex bind to glucocorticoid receptors in the anterior pituitary, hypothalamus, and the hippocampus to regulate production of CRH and ACTH, creating a negative feedback loop that stabilizes circulating levels of stress hormones within an appropriate physiological range.
Adapted from: Schloesser et al. (2012).
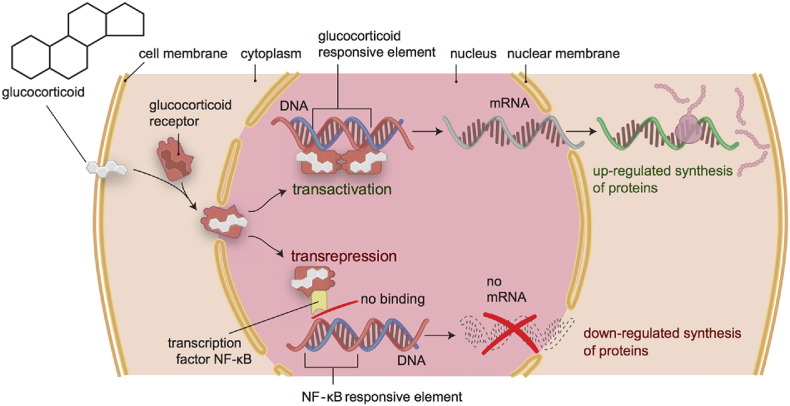
The glucocorticoid receptor (GR) resides in the cell cytoplasm. When bound to glucocorticoids, the GR translocates to the nucleus in a complex of co-chaperones that regulates the synthesis of a number of immune, inflammatory, and metabolic proteins through two genomic mechanisms of action (Fig. 2). First, the GR complex may act as a transcription factor that activates the transcription of immune- and metabolic-related genes by binding directly to glucocorticoid response elements (GREs) in nuclear DNA through a process called transactivation. Second, the GR complex may also repress the transcription of genes that code for immunosuppressive and pro-inflammatory proteins such as cytokines and prostaglandins through a process call transrepression, wherein the GR complex interacts with other transcription factors such as AP-1 and NF-κB to reduce their transcriptional activity without contacting the DNA itself (Ratman et al., 2013, Strehl and Buttgereit, 2013). Binding of the FKBP5 protein to this GR complex lowers the complex's affinity for cortisol and makes nuclear translocation of the GR complex less efficient (Wochnik et al., 2005), while also promoting nuclear translocation of the non-active GRβ splice variant (isoform), and thus decreasing overall GR signaling (Zhang et al., 2008). In this way, overexpression of FKBP5 proteins can lead to glucocorticoid resistance and decreased glucocorticoid sensitivity (Binder, 2009). Expression of FKBP5 mRNA is induced by glucocorticoids, however, leading to an ultra-short negative feedback loop wherein greater circulating levels of glucocorticoids induce greater FKBP5 protein expression, thus regulating GR signaling within an appropriate range (Vermeer et al., 2003).
Fig. 2.
Genomic actions of glucocorticoids (GCs). When bound to GCs, the glucocorticoid receptor (GR) complex translocates to the cell nucleus and modifies the synthesis of a number of immune, inflammatory, and metabolic proteins. This is done through directly binding to glucocorticoid response elements (GREs) in the DNA of genes that code for these proteins (transactivation), and through influencing the activity of transcription factors without contacting the DNA itself (transrepression). Transactivation leads to up-regulated synthesis of immune- and metabolic-related proteins, while transrepression leads to down-regulated synthesis of immunosuppressive and pro-inflammatory proteins.
Source: BioMed Central (van der Goes et al., 2014).
Glucocorticoids also exert non-genomic actions that can occur rapidly within a period of several minutes. This is thought to occur through the activation of signal transduction pathways, or through the interactions of glucocorticoids with cellular membranes (Strehl and Buttgereit, 2013, Kadmiel and Cidlowski, 2013). Through these genomic and non-genomic mechanisms of action, glucocorticoids exert a number of different effects in almost every tissue of the human body. Furthermore, through the above-mentioned interactions and pathways, the HPA axis and glucocorticoids interact with and regulate a number of fundamental physiological systems, including the nervous, cardiovascular, immune, musculoskeletal, visual, reproductive, and integumentary systems, and also play a role in regulating glucose and liver metabolism, mood and cognition, metabolic processes, and maintaining circadian rhythm (Smith and Vale, 2006, Kadmiel and Cidlowski, 2013, Kalsbeek et al., 2012). Not surprisingly, then, the HPA axis has received increasing attention over the past decade due to its critical role in regulating stress and its ability to influence a variety of health outcomes (Moisiadis and Matthews, 2014, Kalsbeek et al., 2012, Turecki, 2014, Eades et al., 2014, Cameron, 2006, Conradt et al., 2013, Edelman et al., 2012, Lee et al., 2014, Wan et al., 2014).
Studies aimed at exploring epigenetic regulation of the genes that code for the hormones, proteins, and receptors within the HPA axis may further our understanding of the pathways through which glucocorticoid dysregulation might increase risk for a number of diseases. Although several studies and reviews have examined the impact of adverse childhood experiences, socioeconomic adversity, and other environmental stressors on epigenetic regulation (most commonly DNA methylation) of individual HPA axis genes, such as NR3C1, AVP, or FKBP5 (Palma-Gudiel et al., 2015, Daskalakis and Yehuda, 2014, Zannas et al., 2016, Needham et al., 2015), no comprehensive review exists that has examined the epigenetic regulation of all HPA axis genes within the entire glucocorticoid regulatory pathway. More importantly, no extant reviews systematically discuss the full range of clinical associations found in relation to epigenetic regulation of these genes. While one recent review has outlined the role of glucocorticoid sensitivity in various diseases (Quax et al., 2013), there also exists no comprehensive review that enumerates the full range of epigenetic pathways that might lead to glucocorticoid dysregulation, and ultimately, disease.
The purpose of this systematic review, therefore, is to critically examine the extant literature on DNA methylation of CRH, CRH-R1/2, CRH-BP, AVP, POMC, ACTH, ACTH-R, NR3C1, FKBP5, and HSD11β1/2 in relation to clinical outcomes in adults. In doing so, our aim is to also highlight current challenges in the field that will need to be addressed in order to develop clinically meaningful prognostic biomarkers, and to outline future research directions needed to create an evidence base that can inform public policy practice.
2. Methods
This systematic review was conducted according to Cochrane PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines (Liberati et al., 2009), and the software used to store information was RevMan 5.3.
2.1. Search Strategy and Selection Criteria
Three databases were used to retrieve studies for this review: PubMed, MEDLINE, and Google Scholar. Our searches were not limited by year of publication because the field of epigenetics is relatively new (the earliest study found was published in 2006).
We searched PubMed for (“methylation”[MeSH Terms] OR “methylation”[All Fields]) in conjunction with the following key search terms: “CRH” (26 results), “CRHR1” (11 results), “CRHR2” (3 results), (“ACTH” OR “adrenocorticotropic”) (68 results), (“ACTHR” OR “MC2”) (2 results), (“GR” OR “hGR” OR “GCR” OR “NR3C1”) (295 results), (“HSD11B1” OR “11β-hydroxysteroid dehydrogenase-1” OR “11-Beta-HSD1” OR “HSD11”) (5 results), (“HSD11B2” OR “11β-hydroxysteroid dehydrogenase-2” OR “11-Beta-HSD2” OR “HSD11”) (25 results), (“Pro-opiomelanocortin” or “POMC”) (100 results), (“tacrolimus binding protein 5”[Supplementary Concept] OR “FKBP5” OR “FK506 binding protein 5” OR “Tacrolimus Binding Proteins/genetics”[Mesh]) (50 results), (“CRHBP” OR “CRH-BP” OR “CRH binding protein”) (4 results), and (AVP OR Vasopressin) (59 results). In total, 648 articles were found on PubMed. No additional studies were found in our Google Scholar of MEDLINE searches. Bibliographic references of retrieved articles were scanned by the authors to identify any final articles to include, although no additional articles were found using this method.
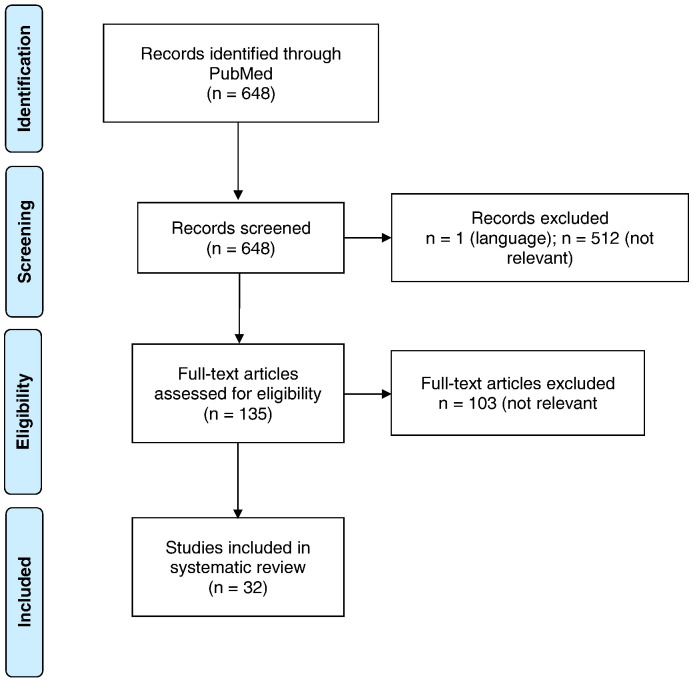
The first and second authors screened the titles and the abstracts of all identified citations and selected potentially eligible studies. Full text articles were then included based on the following inclusion criteria: (1) evidence of quantitatively measured DNA methylation of any of the following: CRH, CRH-R1/2, CRH-BP, AVP, POMC, ACTH, ACTH-R, NR3C1, FKBP5, or HSD11β1/2; (2) recorded evidence of a quantifiable clinical association (e.g., diabetes, hypertension, depression) investigated in direct relation to DNA methylation levels in the above genes; (3) the study was reported in English; and (4) the study included adult human subjects (18 years or older) or used human-derived cell lines. A flow chart of this process is presented in Fig. 3.
Fig. 3.
Flow diagram of study selection.
We excluded studies reported in languages other than English, those using non-human subjects, qualitative studies, duplicates, editorials, case reports, and commentaries. Studies on the mineralocorticoid receptor gene (NR3C2) and both norepinephrine- and epinephrine-related genes were excluded because the focus of our review is on glucocorticoid regulation rather than mineralocorticoid regulation. We have also excluded studies on atrial natriuretic peptide (ANP) in this study, as although it is implicated in HPA activity, it is not directly involved in glucocorticoid regulation (Jessop, 1999). Finally, hormone secreting tumors and paraneoplastic syndromes that naturally dysregulate methylation status in a well-established manner (i.e., pituitary adenomas that secrete ACTH [Ceccato et al., 2015], thymic carcinoids [Mizoguchi et al., 2007], and Cushing syndrome [Newell-Price, 2003]) were excluded because of their ectopic tumor loci, which produce ACTH hormones outside of the HPA axis.
2.2. A Note on the Unique Structure of NR3C1
We did not seek in this review to give a detailed discussion of the methodological differences between studies, given that an insightful examination of this matter can be found in other reviews (Palma-Gudiel et al., 2015, Daskalakis and Yehuda, 2014). We also did not seek to give a detailed characterization of the structural classifications of all genes under study in this paper, since these have been dealt with in excellent detail elsewhere (Daskalakis and Yehuda, 2014, Zannas et al., 2016, Alikhani-Koopaei et al., 2004). However, given that NR3C1 has a rather unique structure in relation to other HPA axis genes included in our review, it is worth providing more detailed information here in order to: (1) contextualize the unique gene region nomenclature used for NR3C1 throughout the research presented in this review; and (2) demonstrate how the unique structure of NR3C1 gives it a transcriptional complexity that may cause epigenetic regulation within different promoter regions to engender different responses to glucocorticoids in a tissue-specific manner.
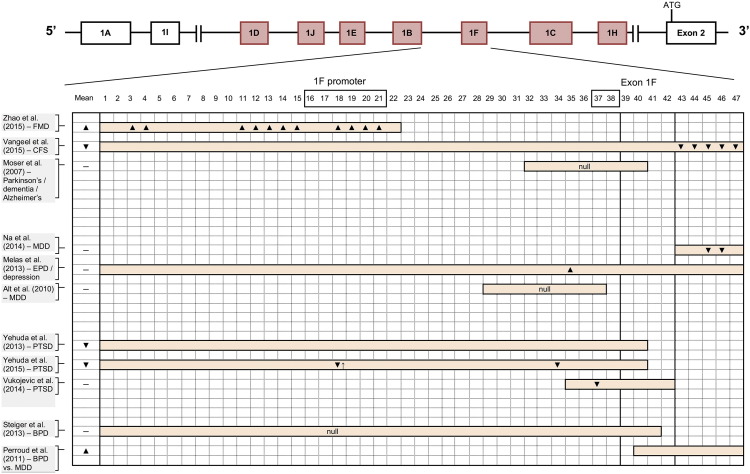
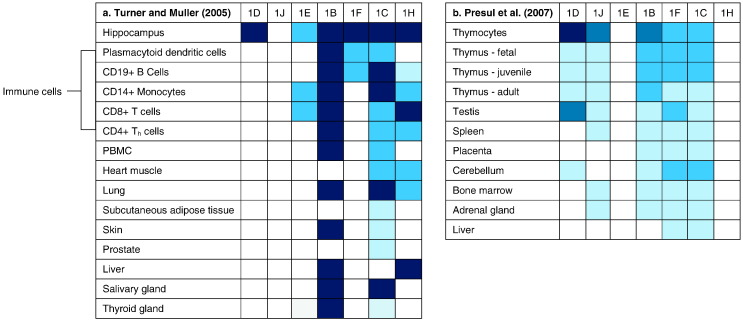
NR3C1 contains 8 coding exons (exons 2–9) and a non-coding 5′ region consisting of 9 non-coding alternative first exons (1A–1J, excluding G) (Fig. 4, Fig. 5). Each of these alternative first exons has its own promoter, which is immediately upstream from the exon itself, and the entire proximal promoter (highlighted in red in Fig. 4, Fig. 5, encompassing exons 1D, 1J, 1E, 1B, 1F, 1C, and 1H) is located within a 3 kb CpG island. This non-coding 5′ region of NR3C1 is largely thought to be responsible for transcriptional regulation of GR protein levels (Turner et al., 2010). Epigenetic research on NR3C1 to date has tended to focus exclusively on the CpG island within the proximal promoter region, with some notable exceptions (Shields et al., 2016). Although the GR protein is ubiquitously expressed throughout all tissues in humans, the different non-coding first exons within the 5′ region of NR3C1 are differentially expressed through alternative gene splicing within various tissues in the body (Fig. 6) (Turner and Muller, 2005, Presul et al., 2007, Cao-Lei et al., 2011). This suggests that these alternative first exons play a role in regulating tissue- and organ-specific responses to glucocorticoids, although the exact role of these first exons remains unknown (Turner et al., 2014).
Fig. 4.
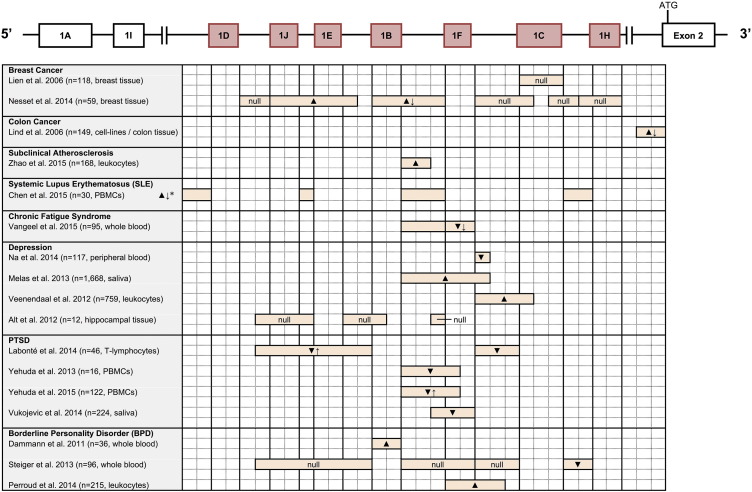
Mean and CpG site-specific methylation results reported for exon 1F and its promoter in NR3C1. First exon variants in red represent the proximal promoter region. ▼ or ▲ denote hypomethylation or hypermethylation, respectively. ↓ or ↑ arrows denote a correlation between observed methylation at a specific CpG site and decreased or increased NR3C1 expression levels, respectively. “Null” denotes no methylation found, or no difference found in methylation between healthy controls and those with the disease under investigation. Boxes around CpG site numbers represent NGFI-A transcription factor binding sites according to McGowan et al. (2009). CpG numbering taken from Palma-Gudiel et al. (2015). Figure is not to scale.
Fig. 5.
Significant methylation results reported across the entire NR3C1 proximal promoter. Methylation results reported only for clinical outcomes that have been significantly associated with methylation status in at least one study. Diseases for which there have only been non-significant findings or for which the authors concluded that NR3C1 was not a likely biomarker are not included. First exon variants in red represent the proximal promoter region. ▼ or ▲ denote hypomethylation or hypermethylation, respectively. ↓ or ↑ arrows denote a correlation between observed methylation and decreased or increased NR3C1 expression levels, respectively. Methylation shown here represents either mean or CpG site-specific methylation, depending on the study method used, and arrows shown here are not meant to indicate methylation at specific CpG loci. Figure is not to scale.
*Mean methylation levels only reported across all 4 first exon promoters, and not within each promoter.
Fig. 6.
Human tissue-specific gene expression of non-coding first exons within the NR3C1 proximal promoter, as reported by Turner and Muller (2005) and Presul et al. (2007). Dark blue to light blue gradients represent strong to weak expression, respectively. Note: exon 1J was discovered by Presul et al., and thus was not measured in Turner and Muller. Presul et al. did not measure exons 1E and 1H.
3. Results
Thirty-two articles were identified as meeting our review inclusion and exclusion criteria. Full details of all studies retrieved in our review are given in Table 1, Table 2, Table 3, Table 4, Table 5; however, we have limited our more detailed discussions in these sections to the high-quality and promising extant research that emerged from our review. Given the exceptional complexity of the HPA axis and its far-reaching interactions with a number of physiological systems, the plausible mechanisms and pathways through which epigenetic regulation of the HPA axis might contribute to disease are understandably varied. We will therefore also provide a critical discussion of the pathways implicated in the results reported across studies, where evidence exists, in order to contextualize the results reported for the many divergent conditions included in this review.
Table 1.
Genomic regions investigated in reviewed studies.
| Authors | Gene | Genomic region tested | Nucleotide position | Exposure/Outcome |
|---|---|---|---|---|
| Friso et al. (2008) | HSD11β2 | Promoter and exon 1 (2 CpG islands analyzed) | Promoter: − 633 to − 97a Exon 1: − 77 to + 460a |
Glucocorticoid treatment/hypertension |
| Pizzolo et al. (2015) | HSD11β2 | Promoter (2 regions analyzed) | − 692 to − 595, and − 419 to − 177 |
AME/hypertension |
| Zhao et al. (2015) | NR3C1 | Exon 1F promoter (22 CpG sites analyzed) | − 3480 to − 3352 | Combat-related PTSD/subclinical atherosclerosis |
| Nesset et al. (2014) | NR3C1 | 6 regions in the proximal promoter:
|
1J: − 4287 to − 4027 1J-E: − 4046 to − 3764 1B: − 3549 to − 3339 1C: − 2868 to − 2665 1C–H: -2294 to − 1997 1H: − 2017 to − 1796 |
None/breast cancer |
| Lien et al. (2006) | NR3C1 | Exon 1C | − 2677 to − 2371 | None/breast cancer |
| Kay et al. (2011) | NR3C1 |
|
(not reported) | None/small cell lung cancer |
| Ahlquist et al. (2008) | NR3C1 | Promoter | (not reported) | None/colorectal cancer |
| Lind et al. (2006) | NR3C1 | Proximal promoter | − 477 to − 25 | None/colon cancer |
| Wu et al. (2007) | NR3C1 | Promoter | (not reported) | None/ovarian cancer |
| Kang et al. (2008) | NR3C1 | Exon 2 | + 168 to + 242 | None/gastric cancer |
| Hiraki et al. (2011) | NR3C1 | Promoter | (not reported) | None/gastric cancer |
| Sanchez-Vega and Gandhi (2009) | NR3C1 | 1B and 1C promoters | (not reported) | None/multiple myeloma |
| Ronco et al. (2010) | HSD11β2 | Promoter (2 CpG islands analyzed) | (not reported) | Cadmium exposure/choriocarcinoma |
| Smyth et al. (2014) | FKBP5 | Epigenome-wide (485,577 unique sites) |
b3 top-ranked CpGs in FKBP5: 5’UTR CpG island south shore: chr6:35,657,202 5’UTR CpG island north shore: chr6:35,654,363 Enhancer: chr6:35,570,224 (CpG island: chr6:35,655,607–35,656,856) |
None/chronic kidney disease |
| Vangeel et al. (2015) | NR3C1 | Exon 1F and 1F promoter (47 CpG sites analyzed) | − 3471 to − 3101 | Childhood trauma/chronic fatigue syndrome |
| Chen et al. (2015) | NR3C1 | Exon 1D, 1E, 1F, and 1H promoters | (not reported) | None/SLE |
| Klengel et al. (2013) | FKBP5 |
|
cCpG island near TSS: chr6:35,763,522–35,764,770 Promoter: chr6:35,798,260–35,798,490 Intron 2: chr6:35,715,732–35,716,027 Intron 5: chr6:35,686,677–35,687,097 Intron 5: chr6:35,677,658–35,677,924 Intron 7: chr6:35,666,288–35,666,763 |
Childhood physical and sexual abuse/PTSD |
| Labonte et al. (2014) | NR3C1 |
|
1B: 479 bp 1C: 364 bp (exact positions not reported) |
None/PTSD |
| Yehuda et al. (2013) |
NR3C1 FKBP5 |
1F promoter and exon 1F (39 CpG sites analyzed)Proximal promoter (38 CpG sites analyzed) | − 3521 to − 3156 − 266 to + 54 |
PE/PTSD |
| Yehuda et al. (2015a) | NR3C1 | 1F promoter and exon 1F (39 CpG sites analyzed) | − 3521 to − 3156 | DEX treatment/PTSD |
| Vukojevic et al. (2014) | NR3C1 | 1F promoter and exon 1F (8 CpG sites analyzed) | − 3236 to − 3030 | Rwandan genocide/PTSD |
| Blair et al. (2013) | FKBP5 |
|
cPromoter: chr6:35,798,260–35,798,490 Intron 2: chr6:35,715,732–35,716,027 Intron 7: chr6:35,666,288–35,666,763 |
None/Alzheimer's disease |
| Moser et al. (2007) | NR3C1 | Small piece of 1F promoter and exon 1F 104 bp around the NGFI-A binding site (9 CpG sites analyzed) | − 3281 to − 3177 | None/Parkinson's disease, presenile and senile dementia-Alzheimer's type, dementia |
| Na et al. (2014) | NR3C1 | 1F promoter and exon 1F (5 CpG sites analyzed) | − 3279 to − 3039 | None/MDD |
| Melas et al. (2013) | NR3C1 | 1F promoter and exon 1F (47 CpG sites analyzed) | − 3480 to − 3126 | Early parental death/depression |
| Veenendaal et al. (2012) | NR3C1 | 1C promoter and partial exon 1C | − 3035 to − 2704 | Prenatal exposure to famine/depression, diabetes, cardiovascular disease |
| Alt et al. (2010) | NR3C1 | 3 regions in the proximal promoter:
|
294 bp (exact position not reported) |
None/MDD |
| Höhne et al. (2015) | FKBP5 | Intron 7 (7 CpG sites analyzed) | chr6:35,666,288–35,666,763c | Childhood averse events, lifetime history of depression/MD |
| Fries et al. (2015) | FKBP5 | Area surrounding glucocorticoid response elements found in 3 regions:
|
cPromoter: chr6:35,798,260–35,798,490 Intron 2: chr6:35,715,732–35,716,027 Intron 7: chr6:35,666,288–35,666,763 |
None/bipolar disorder |
| Dammann et al. (2011) | NR3C1 | Exon 1B | (not reported) | None/borderline personality disorder (BPD) |
| Perroud et al. (2011a) | NR3C1 | 1F promoter and exon 1F (first 8 CpG sites of exon 1F analyzed) |
(not reported) | Childhood maltreatment/BPD, MDD |
| Steiger et al. (2013) | NR3C1 |
|
1B: − 4119 to − 3668 1F: − 3563 to − 3166 1C: − 3035 to − 2692 1H: − 2320 to − 1781 |
None/BN, BPD, Suicidality |
Note: NR3C1 base pair numbering reflects position relative to ATG start codon in exon 2. HSD11β2 and FKBP5 base pair numbering is relative to the transcription start site in exon 1.
Exact base pair locations not given directly in article, but CpG island positions are given in Alikhani-Koopaei et al. (2004).
Reference genome: GRCh37/hg19.
Reference genome: NCBI36/hg18.
Table 2.
Associations between HSD11β2/NR3C1 methylation and cardiovascular diseases.
| Authors | Study design | Gene | Method | Tissue type | Exposure/Outcome | Sample | Results |
|---|---|---|---|---|---|---|---|
| Friso et al. (2008) | Pro-spective | HSD11β2 | Methylation-specific PCR | Peripheral blood (PBMC) | Glucocorticoid treatment/hypertension | N = 57 Italian subjects. 32 glucocorticoid-treated patients normotensive at baseline (mean age: 54.56 years; 4 men). 25 essential hypertensive patients (EH) (mean age: 51.92 years; 14 men) | Glucocorticoid-treated patients who developed hypertension had:
|
| Pizzolo et al. (2015) | Cross-sectional | HSD11β2 | Pyrosequencing | Peripheral blood (PBMC) | AME/hypertension | N = 12 members of the same Italian family. 2 proband brothers (7 and 13 years old) with apparent mineralocorticoid excess (AME; A221G missense mutation) and 10 relatives (age range: 46–71 years), 6 of whom had the A221G mutation | Among the 6 family members heterozygous for 221AG:
|
| Zhao et al. (2015) | Cross-sectional | NR3C1 | Pyrosequencing | Peripheral blood (leukocytes) | Combat-related PTSD/Subclinical atherosclerosis | N = 168 US veterans (84 monozygotic [MZ] twin pairs) (mean age: 55.1 years). The majority of twins were Caucasian (94%) and free of history of cardiovascular disease at enrollment |
|
Table 3.
Associations between NR3C1/HSD11β2 methylation and cancers.
| Authors | Study design | Gene | Method | Tissue type | Exposure/Outcome | Sample | Results |
|---|---|---|---|---|---|---|---|
| Nesset et al. (2014) | Case-control; in vitro cell lines | NR3C1 | Methylation-specific PCR | Breast tissue | None/breast cancer | N = 59 breast cancer patients from Ontario, Canada Cases: tumor tissue samples. Controls: matched normal breast clinical tissue |
|
| Lien et al. (2006) | Cross-sectional | NR3C1 | Methylation-specific PCR | Breast tissue | None/breast cancer | N = 118 GR-immunonegative carcinoma specimens (106 IDCs, 8 ILCs, 1 mucinous carcinoma, 3 DCIS) taken from human breast samples in Taipei City, Taiwan |
|
| Kay et al. (2011) | Case-control (cell line and tissue); prospective | NR3C1 | Bisulfate sequencing | SCLC cell lines | Small cell lung cancer (SCLC) |
Cases: 14 Small Cell Lung Cancer (SCLC) cell lines: Cor L24, Cor L27, Cor L31, Cor L32, Cor L42, Cor L47, Cor L51, Cor L88, Cor L99, Cor L103), DMS 79, DMS 153, HC12, and H 148 Cell line controls: human lung epithelial carcinoma cells A549, human embryonic kidney cells HEK-293, human cervical carcinoma cells HeLa, and human osteosarcoma cells U20S (European Collection of Cell Cultures, Wiltshire, UK). Non-small cell lung cancer (NSCLC) lines NCI-H358 and -H727 (European Collection of Cell Cultures, Wiltshire, UK) and NCI-H23, -H441, -H1299 (American Type Culture Collection, USA) Peripheral blood controls: human peripheral blood mononuclear cells (PBMC) from health donors |
|
| Ahlquist et al. (2008) | Case-control | 11 genes, including NR3C1 | Quantitative methylation-specific PCR | Colorectal cancer (CRC) and normal colon mucosa tissue | None/colorectal cancer |
Cases: 63 adenomas from 52 Norwegian individuals (median age: 67), and 52 carcinomas from 51 patients in 7 hospitals in Oslo, Norway (median age: 70 years) Controls: 21 normal colon mucosa samples (N1) from 20 deceased, cancer-free individuals (median age: 52.5 years), and 18 normal colon mucosa samples (N2) from 18 CRC patients in 7 hospitals in Oslo, Norway (median age: 70.5 years) |
|
| Lind et al. (2006) | Cross-sectional (cell lines); case-control (tissue) | Multiple genes, including NR3C1 | Methylation-specific PCR, bisulfate sequencing | Cell lines, CRC tissue, and normal colon mucosa tissue | Colon cancer |
Cell lines: 20 colon cancer cell lines were included: 9 from microsatellite instable (MSI) tumors (Co115, HCT15, HCT116, LoVo, LS174T, RKO, SW48, TC7, and TC71); 11 from microsatellite stable (MSS) tumors (ALA, Colo320, EB, FRI, HT29, IS1, IS2, IS3, LS1034, SW480, and V9P). 10 cell lines from prostate tissue (N = 3), testicular germ cell tumors (N = 3), and ovary tissues (n = 4) were included, as well as a clinical series of prostate (N = 20) and kidney (N = 20) carcinomas from the University Hospital of Porto, and a series of testicular germ cell tumors (N = 42) from patients admitted to the Norwegian Radium Hospital, Oslo Tissue sample cases: DNA from patients in Norwegian hospitals: 53 colorectal carcinomas (25 MSS and 28 MSI) from 52 patients (mean age 68 years), 63 adenomas (61 MSS and 2 MSI) from 52 patients (mean age 67 years) Tissue sample controls: Normal mucosa samples from 22 colorectal cancer patients (mean age 64 years) taken from distant sites from the primary carcinoma, and 22 normal colorectal mucosa samples from cancer-free individuals (mean age 54 years, including 8 individuals ≥ 60 years) |
Cell line results:
|
| Wu et al. (2007) | Cross-sectional (cell line and tissue) | 13 genes, including NR3C1 | Methylation-specific PCR, bisulfate sequencing | Ovarian cancer cell lines and ovarian tumor tissue | Ovarian cancer |
Cell lines: 4 ovarian carcinoma cell lines, ES-2, OV-90, OVCAR-3, and SKOV-3 (American Type Culture Collection, Manassas, USA) Tissue sample: primary ovarian carcinomas from women (n = 52; mean age: 58 years) displaying a similar distribution regarding FIGO stage and histology. Taken from a tissue bank in Oslo, Norway. Included 19 serous, 5 mucous, 5 clear cell, 17 endometrioid, and 6 of mixed histotype tumors. 2 benign and 2 borderline ovarian tumors were also included |
NR3C1 promoter was unmethylated in ovarian carcinoma tissues and in all cell lines |
| Kang et al. (2008) | Case-control (cell line and tissue) | 17 genes, including NR3C1 | Quantitative methylation-specific PCR (MethyLight) | Gastric cancer cell lines and normal gastric mucosa tissue | Gastric cancer |
Cell line cases: 8 gastric cancer (GC) cell lines treated with 5-aza-deoxycytidine and or Trichostatin A Tissue sample controls: gastric carcinoma-associated non-neoplastic mucosae tissues (GCN) from 33 gastric carcinoma patients in Seoul, Korea (median age: 63 years; male to female ratio 4:1). Normal mucosa tissue from 27 chronic gastritis (CG) patients in Seoul, Korea (median age: 53 years; male to female ratio 19:10). 13 samples were H·Pylori (HP)-negative and 14 samples were HP-positive |
|
| Hiraki et al. (2011) | Case-control (cell line and tissue) | Multiple genes, including NR3C1 | Quantitative methylation-specific PCR (MethyLight) | Gastric cancer cell lines and normal gastric mucosa tissue | Gastric cancer |
Cell lines: 8 gastric cancer cell lines, HSC45, HSC57, KATO III, MKN1, MKN7, MKN28, MKN45, and MKN74 Tissue samples: 20 pairs of adjacent normal gastric mucosa and primary gastric cancer samples. Peritoneal fluid (PF) from 107 patients who underwent surgery in Saga, Japan (median age: 66·5 years; 37 female, 70 male; intestinal type gastric cancer: 46; diffuse type gastric cancer: 61) |
|
| Sanchez-Vega and Gandhi (2009) | Cross-sectional (cell line only) | NR3C1 | 5-aza-deoxycytidine (decitabine) treatment | Myeloma cell lines | Multiple Myeloma | Myeloma cell line (MM.1) taken from peripheral blood cells of a US patient in the leukemic phase of multiple myeloma (MM) being treated with glucocorticoids (GCs). 1 GC-sensitive cell line, MM.1S, and two GC-resistant cell lines, MM.1RE (early phenotype) and MM.1RL (late phenotype) |
|
| Ronco et al. (2010) | Case-control (cell line only) | HSD11β2 | Bisulfate sequencing | Choriocarcinoma cell lines | Chorio-carcinoma | Cases: human choriocarcinoma cells, JEG-3 (HTB-36), from The American Type Culture Collection (ATTC, Manassas, VA) exposed to a low dose of cadmium (Cd2 +)Controls: JEG-3 cells not exposed to Cd2 + |
|
Table 4.
Associations between FKBP5/NR3C1 methylation and renal, metabolic, and inflammatory diseases.
| Authors | Study design | Gene | Method | Tissue type | Exposure/Outcome | Sample | Results |
|---|---|---|---|---|---|---|---|
| Smyth et al. (2014) | Case-control | Epigenome-wide (FKBP5) | Illumina methylation assay (Infinium 450 K BeadChip) | Peripheral blood (leukocytes) | None/chronic kidney disease | N = 407 individuals of white ancestry from the United Kingdom Cases: 255 individuals with chronic kidney disease (CKD) (median age: 36.2 years; 132 male) Controls: 152 individuals with no evidence of renal disease (median age: 36 years; 76 male) |
|
| Vangeel et al. (2015) | Case-control | NR3C1 | Base-specific cleavage/MALDI-TOF mass spectrometry (EpiTYPER) | Peripheral blood (whole blood) | Childhood trauma/chronic fatigue syndrome | N = 95 women from Antwerp, Belgium Cases: 76 female patients with chronic fatigue syndrome (CFS) (mean age: 44.2 years). 46 had no/mild childhood trauma, and 30 with moderate/severe childhood trauma Controls: 19 healthy controls (mean age: 43.2 years) |
|
| Chen et al. (2015) | Case-control | NR3C1 | Methylation-specific PCR | Peripheral blood (PBMC) | None/SLE | N = 30 Chinese patients Cases: 15 newly emerging systemic lupus erythematosus (SLE) patients (mean age: 28·2 years; 13 female) recruited from a Chinese hospital. All SLE patients were steroid-naïve Controls: 15 healthy volunteers (mean age: 27·9 years) |
|
Table 5.
Associations between FKBP5/NR3C1 methylation and mental health outcomes.
| Authors | Study design | Gene | Method | Tissue type | Exposure/Outcome | Sample | Results |
|---|---|---|---|---|---|---|---|
| Klengel et al. (2013) | Case-control; in vitro cell lines | FKBP5 | Pyrosequencing | Peripheral blood (whole blood) | Childhood abuse/PTSD | N = 76 mostly African American U.S. adults from the Grady trauma project Cases: 30 individuals that experienced both physical and sexual child abuse (mean age: 41.46 years; 28 females; 27 African American, 2 white, 1 mixed ethnicity) Controls: 46 individuals without any childhood trauma (mean age: 40.97 years; 36 females; 45 African American, 1 other ethnicity) |
|
| Labonte et al. (2014) | Case-control | NR3C1 | Base-specific cleavage/MALDI-TOF mass spectrometry (EpiTYPER) | Peripheral blood (T-lymphocytes) | None/PTSD |
N = 46 individuals in Montréal, Canada Cases: 30 individuals meeting DSM-IV criteria for lifetime PTSD (average age: 43.4 years; 15 female) Controls: 16 individuals with a negative trauma history (average age: 36.9 years; 8 female), but no lifetime trauma, and no current psychiatric diagnosis |
|
| Yehuda et al. (2013) | Pro-spective | NR3C1, FKBP5 | Bisulfate sequencing (nested PCR) | Peripheral blood (PBMC) | PE/PTSD | N = 16 combat veterans (14 men, 2 women) being treated for PTSD in the Bronx, NY who completed prolonged exposure (PE) psychotherapy. Participants had no significant illness that would interfere with interpretation of biological data, no regular use of benzodiazepines or oral steroids, a BMI < 40, smoked less than two packs per day, no substance abuse or dependence within the last 6 months, and no lifetime history of schizophrenia, schizoaffective disorder, bipolar disorder, obsessive compulsive disorder |
NR3C1:
FKBP5:
|
| Yehuda et al. (2015a) | Case-control | NR3C1 | Bisulfate sequencing (nested PCR) | Peripheral blood (PBMC) | DEX treatment/PTSD |
N = 122 male combat veterans recruited from two hospitals in New York City Cases: 61 veterans with PTSD (mean age: 34.2 years) Controls: 61 veterans without PTSD (mean age: 33 years) |
|
| Vukojevic et al. (2014) | Case-control | NR3C1 | Pyrosequencing | Saliva DNA | Rwandan genocide/PTSD |
N = 224 adults from Rwanda and Switzerland Cases: 152 survivors from the 1994 Rwandan genocide (69 females, 83 males; median age: 35 years) who lived as refugees in Uganda (93 with lifetime PTSD, and 59 individuals without PTSD) Controls: 72 healthy young Swiss subjects (47 females, 25 males; median age: 23 years) |
|
| Blair et al. (2013) | Case-control | FKBP5 | Pyrosequencing | Brain tissue (medial temporal gyrus) | Alzheimer's disease |
N = 15 U.S. elderly adults. Cases: 10 elderly adults with Alzheimer's disease (AD; mean age: 85,6 years; 5 males) Controls: 5 elderly adults without AD (mean age: 92,2 years; 4 males) |
|
| Moser et al. (2007) | Case-control | NRC31 | Methylation-specific PCR | Post-mortem hippocampal tissue | None/Parkinson's disease, presenile and senile dementia-Alzheimer's type, dementia |
N = 32 subjects from Germany Cases: 27 subjects (12 males/20 females, mean age 80·1 years/82·6 years) with various diagnoses: Parkinson's disease (nine), presenile dementia-Alzheimer's type (two), senile dementia- Alzheimer's type (SDAT; eight), dementia (eight) Controls: 5 subjects without any diagnoses |
|
| Na et al. (2014) | Case-control | NR3C1 | Pyrosequencing | Peripheral blood (doesn't specify) | MDD, structural brain alterations | N = 117 participants from Soeul, Korea with no history of comorbid axis I or II disorders, no psychotic features, no history of taking antidepressants or history of primary neurologic diseases, and no organic brain lesions Cases: 45 individuals with major depressive disorder (MDD) (34 female). Controls: 72 healthy individuals (51 male) |
|
| Melas et al. (2013) | Nested case-control | NR3C1 | Base-specific cleavage/MALDI-TOF mass spectrometry (EpiTYPER) | Saliva DNA | Early parental death/depression |
N = 1668 participants from the PART study in Stockholm County, Sweden (male median age: 56; female median age: 58) Cases: 392 individuals diagnosed with major depression, mixed anxiety depression, or dysthymia (285 females and 107 males) Controls: 1276 individuals with no symptom of depression, anxiety (708 females and 568 males) |
|
| Veenendaal et al. (2012) | Case-control | Four genes, including NR3C1 | Methylation-specific PCR | Peripheral blood (leukocytes) | Prenatal exposure to famine/depression, diabetes, cardiovascular disease | N = 759 participants from the Dutch famine birth cohort (mean age: 58 years; 349 [46%] men) Cases: 319 participants who had been exposed to famine in utero Controls: 440 participants who hadn't been exposed to famine |
|
| Alt et al. (2010) | Case-control | NR3C1 | Pyrosequencing | Hippocampus | None/MDD |
N = 12 brain tissue donors from Amsterdam, Netherlands Cases: 6 brain samples from patients with MDD and without documented history of childhood abuse (median age: 70.83 years; 2 females) Controls: 6 brain samples from donors without diagnosed central nervous system (CNS) disease or long-term psychotropic medication (median age: 72.5 years; 3 females) |
|
| Höhne et al. (2015) | Case-control; pro-spective | FKBP5 | Base-specific cleavage/MALDI-TOF mass spectrometry (EpiTYPER) | Peripheral blood (whole blood) | Childhood averse events, lifetime history of depression/major depression |
N = 96 white German participants in the Early Developmental Stages of Psychopathology (EDSP) study Cases: 50 participants with a lifetime DSM-IV diagnosis of major depression (MD) who were in full remission for at least 6 months and without lifetime diagnoses of dysthymia, schizophrenia, substance use disorder, social phobia, or the specific phobia of a blood-injection-injury type Controls: 46 subjects with a negative history of any affective disorder, general anxiety disorder, or any other mental disorder mentioned within the exclusion criteria of the MD sample |
|
| Fries et al. (2015) | Case-control | FKBP5 | Pyrosequencing | Peripheral blood (whole blood) | None/Bipolar disorder | N = 68 Brazilian adults without history of autoimmune diseases, chronic infection/in ammatory disorders, or any severe systemic disease, and use of immunosuppressive therapy Cases: 24 euthymic patients diagnosed with bipolar disorder (BD) according to DSM-IV Axis I criteria (mean age: 46·9 years; 7 males) Controls: 18 siblings of patients with BD (mean age: 51·1 years; 6 males) and 26 non-related healthy controls (mean age: 46·9 years; 8 males) |
|
| Dammann et al. (2011) | Case-control | 14 candidate genes, including NR3C1 | Pyrosequencing | Peripheral blood (whole blood) |
None/BPD |
N = 36 patients from Frauenfeld, Switzerland: Cases: 26 borderline personality disorder (BPD) patients (24 female; mean age 33). Controls: 11 non-BPD individuals (11 female; mean age 32). |
|
| Perroud et al. (2011b) | Cross-sectional, Case-control | NR3C1 | Pyrosequencing | Peripheral blood (leukocytes) | Childhood maltreatment/BPD, MDD |
N = 215 subjects from Geneva, Switzerland Cases: 101 subjects with borderline personality disorder (BPD). These subjects were expected to have a high rate of abuse and maltreatment (mean age: 30.76 years; 95 female [94.06%]) Controls: 99 major depressive disorder (MDD) subjects without past/current PTSD who reported no sexual abuse, no physical abuse and neglect, and no emotional abuse (mean age: 41.63 years; 64 female [64.65%]). 15 MDD subjects with comorbid PTSD (mean age: 37.33 years; 11 female [73.33%]) |
|
| Steiger et al. (2013) | Case-control | NR3C1 | Base-specific cleavage/MALDI-TOF mass spectrometry (EpiTYPER) | Peripheral blood (whole blood) | None/BN, BPD, Suicidality | N = 96 US women between the ages of 17 and 48: Cases: 64 women displaying a Bulimia nervosa (BN)-spectrum disorder Controls: 32 non-eating disordered (NED) women from the same age group with no history of childhood abuse or borderline personality disorder (BPD) |
|
Potential associations between epigenetic regulation of HPA axis genes and clinical outcomes still remain relatively unmapped, as only DNA methylation of NR3C1, HSD11β2, and FKBP5 has been studied in direct relation to risk for any human disease, with the majority of research focusing on epigenetic regulation of NR3C1. Furthermore, the majority of studies vary in terms of gene region(s) or CpG loci selected for DNA methylation analysis (Table 1), although exon 1F or the 1F promoter have been investigated most frequently among NR3C1 methylation studies (17/24; Table 1, Fig. 4, Fig. 5). No studies relevant to our inclusion criteria examined the relationship between DNA methylation of POMC, ACTH, ACTH-R, AVP, CRH, CRH-R1/2, or CRH-BP genes in relation to any clinical outcomes. Notably, three studies were retrieved on POMC methylation in relation to alcohol craving (Muschler et al., 2010), weight regain (Crujeiras et al., 2013), and malnutrition (Ehrlich et al., 2010), but these studies were excluded from the current review because they focused on intermediate outcomes instead of clinically-defined diseased states.
3.1. Cardiovascular Diseases
Three studies identified in our systematic review examined the associations between DNA methylation of either HSD11β2 or NR3C1 and cardiovascular diseases (Table 2).
3.1.1. Hypertension
In a sample of 32 Italian patients who were treated with glucocorticoids, those who developed clinically-diagnosed hypertension had greater HSD11β2 promoter methylation, and a higher urinary tetrahydrocortisol/tetrahydrocortisone (THF/THE) ratio, both of which indicate lower HSD11β2 protein activity (Friso et al., 2008). This prospective study may highlight a potential new mechanism in the pathogenesis of glucocorticoid-induced hypertension, and may furthermore indicate HSD11β2 promoter methylation as a potentially useful biomarker to characterize hypertensive patients. Excess levels of cortisol can effect regulation of sodium absorption in the kidneys, alongside aldosterone, and thus can have a direct impact on salt-induced hypertension (Hunter et al., 2014). Hypermethylation of the HSD11β2 promoter, however, disrupts its ability to carry out cortisol to cortisone conversion through decreasing HSD11β2 protein expression, which leads to greater levels of cortisol relative to cortisone (i.e., a higher tetrahydrocortisol [THF] to tetrahydrocortisone [THE] ratio) and promotes hypertension (Ferrari et al., 2001, Udali et al., 2013).
Indeed, related research has shown that changes in HSD11β2 protein activity significantly affects blood pressure levels in healthy adults (Ferrari et al., 2001), and that HSD11β2 protein activity is decreased in the presence of greater HSD11β2 promoter methylation in human cells (Alikhani-Koopaei et al., 2004). Future prospective research in healthy adults is needed, however, to more precisely characterize the significance of this relationship in the development of hypertension. Furthermore, it will be important for future research to investigate whether early life or current stress play a role in the epigenetic regulation of HSD11β2 in relation to hypertension.
3.1.2. Subclinical Atherosclerosis
Among 84 American veteran monozygotic twin pairs (168 participants in total), intra-pair increases in flow-mediated dilation (FMD; determined using bi-mode ultrasound), a marker of subclinical atherosclerosis, was associated with both mean NR3C1 1F promoter hypermethylation and site-specific methylation at 12 out of 22 CpG sites investigated (Zhao et al., 2015). Even after adjustment for post-traumatic stress disorder (PTSD) symptoms, a 1% increase in the intra-pair difference in mean DNA methylation was associated on average with a 2.83% increase in the intra-pair difference in FMD. Another study included in our review, however, found no association between NR3C1 1C promoter methylation and a range of other factors associated with coronary heart disease, including blood pressure, as well as glucose and insulin levels (Veenendaal et al., 2012). Future research on subclinical atherosclerosis and other risk factors for cardiovascular disease may do well to investigate the 1F promoter, specifically, in NR3C1.
3.2. Cancer
Identifying epigenetic modifications associated with cancer is a rapidly expanding field of study. Our review shows that DNA methylation of NR3C1 has been associated with a variety of cancer outcomes (Table 3), although there is a lack of in vivo studies that could confirm the preliminary conclusions reported by studies using in vitro cell lines. Furthermore, corroboration between observed tumor tissue methylation and methylation levels in peripheral blood, which would be needed to implement non-invasive testing, has not yet been established. These results could then be more appropriately used towards the identification of biomarkers and the development of interventions. Among the studies reviewed, there is currently only strong evidence that NR3C1 methylation might be implicated in breast cancer (Nesset et al., 2014) and small cell lung cancer (SCLC) (Kay et al., 2011). Evidence from these studies indicates that promoter methylation of NR3C1 may contribute to tumorigenesis through decreasing NR3C1 expression, leading to downstream inhibition of the tumor suppressing and anti-proliferative capabilities of the GR for these cancers.
3.2.1. Breast Cancer
One study using matched normal and tumor breast tissue from 59 Canadian breast cancer patients examined methylation in a genomic region spanning almost the entire proximal promoter of NR3C1 (Nesset et al., 2014). The authors found that while non-tumor tissues were uniformly unmethylated, 15% (8/59) of breast cancer tumor tissues were methylated. Specifically, one region containing exons 1J–E and their promoters, as well as a second region containing exon 1B and the 1F promoter, both showed methylation in the highest number of tumors (7/59 and 5/59, respectively). There was also a statistically significant 4.6-fold decrease in NR3C1 expression in tumors methylated at exon 1B and the 1F promoter, compared with tumors unmethylated at this region. A previous study, however, found NR3C1 exon 1C to be uniformly unmethylated in breast tissue samples taken from 118 GR-immunonegative breast cancer carcinoma subjects from Taiwan (Lien et al., 2006), although this study only investigated a small gene region (exon 1C), which is not as heavily expressed in breast tissue, and did not study any of the regions shown to be methylated in the more recent paper.
DNA methylation of exon 1B within the proximal promoter of NR3C1 therefore seems to be a promising early candidate for a possible breast cancer biomarker. This is all the more biologically plausible because Nesset et al. (2014) experimentally determined that exon 1B is the predominant NR3C1 first exon variant expressed in human breast tissue. Importantly, high levels of NR3C1 protein (GR) expression and high concentrations of cortisol in breast tissue have been shown to have an anti-proliferative effect in cancerous breast tissue (Vilasco et al., 2011). Accordingly, Nesset et al. demonstrated that in all patients except one (58/59) from their study, all breast cancer tumors showed lower NR3C1 expression compared with normal tissue, with an average decrease in expression of 13.81-fold. This decrease in NR3C1 expression was also observed in the study by Lien et al. (2006) included in this review. Combined, these findings suggest that decreased NR3C1 expression appears to be a common and important factor in breast cancer tumor development. Furthermore, the results reported by Nesset et al. indicate that promoter methylation of NR3C1 might be one significant mechanism that leads to decreases in gene and GR protein expression, thus disrupting the anti-proliferative capabilities of the GR and contributing to breast cancer tumorigenesis.
Nesset et al. also note that the levels of methylation that they found in NR3C1 (15%) were comparable to promoter methylation levels reported for another known breast cancer tumor suppressor gene, BRCA1 (9–41%) (Birgisdottir et al., 2006, Parrella et al., 2004, Li et al., 2006, Baylin et al., 2001), indicating that future investigation might be warranted into whether NR3C1 itself may act as a tumor suppressor gene for breast cancer. The majority of methylated breast cancer tumors in the sample from Nesset et al. were also ER +/PR +/Her2-, and thus future research will be needed to investigate the role that these subtypes might play in the interaction between breast cancer tumorigenesis and NR3C1 methylation. As a final note, since only 15% of breast cancer tumors shower hypermethylation of NR3C1, while decreased NR3C1 expression was observed in almost all breast cancer tumors, future research should also aim to further identify other mechanisms through which NR3C1 and GR expression are downregulated in breast cancer tumors.
3.2.2. Small Cell Lung Cancer
Kay et al. (2011) conducted an analysis of 14 SCLC cell lines in comparison with a variety of other cancer control cell lines, including non-small cell lung cancer (NSCLC) cells and peripheral mononuclear blood cells taken from healthy donors. They found that there was a significant difference in methylation levels between SCLC cases and controls at several CpG sites in the 1C promoter, as well as a significant difference in methylation levels across the whole 1C promoter region. They also observed a significant association between the number of methylated CpGs and GR expression within the panel of 14 SCLC cell lines. As the only cell line study in our review to employ prospective methods, Kay et al. were able to provide causal support of this association by demonstrating that after treatment with a demethylating agent (decitabine), NR3C1 mRNA and GR protein levels increased dramatically in SCLC DMS79 cells, but not in controls or NSCLC cells. The authors therefore suggest that DNA methylation plays a role in regulating NR3C1 expression in human SCLC cells, but not NSCLC cells, and predict that this increase in expression would restore glucocorticoid sensitivity to these cells. Decitabine has been actively employed, along with its sister drug azacitidine, in the treatment of other cancers that rapidly divide such as acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS) (Tseng et al., 2015, Falini et al., 2015). As such, further studies are needed to confirm the benefits of decitabine to regulate NR3C1 expression in SCLC cancers in vivo.
Importantly, Kay et al. also showed that this increase in NR3C1 and GR expression after decitabine treatment led to several further changes in the SCLC cell lines, including increased cortisol sensitivity, and increases in cleaved caspase-3 activity and decreases in BCL-2 gene expression, both of which indicate cell apoptosis. Indeed, after 5 days of decitabine treatment, the SCLC cell lines underwent cell death. Combined, these data suggest that demethylation of NR3C1 leads to increased gene and protein expression, which in turn promote SCLC cell death in a similar manner to that for conventional SCLC tumor suppressor genes. Although demethylating agents are currently an underutilized spectrum of drugs, we anticipate that targeted use of demethylating agents for specific CpG sites within glucocorticoid regulatory genes may offer novel avenues to lyse rapidly growing tumor cells.
3.2.3. Colorectal and other cancers
One final study showed an increasing methylation frequency of NR3C1 from colorectal adenomas to carcinomas among 103 Norwegian patients, and demonstrated that microsatellite instable (MSI) carcinomas had significantly increased NR3C1 promoter methylation in comparison to microsatellite stable (MSS) tumors (Ahlquist et al., 2008). However, NR3C1 was not among the genes identified as the most promising biomarkers for early detection of colorectal tumors. Unlike the results reported on breast cancer, above, this study did not find NR3C1 to be differentially methylated between colorectal tumors and normal colon mucosa in healthy individuals. They did find several other genes (e.g., Vimentin, ADAMTS1, and MAL) that were methylated in carcinomas and completely unmethylated in normal mucosa of healthy individuals, thus making methylation of those genes more promising biomarker candidates for early, non-invasive detection of colorectal cancer tumors. In another study included in our review, NR3C1 promoter methylation was similarly observed in colon cancer cell lines and colon cancer tissues (while also shown to be unmethylated in non-cancerous tissue from the same individuals), and this methylation was significantly associated with reduced gene expression (Lind et al., 2006). However, the authors were unable to determine if the observed methylation was specific to colon cancer, and two other genes, ADAMTS1 and CRABP, showed much higher methylation frequencies. Therefore, future research is needed to confirm the extent to which NR3C1 methylation is involved in colon cancer tumorigenesis.
NR3C1 methylation does not seem likely to serve as an early detection biomarker for ovarian (Wu et al., 2007) or gastric cancers (Kang et al., 2008, Hiraki et al., 2011) because of non-significant methylation patterns found, or because other genes have been identified that show much more significant patterns of DNA methylation. Interestingly, however, Sanchez-Vega and Gandhi (2009) showed that histone H3 was trimethylated on Lys4 (3 mK4) in exons 1C, 2, and 3, and throughout intron B of NR3C1 RNA in glucocorticoid-sensitive myeloma cells, but not glucocorticoid resistant cells. Thus, the authors concluded that while NR3C1 promoter methylation is likely not implicated in the development of glucocorticoid resistance in myeloma cells, histone modification—another epigenetic mechanism—likely plays a role, and may influence glucocorticoid resistance.
3.3. Renal, Metabolic, and Inflammatory Diseases
Table 4 summarizes the 3 studies included in our review that investigated FKBP5 and NR3C1 methylation in relation to renal, metabolic, and inflammatory diseases.
3.3.1. Chronic Kidney Disease
In an epigenome-wide methylation analysis of 407 white participants from the UK (Smyth et al., 2014), FKBP5 emerged as a strong biomarker candidate for chronic kidney disease (CKD), with significantly lower methylation observed in CKD cases versus healthy controls at three CpG sites (one in an enhancer in intron 4, and two in the 5′UTR region in the north and south shores of a CpG island). Hypomethylation of FKBP5 was not associated with a change in gene expression, although this study had an extremely small subsample for expression analyses (2 CKD cases versus 2 healthy controls). The Klengel et al. (2013) PTSD study described later in this review, however, has documented an association between decreased FKBP5 methylation and increased stress-dependent gene transcription of FKBP5.
3.3.2. Other Inflammatory Diseases
NR3C1 methylation has also been investigated in relation to several inflammatory diseases. In a study of 95 women from Belgium (Vangeel et al., 2015), mean and CpG site-specific NR3C1 1F promoter hypomethylation was seen in those with chronic fatigue syndrome (CFS; clinically diagnosed according to U.S. CDC criteria [Fukuda et al., 1994]) compared with healthy controls. In a study of 30 Chinese patients (Chen et al., 2015), there were significantly higher mean methylation levels across four NR3C1 first exon promoters (1D, 1E, 1F, and 1H) between systemic lupus erythematosus (SLE; clinically diagnosed according to American College of Rheumatology revised criteria [Hochberg, 1997]) and healthy control patients, but no results were reported for each of the individual promoters. Site-specific CpG methylation varied greatly between SLE individuals in this region, with no patterns of common CpG site methylation emerging among the entire SLE group. There was also an inverse association in the SLE group between the overall mean methylation status of the 4 promoters and GRα mRNA expression, which is the active 3′ GR isoform. Caution should be taken in interpreting these associations, however, as a recent epigenome-wide methylation analysis of Japanese monozygotic twins discordant for SLE identified differential methylation and expression of 5 genes that may be important in the pathogenesis of SLE, but NR3C1 was not among these (Furukawa et al., 2013).
3.4. Mental Health Outcomes
Table 5 summarizes the 16 studies that have investigated NR3C1 and FKBP5 methylation status in relation to mental health outcomes. To date, DNA methylation of other genes along the HPA axis has not been investigated in relation to any mental health outcomes.
3.4.1. Post-traumatic Stress Disorder
One study showed a strong association between FKBP5 methylation and PTSD (Klengel et al., 2013). Specifically, those from the U.S. Grady Trauma Project Cohort (N = 1963) who experienced child abuse (measured via self-report by the CTQ [Fink et al., 1995]) with a known risk allele (rs1360780) for PTSD, but not those with the protective allele, showed greater adult PTSD symptoms (measured via self-report by the mPSS scale [Falsetti et al., 1993, Foa and Tolin, 2000]), and also greater lifetime PTSD (clinically-diagnosed via the CAPS scale [Weathers et al., 2001]; N = 519). In subsequent analyses among a mostly African American subsample of 76 adults from the Grady Cohort, child abuse-exposed risk allele carriers showed an average decrease of 12.3% in DNA methylation in a bin of 3 CpGs arranged around glucocorticoid response elements in intron 7 (bin 2) compared to those abused without the risk allele or those not abused with or without the risk allele. This same site showed no association with adult trauma (measured by the TEI [Binder et al., 2008]), pointing to a sensitive window in childhood for such epigenetic programming. These results were further replicated in a smaller cohort of African American women (N = 57).
The authors of this study also determined that childhood abuse was associated with increased expression of 76 GR-responsive immune system genes, and that FKBP5 demethylation at intron 7 bin 2 was associated with altered GR sensitivity in peripheral blood. This latter result suggested an enhancement of the ultra-short feedback loop between the GR and FKBP5, which leads to increased GR resistance. Lastly, demethylation of FKBP5 intron 7 in a subset of the replication cohort was also associated greater volume of the right hippocampal head—a change that indicates greater stress hormone system reactivity—thus showing a connection between abuse-related methylation changes in peripheral blood and effects in the central nervous system (greater detail given in Table 5).
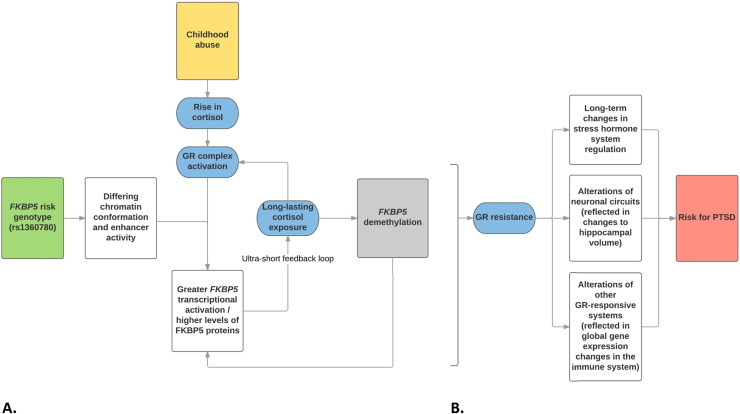
This paper represents a serious advancement in understanding the functional epigenetic pathways contributing to PTSD. Based on these results, the authors have put forward a model of PTSD pathogenesis in which FKBP5 intron 7 demethylation—induced by long-lasting cortisol exposure (Yang et al., 2012) as a result of childhood abuse and risk allele status—leads to a tightening of the ultra-short feedback loop between the GR and FKBP5 by local activation of the GR complex in peripheral tissue (which prolongs the stress cascade). On a systems level, these changes lead to GR resistance, alterations to stress hormone, neuronal, and other GR-responsive systems, and ultimately, higher risk for PTSD (see Fig. 7).
Fig. 7.
Model of (A) epigenetic and (B) subsequent systems-level PTSD pathogenesis proposed by Klengel et al. (2013). Note: Klengel et al. observed that FKBP5 methylation only leads to higher levels of FKBP5 expression in the presence of GR complex activation.
Although not as rich in functional analyses, the results reported on NR3C1 methylation levels and PTSD show the most uniform trend for NR3C1 methylation in relation to any mental health outcome in the literature—all studies show lower methylation despite examining different NR3C1 promoter regions (see Fig. 4, Fig. 5).
One study of 46 individuals from Canada showed that those with lifetime PTSD (assessed via the CAPS scale) showed lower overall NR3C1 methylation at the 1B and 1C promoters compared with controls, with site-specific hypomethylation found in the 1B and 1C promoters (Labonte et al., 2014). One CpG site in the 1C promoter region, however, was significantly hypermethylated. Furthermore, an association was found between overall and site-specific hypomethylation in the NR3C1 1B promoter and both higher total mRNA expression levels for NR3C1 and higher relative mRNA expression levels for the 1B promoter. Cortisol levels were positively associated with methylation levels at CpG sites 11 and 13 in exon 1B. No association was found, however, between total methylation in the 1C promoter and relative NR3C1 1C mRNA expression or cortisol levels.
Another study of 16 racially diverse combat veterans being treated for PTSD with prolonged exposure (PE) therapy in New York found that methylation levels could predict treatment response (Yehuda et al., 2013). Specifically, those with higher methylation of the 1F promoter had significantly lower post-treatment PTSD symptom severity (clinician-assessed via CAPS) and a significantly greater reduction in PTSD symptom severity from pre- to post-treatment, suggesting that higher NR3C1 methylation may be a useful marker for better PTSD prognosis and may even mediate disease progression. The authors also investigated FKBP5 methylation, although unlike NR3C1, FKBP5 promoter methylation did not predict treatment outcome or symptom severity. However, higher FKBP5 promoter methylation associated cross-sectionally with measures of cortisol at pre-treatment and ACTH at post-treatment. This led the authors to conclude that FKBP5 promoter methylation may be associated with changes in HPA axis activity connected to changes in PTSD symptom expression, rather than upstream regulation of cortisol.
Furthermore, among a racially diverse sample of 122 combat veterans recruited from two hospitals in New York, those with PTSD (clinician-assessed via CAPS) showed significantly lower overall methylation of the NR3C1 1F promoter, compared with those without PTSD, with site-specific significant differences at CpGs 23 and 39 (Yehuda et al., 2015a). 1F promoter methylation was not associated with NR3C1 1F expression, except at CpG site 23, however lower 1F promoter methylation levels were associated with greater cortisol decline in response to dexamethasone treatment, and associated with poorer sleep quality, higher peritraumatic dissociation, and higher psychiatric distress. Early life trauma exposure (measured by the ETI [Bremner et al., 2000]) was not associated with 1F promoter methylation.
Lastly, in a study of 224 survivors of the Rwandan genocide (Vukojevic et al., 2014), lower methylation levels at a CpG site embedded in a transcription factor (NGFI-A) binding site of the NR3C1 1F promoter were significantly associated with greater PTSD symptom severity (assessed via the PDS [Foa et al., 1997]) related to re-experiencing traumatic events (intrusive memories) in men but not in women. This methylation was not associated with the severity of PTSD symptoms related to avoidance and hyperarousal, indicating that the methylation changes at this site were likely related to the memory aspects of PTSD. Higher methylation levels at this site were also significantly associated with a lower lifetime PTSD risk in men but not in women. Importantly, however, the authors did not find a significant difference in methylation at this CpG site between the Rwandan genocide and healthy Swiss samples, meaning that methylation at this site may be a more stable trait that preexisted genocide trauma, or reflects a trauma or stress earlier in life.
Although the literature on PTSD from this review lends the strongest support to a connection between NR3C1 methylation and mental health, the results reported are mixed on the connection between this methylation and adverse childhood experiences. For instance, Yehuda et al. (2015a) reported no association between NR3C1 1F promoter methylation and early life trauma exposure (measured by the ETI) in combat veterans with PTSD, and Vukojevic et al. (2014) found no difference in NR3C1 1F promoter methylation between those who had survived the Rwandan genocide and Swiss controls. These results indicate that great care needs to be taken in future research to identify if trauma at different points in life or slightly different forms of trauma have differential effects on NR3C1 methylation and development of PTSD.
3.4.2. Alzheimer's Disease
One study examining medial temporal gyrus brain tissue from 15 elderly U.S. adults found that one CpG site in intron 7 and two CpGs in the promoter of FKBP5 showed significant hypomethylation in Alzheimer's disease (AD) cases versus healthy controls (Blair et al., 2013). One CpG in intron 2, however, showed significant hypermethylation. mRNA expression analyses in a separate and larger sample (N = 59) showed that FKBP5 expression was significantly higher in AD cases versus healthy controls, and that increased FKBP5 expression was significantly associated with Braak staging—a clinical tool used to assess the degree of pathology in AD (Braak and Braak, 1991)—even when evaluating the oldest samples alone. These results show a promising initial association between altered FKBP5 methylation levels and AD. Further research in larger, more adequately powered samples is needed, however, to directly identify the genomic region of FKBP5 wherein methylation is functionally associated with the differing FKBP5 expression levels observed between AD cases and healthy controls. This same study also experimentally determined in mouse models that overexpression of FKBP5 preserves the soluble species of tau proteins that have been linked to AD (Lasagna-Reeves et al., 2012, de Calignon et al., 2010, Santacruz et al., 2005, de Calignon et al., 2012), and so if FKBP5 methylation levels at certain sites can be linked with FKBP5 expression, then these results would point towards a biologically plausible role of FKBP5 methylation in leading to AD and expediting pathogenesis through encouraging the production and accumulation of toxic tau proteins.
Research to date on NR3C1 and AD has showed no differences in methylation between healthy controls and those with AD, Parkinson's disease, or dementia (Moser et al., 2007). This study, however, suffered from a small sample size (5 controls vs. 27 cases).
3.4.3. Depression
Results reported on depression are mixed (see Fig. 4, Fig. 5), although several different types of tissue were analyzed between studies. Among 117 participants from Korea (Na et al., 2014), those with major depressive disorder (MDD; clinically-diagnosed using the Korean version of the SCID-IV [Hahn et al., 2000]) had significantly lower blood NR3C1 methylation at CpGs 3–4 of the 1F promoter compared with healthy controls, and among those with MDD, those with higher perceived stress showed even lower methylation at CpG3 of the 1F promoter.
Conversely, in an analysis of 1668 Swedish women with a low expression variant of MAOA (MAOA-L) (Melas et al., 2013), parental loss in early in life was associated with increased saliva DNA methylation levels at CpG site 35 of the 1F promoter, and increased risk of developing depression (self-reported using the MDI [Bech et al., 2001]) compared to those with the MAOA-H genotype. A study of 759 participants from the Dutch famine birth cohort (Veenendaal et al., 2012) also found that increased blood NR3C1 methylation was associated with higher levels of anxiety and depression (self-reported using the anxiety and depression subscales from a Dutch translation of the HADS [Zigmond and Snaith, 1983]), but at the 1C promoter.
In contrast to the above studies, another study using hippocampal tissue from 12 brain tissue donors in the Netherlands found no difference in tissue methylation levels between MDD donors (confirmed post-mortem by a psychiatrist examining medical records according to DSM-IV criteria) and healthy controls across a number of regions in NR3C1 (exon 1J and 1B, and promoters 1J, 1E, 1B, and 1F) (Alt et al., 2010).
To summarize, then, depression has been associated with: (a) lower blood methylation of the exon 1F promoter (Na et al., 2014); (b) higher saliva methylation of the exon 1F promoter (Melas et al., 2013) and higher blood methylation of the 1C promoter (Veenendaal et al., 2012); and (c) no change in hippocampal methylation at exon 1J, and the 1B and 1F promoters (Alt et al., 2010). The discrepancies in findings between studies, however, may be largely driven by the different forms of stress or adversity that were analyzed, the different promoter regions analyzed, or perhaps even the different tissue types analyzed. While Na et al. (2014) demonstrated that current perceived stress (measured via the PSS [Cohen et al., 1983]) in depressive patients was associated with lower NR3C1 1F promoter methylation, Melas et al. (2013) found that early childhood trauma, measured as early parental death, parental divorce, or early childhood financial constraints (assessed via original measures in the PART questionnaire), was associated with increased methylation of the 1F promoter and depression. Veenendaal et al. (2012), on the other hand, found an association between increased NR3C1 1C promoter methylation and depression, but no association between NR3C1 1C promoter methylation and early childhood adversity when looking at gestational exposure to famine as a measure of early childhood adversity.
It is important to point out here that none of the above studies on depression measured NR3C1 expression or cortisol levels. Although Na et al. (2014) suggest that NR3C1 methylation may occur in a compensatory pattern opposite of HPA activity, the authors could not determine whether the HPA axis was hypoactive in their MDD sample. Therefore, the functional and biological mechanisms connecting HPA axis gene methylation levels and depression remain largely unknown. This may also account for the discrepancy in the results reported, as methylation of specific CpG sites in NR3C1 may simply prove to have no functional contribution towards depression. Future research on depression should aim to elucidate which regions of NR3C1 play a functional role in the relationship between early childhood adversity, NR3C1 methylation, and depression through at least measuring cortisol and NR3C1 expression levels.
One study examined FKBP5 methylation in relation to depression (Höhne et al., 2015). Among 96 German participants from the Early Developmental Stages of Psychopathology (EDSP) study, subjects with the FKBP5 rs1360780 T risk allele genotype and a lifetime history of major depression (MD; self-reported via the M-CIDI [Wittchen et al., 1998]) had a 10% higher DNA methylation rate in intron 7 of FKBP5 than healthy controls with the same genotype, although posthoc comparisons did not reach significance and only showed a non-significant trend. The authors note that this was likely due to a small, inadequately powered sample. The same study found altered cortisol and expression levels after a stress test in healthy adults but not in depressed cases, which suggests that methylation of another region of FKBP5 may be more functionally important in depression. Future research should investigate early life stress (and, if necessary, also measure and compare levels of current stress), use greater sample sizes, and explore different regions of FKBP5 known to be sensitive to psychosocial or early life stress.
3.4.4. Bipolar Disorder
One study examined the role of FKBP5 methylation in bipolar disorder (BD) (Fries et al., 2015). Among 68 Brazilian adults, those with BD (clinically diagnosed according to DSM-IV Axis I criteria) showed significantly higher methylation of one CpG in intron 7 of FKBP5 compared with their siblings, and significantly higher methylation of one CpG in intron 2 compared with non-related healthy controls. No association was found, however, between DNA methylation and basal FKBP5 mRNA and protein levels.
3.4.5. Borderline Personality Disorder and Eating Disorders
The results concerning NR3C1 methylation and borderline personality disorder (BPD) are also mixed. In a sample of 36 patients from Switzerland, significantly increased methylation of NR3C1 exon 1B was found at CpG1 and CpG5 in BPD patients (clinically diagnosed according to DSM IV criteria), but the low level of methylation changes observed relative to other candidate genes led the authors to conclude that NR3C1 methylation is likely not a suitable biomarker for BPD (Dammann et al., 2011). A further study of 101 Swiss subjects with BPD (clinically assessed via the SCID-II [First et al., 1994]) and 99 MDD controls (clinically assessed via the French version of the DIGS [Preisig et al., 1999]) showed that those with BPD and no sexual abuse had significantly greater NR3C1 exon 1F methylation compared with those with MDD and no sexual abuse (Perroud et al., 2011a).
In another sample of 96 U.S. women between the ages of 17 and 48, women with bulimia nervosa (BN; self-reported according to DSM-IV-TR criteria) with comorbid BPD (self-reported via the SCID-II) showed elevated, but non-significant methylation at CpG sites 10 and 21 in the promoter of NR3C1 exon 1C, compared with non-eating disordered controls (Steiger et al., 2013). BN women with comorbid suicidality, on the other hand, showed significantly elevated methylation in CpG sites 10, 22, and 29 in the 1C promoter, compared to non-eating disordered controls. BN women with comorbid BPD also displayed significantly lower methylation at the 1H promoter when compared to BN women with no BPD or non-eating disordered individuals. These results suggest that NR3C1 methylation may act as an epigenetic process regulating comorbid BPD and suicidality expression in BN, rather than being directly implicated in the pathogenesis of BPD, although they do provide indication that 1H hypomethylation, specifically, may be associated with BPD.
A recent epigenome-wide methylation study of 96 Swiss subjects with BPD did not find high DNA methylation levels of NR3C1 in relation to methylation levels of 14 other candidate genes identified (Prados et al., 2015), however future research should try to determine if NR3C1 methylation plays a more prominent role in development of BPD among those who have experienced early childhood adversity. One study not included in this review (Martin-Blanco et al., 2014) found that among 281 Spanish subjects with BPD (clinically-assessed via a Spanish version of the SCID-II [Gómez-Beneyto et al., 1994]), there was a positive association between mean DNA methylation of NR3C1 exon 1F and childhood physical abuse (measured via the CTQ). Site-specific methylation at three CpGs also showed a significant association with physical abuse and emotional neglect in childhood (measured via the CTQ), and with unemployment. Greater BPD severity scores were also associated with exon 1F methylation at 4 different CpG sites. The study by Perroud et al. (2011a), included in our review, also showed that among those with BPD, increased severity of childhood sexual abuse and childhood physical neglect (measured by the CTQ) were significantly associated with increased NR3C1 1F promoter methylation, and that those with one reported experience of abuse had less methylation than those reporting two or more experiences of abuse. In fact, all CpGs investigated in this study except one showed a positive association between methylation and number of types of abuse and neglect at a level of significance of P < 10− 6.
Lastly, given the significant results reported by Steiger et al. (2013) at the 1H promoter but not at the 1F promoter, future research investigating NR3C1 methylation as a potential biomarker for BPD in those with childhood trauma should also investigate whether the 1H promoter region is a stronger candidate than the 1F promoter, which may perhaps be more significantly implicated in MDD or PTSD.
4. Discussion
This is the first systematic review to examine DNA methylation status for multiple genes along the HPA axis that are involved in glucocorticoid regulation. While other reviews have examined the association between early life stressors and HPA axis gene methylation, this review critically maps the terrain of clinical relevance for emerging epigenetics research concerning HPA axis genes. In this critical review, we synthesize current, clinically important epigenetics research in order to highlight the potential utility of using DNA methylation of HPA axis genes as a potential biomarker useful to advance early detection methods for key conditions, including several cancers, and to help inform the development of more effective prevention strategies aimed at reducing the burden of chronic illness.
There are many limitations that stand out among the studies reviewed here. Most notably, only 4 of the studies reviewed used prospective methods (Friso et al., 2008, Kay et al., 2011, Yehuda et al., 2013, Höhne et al., 2015), whereas the remaining were cross-sectional or case-control. The majority of extant studies thus cannot adequately indicate whether gene methylation is a predisposing factor for the outcomes reported, or whether it is a consequence of developing a specific pathology. Second, only a handful of studies included in our review stratified their analyses by sex (Yehuda et al., 2015a, Vukojevic et al., 2014, Melas et al., 2013, Steiger et al., 2013), even though HPA activity has been shown to vary by sex (Kirschbaum et al., 1999, Kunzel et al., 2003).
In the remainder of this review, we highlight key challenges and future research directions that will need to be addressed in order to develop both clinically meaningful prognostic biomarkers and an evidence base that can inform public policy practice.
4.1. Poor Socio-demographic Characterization of Study Populations
One of our deepest concerns about current epigenetics study design, at least within the studies included in this review, is a general lack of detailed reporting regarding the socio-demographic characteristics of study populations. This is an important point, since epigenetic modifications of HPA axis genes are highly dependent on specific socioeconomic, social, and psychosocial factors (Needham et al., 2015, Yehuda et al., 2015b). Of the 30 articles reviewed here that used blood or tissue samples for their methylation analyses (excluding 2 in vitro only studies), 23 did not report any detailed information about the race/ethnicity of their sample populations. Furthermore, only half of the studies conducted within the U.S. reported information on race/ethnicity. Given the findings reported in this review on hypertension and cancer, however, such information is clinically relevant. African Americans bear a disproportionate burden of hypertension (Yoon et al., 2015), and African American women have a prevalence of hypertension 2–3 times greater than white women (Gillum, 1996, Nwankwo et al., 2013). There is also a significant difference in cancer mortality between African American and white populations in the U.S. (Siegel et al., 2015, NCI, 2008). Future researchers should therefore endeavor to design studies with samples from minority and disadvantaged populations in order to better understand whether the more severe clinical disease risk that these populations experience is a result of variable epigenetic regulation in the face of unique environmental and psychosocial stressors. Similar efforts should be undertaken to better characterize the socioeconomic characteristics of study samples.
4.2. Lack of Consistency in Gene Regions Studied and CpG Nomenclature
To date, the full potential of existing epigenetics research on HPA axis genes has been limited by several factors that need to be addressed in order to realize the full clinical and public health benefit of this work. These include: (1) investigating different and non-overlapping gene regions between studies on the same disease, without also replicating results of prior studies on previously measured CpG sites, creating a patchwork of somewhat unrelated results; (2) inconsistent CpG numbering and nomenclature between studies, making it difficult to easily compare data across studies; and (3) failure to select gene regions to study based on theoretical grounds pertinent to the disease phenotype studied (as opposed to merely studying the most cited, but perhaps less relevant region).
Regarding our first concern, the exact gene regions examined for DNA methylation analyses varied significantly between the studies on NR3C1 reviewed here. Some looked at the entire promoter of a gene, while others focused on only 5 CpG sites with a single promoter region, making comparison between findings difficult. Furthermore, some studies examining the same outcome or disease didn't look at overlapping promoter regions within the same genes at all (Fig. 4, Fig. 5), making comparisons and identifying trends a tenuous exercise, at best. There is, however, consistency within studies investigating methylation of FKBP5, as nearly all studies included in our review investigated the same regions outlined in Klengel et al. (2013) (Table 1).
Second, the inconsistency between studies on NR3C1 methylation regarding how CpG sites are identified and labeled makes understanding overlap in CpG sites investigated and synthesizing findings between studies difficult. Future studies would therefore benefit from explicit selection of gene promoter regions or CpG sites that have been studied previously, and from a discussion of how these regions overlap with previous research in order to more easily amass a comprehensive evidence base. If researchers wish to also investigate novel sites, then simultaneously investigating regions that overlap with previous research would give much more contextual significance to any findings observed in these new sites. We believe the field as a whole could benefit from adopting the CpG numbering for NR3C1 found in Palma-Gudiel et al. (2015), as these numberings reflect position within the entire NR3C1 proximal promoter, and have been assigned according to their review of existing research on epigenetic modifications of NR3C1.
Our third concern stems from the tissue-specificity observed between expression of the various NR3C1 first exons (Fig. 6). Future studies looking at NR3C1 methylation should be sure to investigate the most pertinent region of the NR3C1 promoter for the disease in question, instead of merely looking at mean methylation across the entire promoter or simply looking at exon 1F or its promoter because it is the most widely cited in the literature.
4.3. Measuring the Functional Effects of DNA Methylation
It is crucial that all future studies measure NR3C1 expression and glucocorticoid hormone levels, at least, in order to determine which CpG sites are functionally sensitive in relation to a given pathology. Further functional studies of more pathology-specific markers similar to those done in the studies included in this review on hypertension (Friso et al., 2008), small cell lung cancer (Kay et al., 2011), Alzheimer's (Blair et al., 2013), and PTSD (Klengel et al., 2013) will be especially useful. This will also be important for understanding the contradictory results found in studies on depression and other mental health outcomes. Furthermore, given that one of the prominent mechanisms through which DNA methylation silences gene expression is inhibition of transcription factor binding, future research on any of the genes reviewed here should also be certain to include specific CpG loci known to be located in transcription factor binding sites throughout the NR3C1 proximal promoter (Turner et al., 2010), the 4 CpG islands found in HSD11β2 (Alikhani-Koopaei et al., 2004), or in distal intronic glucocorticoid response elements (GREs) in FKBP5 through which the GR complex regulates FKBP5 expression (Paakinaho et al., 2010).
4.4. Accounting for Genotype-specific Associations and Cell Type Heterogeneity in Blood Samples
Several studies included in our review showed that adverse childhood experiences only associated with depression (Melas et al., 2013, Höhne et al., 2015) or PTSD (Klengel et al., 2013) among those with a known risk genotype. Furthermore, DNA methylation of FKBP5 and NR3C1 in these studies was only associated with these exposures and outcomes among those with the risk genotype. It will be crucial for future studies to investigate the interaction of known risk genotypes with the exposures and outcomes they are investigating, and caution should be taken in interpreting the methylation results of any studies that have not investigated genotype-specific associations for exposures or outcomes where there is a known risk genotype. In particular, the rs1360780 risk genotype in FKBP5 plays a clear role in modulating stress reactivity (Zannas et al., 2016, Klengel et al., 2013), and further research should endeavor to more precisely elucidate the complex interplay between environmental stress, genotype, and epigenetic regulation of FKBP5.
Cell type heterogeneity may also be an important source of confounding in DNA methylation studies using blood samples (Bock, 2012, Adalsteinsson et al., 2012, Lam et al., 2012), and statistical adjustment for cell composition in blood samples is increasingly becoming a standard methodological characteristic for high-quality studies on DNA methylation of NR3C1, in particular, in relation to environmental exposures (Madrigano et al., 2012, Burris et al., 2013, Kim et al., 2016). Only 3 studies included in our review, however, made statistical adjustments to account for cell composition in their analyses (Smyth et al., 2014, Yehuda et al., 2015a, Höhne et al., 2015). This will be an increasingly crucial methodology to include in future research, although it will require consideration early in the process of planning candidate gene research, since complete blood counts (CBCs) can only be done when blood samples are first drawn. For epigenome-wide and high-throughput studies, however, statistical methods have been developed to determine cell composition in blood samples without the need for a CBC or a reference dataset (Houseman et al., 2014, Houseman et al., 2016, Shannon et al., 2017).
4.5. More Comprehensively Understanding the Epigenetic Plasticity of HPA Axis Genes
Although recent research included in our review has shown that early childhood is a sensitive period in life during which stable epigenetic regulation of FKBP5 can occur and persist into adulthood independent of adulthood stressors (Klengel et al., 2013), the epigenetic plasticity exhibited by NR3C1 needs to be further studied to better understand the contribution of various temporal stressors and influences to the etiology of disease across the lifecourse. Attenuating influences in later life may reverse prior NR3C1 methylation, which in part may explain the divergences in findings for NR3C1 1F promoter methylation and depression among those experiencing current stress (Na et al., 2014) versus early life stress (Melas et al., 2013). This very plasticity, which on the one hand creates methodological challenges, is in fact what makes epigenetics such a promising tool to inform developments in clinical and public health practice.
4.6. Building Consensus Regarding Optimal Social/Psychosocial Measures to be Used in Studies
The link between adverse childhood experiences (ACE) and NR3C1 or FKBP5 methylation has been well established in numerous systematic reviews and recent studies (Palma-Gudiel et al., 2015, Daskalakis and Yehuda, 2014, Needham et al., 2015, Yehuda et al., 2015b, Turecki and Meaney, 2014, Tyrka et al., 2015). Indeed, our review demonstrates that there now exists evidence for an association between such NR3C1 or FKBP5 methylation resulting from early childhood stress and the development of mental health disorders later in life, although the temporality of stressors, as well as the number and type of adversities experienced, seem to have a strong influence on the functional role that NR3C1 or FKBP5 methylation play in the emergence of specific mental health disorders later in life.
Insights gained from epigenetics research on HPA axis genes involved in glucocorticoid regulation could be significantly advanced through building a consensus in the scientific community on which measures, instruments, or scales of early childhood trauma or adversity, adult psychosocial stress, and environmental and social stressors optimally capture critical exposures likely to affect human health across the lifecourse. Our review revealed that NR3C1 methylation can vary significantly by type and number of abuses experienced in childhood, for example, but extant studies used varying measures of childhood trauma or adversity, making comparisons of results across studies difficult. Future studies are needed that might replicate the results reported here using the same combination of outcomes, NR3C1 promoter region(s), and exact measures of childhood trauma or adversity in new populations to test the strength of established associations.
5. Conclusion
We are hopeful that the findings reported in this review will reveal new insights useful to molecular biologists, clinicians, and researchers alike who might consider DNA methylation status as a potential disease- or gene-specific biomarker to advance our understanding of disease etiology and perhaps even treatment. Strong functional relationships, in particular, need to be examined using more robust study designs in order to prevent reporting associations where none actually exist. In order for this to be accomplished, the results reported in this review will require further prospective exploration across diverse populations to form a conclusive evidence base regarding the directionality and magnitude of clinical associations reported to date. Despite these challenges, the results of this review indicate that investigating DNA methylation of HPA axis genes involved in glucocorticoid regulation is an important and growing field for better understanding the etiology—and ultimately, how to detect and treat—of a number of key diseases affecting population health, including hypertension, breast cancer, small cell lung cancer, chronic kidney disease, Alzheimer's, PTSD, depression, and borderline personality disorder.
Sources of Financial Support
The John Templeton Foundation grant #59607 (Alexandra Shields).
Conflicts of Interest
The authors report no conflicts of interest.
References
- Adalsteinsson B.T., Gudnason H., Aspelund T. Heterogeneity in white blood cells has potential to confound DNA methylation measurements. PLoS One. 2012;7(10):e46705. doi: 10.1371/journal.pone.0046705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist T., Lind G.E., Costa V.L. Gene methylation profiles of normal mucosa, and benign and malignant colorectal tumors identify early onset markers. Mol. Cancer. 2008;7:94. doi: 10.1186/1476-4598-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhani-Koopaei R., Fouladkou F., Frey F.J., Frey B.M. Epigenetic regulation of 11 beta-hydroxysteroid dehydrogenase type 2 expression. J. Clin. Invest. 2004;114(8):1146–1157. doi: 10.1172/JCI21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt S.R., Turner J.D., Klok M.D. Differential expression of glucocorticoid receptor transcripts in major depressive disorder is not epigenetically programmed. Psychoneuroendocrinology. 2010;35(4):544–556. doi: 10.1016/j.psyneuen.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Baylin S.B., Esteller M., Rountree M.R., Bachman K.E., Schuebel K., Herman J.G. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 2001;10(7):687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- Bech P., Rasmussen N.A., Olsen L.R., Noerholm V., Abildgaard W. The sensitivity and specificity of the major depression inventory, using the present state examination as the index of diagnostic validity. J. Affect. Disord. 2001;66(2–3):159–164. doi: 10.1016/s0165-0327(00)00309-8. [DOI] [PubMed] [Google Scholar]
- Binder E.B. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34(Suppl. 1):S186–S195. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Binder E.B., Bradley R.G., Liu W. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA. 2008;299(11):1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir V., Stefansson O.A., Bodvarsdottir S.K., Hilmarsdottir H., Jonasson J.G., Eyfjord J.E. Epigenetic silencing and deletion of the BRCA1 gene in sporadic breast cancer. Breast Cancer Res. 2006;8(4):R38. doi: 10.1186/bcr1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair L.J., Nordhues B.A., Hill S.E. Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. J. Clin. Invest. 2013;123(10):4158–4169. doi: 10.1172/JCI69003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C. Analysing and interpreting DNA methylation data. Nat. Rev. Genet. 2012;13(10):705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- Braak H., Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Bremner J.D., Vermetten E., Mazure C.M. Development and preliminary psychometric properties of an instrument for the measurement of childhood trauma: the early trauma inventory. Depress Anxiety. 2000;12(1):1–12. doi: 10.1002/1520-6394(2000)12:1<1::AID-DA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Burris H.H., Braun J.M., Byun H.M. Association between birth weight and DNA methylation of IGF2, glucocorticoid receptor and repetitive elements LINE-1 and Alu. Epigenomics. 2013;5(3):271–281. doi: 10.2217/epi.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron O.G. Anxious-depressive comorbidity: effects on HPA axis and CNS noradrenergic functions. Essent. Psychopharmacol. 2006;7(1):24–34. [PubMed] [Google Scholar]
- Cao-Lei L., Leija S.C., Kumsta R. Transcriptional control of the human glucocorticoid receptor: identification and analysis of alternative promoter regions. Hum. Genet. 2011;129(5):533–543. doi: 10.1007/s00439-011-0949-1. [DOI] [PubMed] [Google Scholar]
- Ceccato F., Lombardi G., Manara R. Temozolomide and pasireotide treatment for aggressive pituitary adenoma: expertise at a tertiary care center. J. Neuro-Oncol. 2015;122(1):189–196. doi: 10.1007/s11060-014-1702-0. [DOI] [PubMed] [Google Scholar]
- Chen H., Fan J., Shou Q., Zhang L., Ma H., Fan Y. Hypermethylation of glucocorticoid receptor gene promoter results in glucocorticoid receptor gene low expression in peripheral blood mononuclear cells of patients with systemic lupus erythematosus. Rheumatol. Int. 2015;35(8):1335–1342. doi: 10.1007/s00296-015-3266-5. [DOI] [PubMed] [Google Scholar]
- Cohen S., Kamarck T., Mermelstein R. A global measure of perceived stress. J. Health Soc. Behav. 1983;24(4):385–396. [PubMed] [Google Scholar]
- Conradt E., Lester B.M., Appleton A.A., Armstrong D.A., Marsit C.J. The roles of DNA methylation of NR3C1 and 11beta-HSD2 and exposure to maternal mood disorder in utero on newborn neurobehavior. Epigenetics. 2013;8(12):1321–1329. doi: 10.4161/epi.26634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crujeiras A.B., Campion J., Diaz-Lagares A. Association of weight regain with specific methylation levels in the NPY and POMC promoters in leukocytes of obese men: a translational study. Regul. Pept. 2013;186:1–6. doi: 10.1016/j.regpep.2013.06.012. [DOI] [PubMed] [Google Scholar]
- Dammann G., Teschler S., Haag T., Altmuller F., Tuczek F., Dammann R.H. Increased DNA methylation of neuropsychiatric genes occurs in borderline personality disorder. Epigenetics. 2011;6(12):1454–1462. doi: 10.4161/epi.6.12.18363. [DOI] [PubMed] [Google Scholar]
- Daskalakis N.P., Yehuda R. Site-specific methylation changes in the glucocorticoid receptor exon 1F promoter in relation to life adversity: systematic review of contributing factors. Front. Neurosci. 2014;8:369. doi: 10.3389/fnins.2014.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Calignon A., Fox L.M., Pitstick R. Caspase activation precedes and leads to tangles. Nature. 2010;464(7292):1201–1204. doi: 10.1038/nature08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Calignon A., Polydoro M., Suarez-Calvet M. Propagation of tau pathology in a model of early Alzheimer's disease. Neuron. 2012;73(4):685–697. doi: 10.1016/j.neuron.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eades L.E., Thiagarajah A.S., Harris J., Jones S.A., Morand E.F., Leech M. GILZ: a new link between the hypothalamic pituitary adrenal axis and rheumatoid arthritis? Immunol. Cell Biol. 2014;92(9):747–751. doi: 10.1038/icb.2014.56. [DOI] [PubMed] [Google Scholar]
- Edelman S., Shalev I., Uzefovsky F. Epigenetic and genetic factors predict women's salivary cortisol following a threat to the social self. PLoS One. 2012;7(11):e48597. doi: 10.1371/journal.pone.0048597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S., Weiss D., Burghardt R. Promoter specific DNA methylation and gene expression of POMC in acutely underweight and recovered patients with anorexia nervosa. J. Psychiatr. Res. 2010;44(13):827–833. doi: 10.1016/j.jpsychires.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Falini B., Sportoletti P., Brunetti L., Martelli M.P. Perspectives for therapeutic targeting of gene mutations in acute myeloid leukaemia with normal cytogenetics. Br. J. Haematol. 2015 doi: 10.1111/bjh.13409. [DOI] [PubMed] [Google Scholar]
- Falsetti S.A., Resnick H.S., Resick P.A., Kilpatrick D. The Modified PTSD Symptom Scale: a brief self-report measure of posttraumatic stress disorder. Behav. Ther. 1993;16:161–162. [Google Scholar]
- Ferrari P., Sansonnens A., Dick B., Frey F.J. In vivo 11beta-HSD-2 activity: variability, salt-sensitivity, and effect of licorice. Hypertension. 2001;38(6):1330–1336. doi: 10.1161/hy1101.096112. [DOI] [PubMed] [Google Scholar]
- Fink L.A., Bernstein D., Handelsman L., Foote J., Lovejoy M. Initial reliability and validity of the childhood trauma interview: a new multidimensional measure of childhood interpersonal trauma. Am. J. Psychiatry. 1995;152(9):1329–1335. doi: 10.1176/ajp.152.9.1329. [DOI] [PubMed] [Google Scholar]
- First M., Gibbon M., Spitzer R., Williams J.B.W., Smith B.L. American Psychiatric Association; New York: 1994. Structured Clinical Interview for DSM-IV Personality Disorders (SCID-II) [Google Scholar]
- Foa E.B., Tolin D.F. Comparison of the PTSD symptom scale-interview version and the clinician-administered PTSD scale. J. Trauma. Stress. 2000;13(2):181–191. doi: 10.1023/A:1007781909213. [DOI] [PubMed] [Google Scholar]
- Foa E., Cashman L., Jaycox L., Perry K. The validation of a self-report measure of PTSD: The Posttraumatic Diagnostic Scale. Psychol. Assess. 1997;9:445–451. [Google Scholar]
- Fries G.R., Vasconcelos-Moreno M.P., Gubert C. Hypothalamic-pituitary-adrenal axis dysfunction and illness progression in bipolar disorder. Int. J. Neuropsychopharmacol. 2015;18(1) doi: 10.1093/ijnp/pyu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso S., Pizzolo F., Choi S.W. Epigenetic control of 11 beta-hydroxysteroid dehydrogenase 2 gene promoter is related to human hypertension. Atherosclerosis. 2008;199(2):323–327. doi: 10.1016/j.atherosclerosis.2007.11.029. [DOI] [PubMed] [Google Scholar]
- Fukuda K., Straus S.E., Hickie I., Sharpe M.C., Dobbins J.G., Komaroff A. The chronic fatigue syndrome: a comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group. Ann. Intern. Med. 1994;121(12):953–959. doi: 10.7326/0003-4819-121-12-199412150-00009. [DOI] [PubMed] [Google Scholar]
- Furukawa H., Oka S., Matsui T. Genome, epigenome and transcriptome analyses of a pair of monozygotic twins discordant for systemic lupus erythematosus. Hum. Immunol. 2013;74(2):170–175. doi: 10.1016/j.humimm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Gillum R.F. Epidemiology of hypertension in African American women. Am. Heart J. 1996;131(2):385–395. doi: 10.1016/s0002-8703(96)90371-3. [DOI] [PubMed] [Google Scholar]
- Gómez-Beneyto M., Villar M., Renovell M. The diagnosis of personality disorder with a modified version of the SCID-II in a Spanish clinical sample. J. Personal. Disord. 1994;8(2):104–110. [Google Scholar]
- Hahn O.S., Ahn J.H., Song S.H. Development of Korean version of structured clinical interview schedule for DSM-IV axis I disorder: interrater reliability. J. Korean Neuropsychiatr. Assoc. 2000;39(2):362–372. [Google Scholar]
- Hiraki M., Kitajima Y., Koga Y. Aberrant gene methylation is a biomarker for the detection of cancer cells in peritoneal wash samples from advanced gastric cancer patients. Ann. Surg. Oncol. 2011;18(10):3013–3019. doi: 10.1245/s10434-011-1636-0. [DOI] [PubMed] [Google Scholar]
- Hochberg M.C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- Höhne N., Poidinger M., Merz F. FKBP5 genotype-dependent DNA methylation and mRNA regulation after psychosocial stress in remitted depression and healthy controls. Int J Neuropsychopharmacol. 2015;18(4) doi: 10.1093/ijnp/pyu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E.A., Molitor J., Marsit C.J. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30(10):1431–1439. doi: 10.1093/bioinformatics/btu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman E.A., Kile M.L., Christiani D.C., Ince T.A., Kelsey K.T., Marsit C.J. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinforma. 2016;17:259. doi: 10.1186/s12859-016-1140-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter R.W., Ivy J.R., Bailey M.A. Glucocorticoids and renal Na(+) transport: implications for hypertension and salt sensitivity. J. Physiol. 2014;592(8):1731–1744. doi: 10.1113/jphysiol.2013.267609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop D.S. Review: central non-glucocorticoid inhibitors of the hypothalamo-pituitary-adrenal axis. J. Endocrinol. 1999;160(2):169–180. doi: 10.1677/joe.0.1600169. [DOI] [PubMed] [Google Scholar]
- Kadmiel M., Cidlowski J.A. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 2013;34(9):518–530. doi: 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsbeek A., van der Spek R., Lei J., Endert E., Buijs R.M., Fliers E. Circadian rhythms in the hypothalamo-pituitary-adrenal (HPA) axis. Mol. Cell. Endocrinol. 2012;349(1):20–29. doi: 10.1016/j.mce.2011.06.042. [DOI] [PubMed] [Google Scholar]
- Kang G.H., Lee S., Cho N.Y. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Lab. Investig. 2008;88(2):161–170. doi: 10.1038/labinvest.3700707. [DOI] [PubMed] [Google Scholar]
- Kay P., Schlossmacher G., Matthews L. Loss of glucocorticoid receptor expression by DNA methylation prevents glucocorticoid induced apoptosis in human small cell lung cancer cells. PLoS One. 2011;6(10):e24839. doi: 10.1371/journal.pone.0024839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorram N.M., Magee T.R., Wang C., Desai M., Ross M., Khorram O. Maternal undernutrition programs offspring adrenal expression of steroidogenic enzymes. Reprod. Sci. (Thousand Oaks, Calif) 2011;18(10):931–940. doi: 10.1177/1933719111404613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Kubzansky L.D., Baccarelli A.A. Psychological factors and DNA methylation of genes related to immune/inflammatory system markers: the VA Normative Aging Study. BMJ Open. 2016;6(1):e009790. doi: 10.1136/bmjopen-2015-009790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschbaum C., Kudielka B.M., Gaab J., Schommer N.C., Hellhammer D.H. Impact of gender, menstrual cycle phase, and oral contraceptives on the activity of the hypothalamus-pituitary-adrenal axis. Psychosom. Med. 1999;61(2):154–162. doi: 10.1097/00006842-199903000-00006. [DOI] [PubMed] [Google Scholar]
- Klengel T., Mehta D., Anacker C. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci. 2013;16(1):33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzel H.E., Binder E.B., Nickel T. Pharmacological and nonpharmacological factors influencing hypothalamic-pituitary-adrenocortical axis reactivity in acutely depressed psychiatric in-patients, measured by the Dex-CRH test. Neuropsychopharmacology. 2003;28(12):2169–2178. doi: 10.1038/sj.npp.1300280. [DOI] [PubMed] [Google Scholar]
- Labonte B., Azoulay N., Yerko V., Turecki G., Brunet A. Epigenetic modulation of glucocorticoid receptors in posttraumatic stress disorder. Transl. Psychiatry. 2014;4 doi: 10.1038/tp.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam L.L., Emberly E., Fraser H.B. Factors underlying variable DNA methylation in a human community cohort. Proc. Natl. Acad. Sci. U. S. A. 2012;109(Suppl. 2):17253–17260. doi: 10.1073/pnas.1121249109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasagna-Reeves C.A., Castillo-Carranza D.L., Sengupta U. Identification of oligomers at early stages of tau aggregation in Alzheimer's disease. FASEB J. 2012;26(5):1946–1959. doi: 10.1096/fj.11-199851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.G., Buckmaster C.L., Yi E., Schatzberg A.F., Lyons D.M. Coping and glucocorticoid receptor regulation by stress inoculation. Psychoneuroendocrinology. 2014;49:272–279. doi: 10.1016/j.psyneuen.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Rong M., Iacopetta B. DNA hypermethylation in breast cancer and its association with clinicopathological features. Cancer Lett. 2006;237(2):272–280. doi: 10.1016/j.canlet.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Liberati A., Altman D.G., Tetzlaff J. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien H.C., Lu Y.S., Cheng A.L. Differential expression of glucocorticoid receptor in human breast tissues and related neoplasms. J. Pathol. 2006;209(3):317–327. doi: 10.1002/path.1982. [DOI] [PubMed] [Google Scholar]
- Lind G.E., Kleivi K., Meling G.I. ADAMTS1, CRABP1, and NR3C1 identified as epigenetically deregulated genes in colorectal tumorigenesis. Cell. Oncol. 2006;28(5–6):259–272. doi: 10.1155/2006/949506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigano J., Baccarelli A., Mittleman M.A. Air pollution and DNA methylation: interaction by psychological factors in the VA normative aging study. Am. J. Epidemiol. 2012;176(3):224–232. doi: 10.1093/aje/kwr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Blanco A., Ferrer M., Soler J. Association between methylation of the glucocorticoid receptor gene, childhood maltreatment, and clinical severity in borderline personality disorder. J. Psychiatr. Res. 2014;57:34–40. doi: 10.1016/j.jpsychires.2014.06.011. [DOI] [PubMed] [Google Scholar]
- McGowan P.O., Sasaki A., D'Alessio A.C. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melas P.A., Wei Y., Wong C.C. Genetic and epigenetic associations of MAOA and NR3C1 with depression and childhood adversities. Int. J. Neuropsychopharmacol. 2013;16(7):1513–1528. doi: 10.1017/S1461145713000102. [DOI] [PubMed] [Google Scholar]
- Mizoguchi Y., Kajiume T., Miyagawa S., Okada S., Nishi Y., Kobayashi M. Steroid-dependent ACTH-produced thymic carcinoid: regulation of POMC gene expression by cortisol via methylation of its promoter region. Horm. Res. 2007;67(5):257–262. doi: 10.1159/000098548. [DOI] [PubMed] [Google Scholar]
- Moisiadis V.G., Matthews S.G. Glucocorticoids and fetal programming part 1: outcomes. Nat. Rev. Endocrinol. 2014;10(7):391–402. doi: 10.1038/nrendo.2014.73. [DOI] [PubMed] [Google Scholar]
- Moser D., Molitor A., Kumsta R., Tatschner T., Riederer P., Meyer J. The glucocorticoid receptor gene exon 1-F promoter is not methylated at the NGFI-A binding site in human hippocampus. World J. Biol. Psychiatry. 2007;8(4):262–268. doi: 10.1080/15622970701429862. [DOI] [PubMed] [Google Scholar]
- Muschler M.A., Hillemacher T., Kraus C., Kornhuber J., Bleich S., Frieling H. DNA methylation of the POMC gene promoter is associated with craving in alcohol dependence. J Neural Transm. (Vienna) 2010;117(4):513–519. doi: 10.1007/s00702-010-0378-7. [DOI] [PubMed] [Google Scholar]
- Na K.S., Chang H.S., Won E. Association between glucocorticoid receptor methylation and hippocampal subfields in major depressive disorder. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0085425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCI Cancer Health Disparities. 2008. http://www.cancer.gov/about-nci/organization/crchd/cancer-health-disparities-fact-sheet
- Needham B.L., Smith J.A., Zhao W. Life course socioeconomic status and DNA methylation in genes related to stress reactivity and inflammation: the multi-ethnic study of atherosclerosis. Epigenetics. 2015;10(10):958–969. doi: 10.1080/15592294.2015.1085139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesset K.A., Perri A.M., Mueller C.R. Frequent promoter hypermethylation and expression reduction of the glucocorticoid receptor gene in breast tumors. Epigenetics. 2014;9(6):851–859. doi: 10.4161/epi.28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Price J. Proopiomelanocortin gene expression and DNA methylation: implications for Cushing's syndrome and beyond. J. Endocrinol. 2003;177(3):365–372. doi: 10.1677/joe.0.1770365. [DOI] [PubMed] [Google Scholar]
- Nwankwo T., Yoon S.S., Burt V., Gu Q. Hypertension among adults in the United States: National Health and Nutrition Examination Survey, 2011-2012. NCHS Data Brief. 2013;(133):1–8. [PubMed] [Google Scholar]
- Paakinaho V., Makkonen H., Jaaskelainen T., Palvimo J.J. Glucocorticoid receptor activates poised FKBP51 locus through long-distance interactions. Mol. Endocrinol. 2010;24(3):511–525. doi: 10.1210/me.2009-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Gudiel H., Cordova-Palomera A., Leza J.C., Fananas L. Glucocorticoid receptor gene (NR3C1) methylation processes as mediators of early adversity in stress-related disorders causality: a critical review. Neurosci. Biobehav. Rev. 2015;55:520–535. doi: 10.1016/j.neubiorev.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Papadopoulou N.G., Oleson L., Kempuraj D., Donelan J., Cetrulo C.L., Theoharides T.C. Regulation of corticotropin-releasing hormone receptor-2 expression in human cord blood-derived cultured mast cells. J. Mol. Endocrinol. 2005;35(3):R1–R8. doi: 10.1677/jme.1.01833. [DOI] [PubMed] [Google Scholar]
- Parrella P., Poeta M.L., Gallo A.P. Nonrandom distribution of aberrant promoter methylation of cancer-related genes in sporadic breast tumors. Clin. Cancer Res. 2004;10(16):5349–5354. doi: 10.1158/1078-0432.CCR-04-0555. [DOI] [PubMed] [Google Scholar]
- Perroud N., Paoloni-Giacobino A., Prada P. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Transl. Psychiatry. 2011;1:e59. doi: 10.1038/tp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud N., Paoloni-Giacobino A., Prada P. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Transl. Psychiatry. 2011;1(12) doi: 10.1038/tp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzolo F., Friso S., Morandini F. Apparent mineralocorticoid excess by a novel mutation and epigenetic modulation by HSD11B2 promoter methylation. J. Clin. Endocrinol. Metab. 2015;100(9):E1234–E1241. doi: 10.1210/jc.2015-1760. [DOI] [PubMed] [Google Scholar]
- Prados J., Stenz L., Courtet P. Borderline personality disorder and childhood maltreatment: a genome-wide methylation analysis. Genes Brain Behav. 2015;14(2):177–188. doi: 10.1111/gbb.12197. [DOI] [PubMed] [Google Scholar]
- Preisig M., Fenton B.T., Matthey M.-L., Berney A., Ferrero F. Diagnostic interview for genetic studies (DIGS): inter-rater and test-retest reliability of the French version. Eur. Arch. Psychiatry Clin. Neurosci. 1999;249(4):174–179. doi: 10.1007/s004060050084. [DOI] [PubMed] [Google Scholar]
- Presul E., Schmidt S., Kofler R., Helmberg A. Identification, tissue expression, and glucocorticoid responsiveness of alternative first exons of the human glucocorticoid receptor. J. Mol. Endocrinol. 2007;38(1–2):79–90. doi: 10.1677/jme.1.02183. [DOI] [PubMed] [Google Scholar]
- Quax R.A., Manenschijn L., Koper J.W. Glucocorticoid sensitivity in health and disease. Nat. Rev. Endocrinol. 2013;9(11):670–686. doi: 10.1038/nrendo.2013.183. [DOI] [PubMed] [Google Scholar]
- Ratman D., Vanden Berghe W., Dejager L. How glucocorticoid receptors modulate the activity of other transcription factors: a scope beyond tethering. Mol. Cell. Endocrinol. 2013;380(1–2):41–54. doi: 10.1016/j.mce.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Ronco A.M., Llaguno E., Epunan M.J., Llanos M.N. Effect of cadmium on cortisol production and 11beta-hydroxysteroid dehydrogenase 2 expression by cultured human choriocarcinoma cells (JEG-3) Toxicol. in Vitro. 2010;24(6):1532–1537. doi: 10.1016/j.tiv.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vega B., Gandhi V. Glucocorticoid resistance in a multiple myeloma cell line is regulated by a transcription elongation block in the glucocorticoid receptor gene (NR3C1) Br. J. Haematol. 2009;144(6):856–864. doi: 10.1111/j.1365-2141.2008.07549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santacruz K., Lewis J., Spires T. Tau suppression in a neurodegenerative mouse model improves memory function. Science. 2005;309(5733):476–481. doi: 10.1126/science.1113694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloesser R.J., Martinowich K., Manji H.K. Mood-stabilizing drugs: mechanisms of action. Trends Neurosci. 2012;35(1):36–46. doi: 10.1016/j.tins.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Shannon C.P., Balshaw R., Chen V. Enumerateblood – an R package to estimate the cellular composition of whole blood from Affymetrix Gene ST gene expression profiles. BMC Genomics. 2017;18:43. doi: 10.1186/s12864-016-3460-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields A.E., Wise L.A., Ruiz-Narvaez E.A. Childhood abuse, promoter methylation of leukocyte NR3C1 and the potential modifying effect of emotional support. Epigenomics. 2016;8(11):1507–1517. doi: 10.2217/epi-2016-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2015. CA Cancer J. Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- Smith S.M., Vale W.W. The role of the hypothalamic-pituitary-adrenal axis in neuroendocrine responses to stress. Dialogues Clin. Neurosci. 2006;8(4):383–395. doi: 10.31887/DCNS.2006.8.4/ssmith. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth L.J., McKay G.J., Maxwell A.P., McKnight A.J. DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics. 2014;9(3):366–376. doi: 10.4161/epi.27161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger H., Labonte B., Groleau P., Turecki G., Israel M. Methylation of the glucocorticoid receptor gene promoter in bulimic women: associations with borderline personality disorder, suicidality, and exposure to childhood abuse. Int. J. Eat. Disord. 2013;46(3):246–255. doi: 10.1002/eat.22113. [DOI] [PubMed] [Google Scholar]
- Strehl C., Buttgereit F. Optimized glucocorticoid therapy: teaching old drugs new tricks. Mol. Cell. Endocrinol. 2013;380(1–2):32–40. doi: 10.1016/j.mce.2013.01.026. [DOI] [PubMed] [Google Scholar]
- Tasker J.G., Herman J.P. Mechanisms of rapid glucocorticoid feedback inhibition of the hypothalamic-pituitary-adrenal axis. Stress. 2011;14(4):398–406. doi: 10.3109/10253890.2011.586446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson J.W., Walker E.A., Bujalska I.J. 11β-Hydroxysteroid dehydrogenase type 1: a tissue-specific regulator of glucocorticoid response. Endocr. Rev. 2004;25(5):831–866. doi: 10.1210/er.2003-0031. [DOI] [PubMed] [Google Scholar]
- Tseng E., Prica A., Zhang L. Monthly blood transfusions decrease after four months of azacitidine. Vox Sang. 2015 doi: 10.1111/vox.12266. [DOI] [PubMed] [Google Scholar]
- Turner J.D., Alt S.R., Cao L. Transcriptional control of the glucocorticoid receptor: CpG islands, epigenetics and more. Biochem. Pharmacol. 2010;80(12):1860–1868. doi: 10.1016/j.bcp.2010.06.037. [DOI] [PubMed] [Google Scholar]
- Turecki G. The molecular bases of the suicidal brain. Nat. Rev. Neurosci. 2014;15(12):802–816. doi: 10.1038/nrn3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecki G., Meaney M.J. Effects of the social environment and stress on glucocorticoid receptor gene methylation: a systematic review. Biol. Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.D., Muller C.P. Structure of the glucocorticoid receptor (NR3C1) gene 5′ untranslated region: identification, and tissue distribution of multiple new human exon 1. J. Mol. Endocrinol. 2005;35(2):283–292. doi: 10.1677/jme.1.01822. [DOI] [PubMed] [Google Scholar]
- Turner J.D., Vernocchi S., Schmitz S., Muller C.P. Role of the 5′-untranslated regions in post-transcriptional regulation of the human glucocorticoid receptor. Biochim. Biophys. Acta. 2014;1839(11):1051–1061. doi: 10.1016/j.bbagrm.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Tyrka A.R., Ridout K.K., Parade S.H., Paquette A., Marsit C.J., Seifer R. Childhood maltreatment and methylation of FK506 binding protein 5 gene (FKBP5) Dev. Psychopathol. 2015;27(4 Pt 2):1637–1645. doi: 10.1017/S0954579415000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udali S., Guarini P., Moruzzi S., Choi S.W., Friso S. Cardiovascular epigenetics: from DNA methylation to microRNAs. Mol. Asp. Med. 2013;34(4):883–901. doi: 10.1016/j.mam.2012.08.001. [DOI] [PubMed] [Google Scholar]
- van der Goes M.C., Jacobs J.W., Bijlsma J.W. The value of glucocorticoid co-therapy in different rheumatic diseases - positive and adverse effects. Arthritis Res. Ther. 2014;16(Suppl. 2):S2. doi: 10.1186/ar4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangeel E., Van Den Eede F., Hompes T. Chronic fatigue syndrome and DNA hypomethylation of the glucocorticoid receptor gene promoter 1F region: associations with HPA Axis hypofunction and childhood trauma. Psychosom. Med. 2015;77(8):853–862. doi: 10.1097/PSY.0000000000000224. [DOI] [PubMed] [Google Scholar]
- Veenendaal M.V., Costello P.M., Lillycrop K.A. Prenatal famine exposure, health in later life and promoter methylation of four candidate genes. J. Dev. Orig. Health Dis. 2012;3(6):450–457. doi: 10.1017/S2040174412000396. [DOI] [PubMed] [Google Scholar]
- Vermeer H., Hendriks-Stegeman B.I., van der Burg B., van Buul-Offers S.C., Jansen M. Glucocorticoid-induced increase in lymphocytic FKBP51 messenger ribonucleic acid expression: a potential marker for glucocorticoid sensitivity, potency, and bioavailability. J. Clin. Endocrinol. Metab. 2003;88(1):277–284. doi: 10.1210/jc.2002-020354. [DOI] [PubMed] [Google Scholar]
- Vilasco M., Communal L., Mourra N., Courtin A., Forgez P., Gompel A. Glucocorticoid receptor and breast cancer. Breast Cancer Res. Treat. 2011;130(1):1–10. doi: 10.1007/s10549-011-1689-6. [DOI] [PubMed] [Google Scholar]
- Vukojevic V., Kolassa I.T., Fastenrath M. Epigenetic modification of the glucocorticoid receptor gene is linked to traumatic memory and post-traumatic stress disorder risk in genocide survivors. J. Neurosci. 2014;34(31):10274–10284. doi: 10.1523/JNEUROSCI.1526-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q., Gao K., Rong H. Histone modifications of the Crhr1 gene in a rat model of depression following chronic stress. Behav. Brain Res. 2014;271:1–6. doi: 10.1016/j.bbr.2014.05.031. [DOI] [PubMed] [Google Scholar]
- Weathers F.W., Keane T.M., Davidson J.R. Clinician-administered PTSD scale: a review of the first ten years of research. Depress Anxiety. 2001;13(3):132–156. doi: 10.1002/da.1029. [DOI] [PubMed] [Google Scholar]
- Westphal N.J., Seasholtz A.F. CRH-BP: the regulation and function of a phylogenetically conserved binding protein. Front. Biosci. 2006;11:1878–1891. doi: 10.2741/1931. [DOI] [PubMed] [Google Scholar]
- Wittchen H.U., Lachner G., Wunderlich U., Pfister H. Test-retest reliability of the computerized DSM-IV version of the Munich-Composite International Diagnostic Interview (M-CIDI) Soc. Psychiatry Psychiatr. Epidemiol. 1998;33(11):568–578. doi: 10.1007/s001270050095. [DOI] [PubMed] [Google Scholar]
- Wochnik G.M., Ruegg J., Abel G.A., Schmidt U., Holsboer F., Rein T. FK506-binding. Proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J. Biol. Chem. 2005;280(6):4609–4616. doi: 10.1074/jbc.M407498200. [DOI] [PubMed] [Google Scholar]
- Wu Q., Lothe R.A., Ahlquist T. DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol. Cancer. 2007;6:45. doi: 10.1186/1476-4598-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Ewald E.R., Huo Y. Glucocorticoid-induced loss of DNA methylation in non-neuronal cells and potential involvement of DNMT1 in epigenetic regulation of Fkbp5. Biochem. Biophys. Res. Commun. 2012;420(3):570–575. doi: 10.1016/j.bbrc.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R., Daskalakis N.P., Desarnaud F. Epigenetic biomarkers as predictors and correlates of symptom improvement following psychotherapy in combat veterans with PTSD. Front. Psychiatry. 2013;4:118. doi: 10.3389/fpsyt.2013.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R., Flory J.D., Bierer L.M. Lower methylation of glucocorticoid receptor gene promoter 1 in peripheral blood of veterans with posttraumatic stress disorder. Biol. Psychiatry. 2015;77(4):356–364. doi: 10.1016/j.biopsych.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Yehuda R., Daskalakis N.P., Bierer L.M. Holocaust exposure induced intergenerational effects on FKBP5 methylation. Biol. Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Yoon S.S., Carroll M.D., Fryar C.D. Hypertension prevalence and control among adults: United States, 2011–2014. NCHS Data Brief. 2015;(220):1–8. [PubMed] [Google Scholar]
- Zannas A.S., Wiechmann T., Gassen N.C., Binder E.B. Gene-stress-epigenetic regulation of FKBP5: clinical and translational implications. Neuropsychopharmacology. 2016;41(1):261–274. doi: 10.1038/npp.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Clark A.F., Yorio T. FK506-binding protein 51 regulates nuclear transport of the glucocorticoid receptor beta and glucocorticoid responsiveness. Invest. Ophthalmol. Vis. Sci. 2008;49(3):1037–1047. doi: 10.1167/iovs.07-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., An Q., Goldberg J., Quyyumi A.A., Vaccarino V. Promoter methylation of glucocorticoid receptor gene is associated with subclinical atherosclerosis: a monozygotic twin study. Atherosclerosis. 2015;242(1):71–76. doi: 10.1016/j.atherosclerosis.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond A.S., Snaith R.P. The hospital anxiety and depression scale. Acta Psychiatr. Scand. 1983;67(6):361–370. doi: 10.1111/j.1600-0447.1983.tb09716.x. [DOI] [PubMed] [Google Scholar]