ABSTRACT
In Gram-negative bacteria, the cytoplasmic membrane protein TonB transmits energy derived from proton motive force to energize transport of important nutrients through TonB-dependent transporters in the outer membrane. Each transporter consists of a beta barrel domain and a lumen-occluding cork domain containing an essential sequence called the TonB box. To date, the only identified site of transporter-TonB interaction is between the TonB box and residues ∼158 to 162 of TonB. While the mechanism of ligand transport is a mystery, a current model based on site-directed spin labeling and molecular dynamics simulations is that, following ligand binding, the otherwise-sequestered TonB box extends into the periplasm for recognition by TonB, which mediates transport by pulling or twisting the cork. In this study, we tested that hypothesis with the outer membrane transporter FepA using in vivo photo-cross-linking to explore interactions of its TonB box and determine whether additional FepA-TonB interaction sites exist. We found numerous specific sites of FepA interaction with TonB on the periplasmic face of the FepA cork in addition to the TonB box. Two residues, T32 and A33, might constitute a ligand-sensitive conformational switch. The facts that some interactions were enhanced in the absence of ligand and that other interactions did not require the TonB box argued against the current model and suggested that the transport process is more complex than originally conceived, with subtleties that might provide a mechanism for discrimination among ligand-loaded transporters. These results constitute the first study on the dynamics of TonB-gated transporter interaction with TonB in vivo.
IMPORTANCE The TonB system of Gram-negative bacteria has a noncanonical active transport mechanism involving signal transduction and proteins integral to both membranes. To achieve transport, the cytoplasmic membrane protein TonB physically contacts outer membrane transporters such as FepA. Only one contact between TonB and outer membrane transporters has been identified to date: the TonB box at the transporter amino terminus. The TonB box has low information content, raising the question of how TonB can discriminate among multiple different TonB-dependent transporters present in the bacterium if it is the only means of contact. Here we identified several additional sites through which FepA contacts TonB in vivo, including two neighboring residues that may explain how FepA signals to TonB that ligand has bound.
KEYWORDS: TonB, FepA, cork domain, p-benzoyl-l-phenylalanine, enterochelin, TonB box, energy transduction
INTRODUCTION
Iron is essential for growth of Gram-negative bacteria but can be difficult to acquire because of its insolubility under aerobic conditions, its scarcity in hosts, and the protective nature of the Gram-negative bacterial outer membrane (OM) (1). The majority of Gram-negative bacteria synthesize and secrete siderophores, which are high-affinity iron chelators that are then transported into the bacteria with a captured iron atom. Consequently, Gram-negative bacteria use the TonB system to energize the active transport of large, scarce, important nutrients, such as iron siderophores and vitamin B12, across the essentially unenergized OM. Nutrients bind to OM TonB-dependent transporters, and a complex of the cytoplasmic membrane (CM) proteins ExbB, ExbD, and TonB transduces energy derived from CM proton motive force to promote passage into the periplasmic space (2). Of the three integral CM proteins, TonB (239 residues) is anchored in the cytoplasmic membrane by an amino-terminal transmembrane domain and extends its carboxy terminus across the periplasmic space to directly interact with ligand-loaded transporters (3–5). ExbD appears to determine the conformation of TonB required for productive interaction with OM transporters, and ExbB scaffolds both TonB and ExbD (6). Escherichia coli K-12 produces 7 different OM transporters for binding of various siderophores, other iron-containing compounds, and vitamin B12. Other bacteria, such as Bacteroides thetaiotaomicron, encode up to 120 putative transporters, some of which are dedicated to uptake of plant carbohydrates and starch (7). Many pathogenic Gram-negative bacteria use the TonB system to acquire host heme as a source of iron (8).
Each OM transporter consists of two domains: a 22-strand beta barrel and an essential globular domain of ∼150 residues, referred to as a cork (or hatch), that completely blocks the lumen of the barrel (for a review, see reference 9). There is general agreement in the field that the cork must undergo conformational changes both to signal that a transport ligand has been bound to external surface loops with nanomolar affinities and to subsequently release the transport ligand into the periplasm (10–13). The conservation of the 4-strand beta sheet within the cork domain suggests a similarity of mechanism, but its role is unknown (9, 14, 15). Study of the means by which the cork moves and the extent of movement has thus far been focused entirely on a semiconserved, and essential, sequence at the amino terminus of the cork where TonB interacts, called the TonB box (Fig. 1C) (4, 5, 16–18). The TonB boxes of transporters are interchangeable, confirming similarity of transport mechanisms (5, 16). TonB Cys substitutions in the region of Q160 form disulfide bonds in vivo with Cys substitutions in the TonB boxes of FhuA and BtuB (4, 5). In vitro, TonB residues ∼153 to 235 cocrystalize with transporters BtuB and FhuA such that the region around Q160 likewise binds to the TonB box but to slightly different regions of the TonB box, depending on the cocrystal structure (19, 20).
FIG 1.
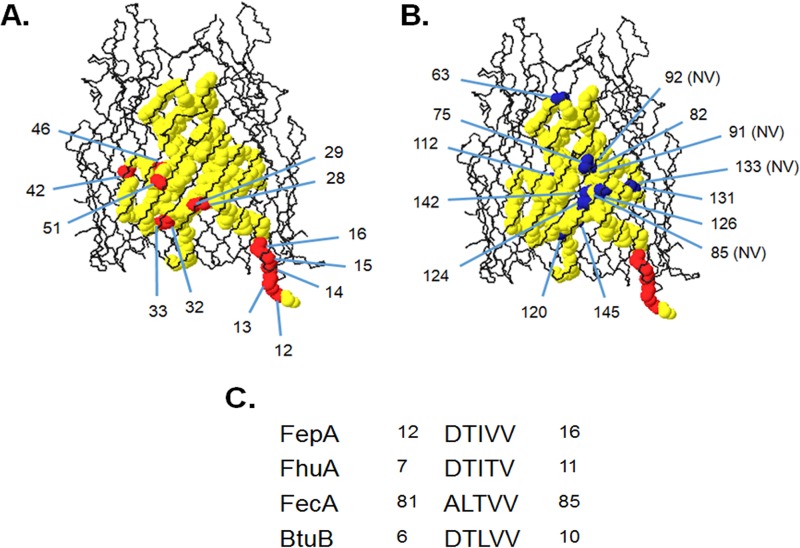
Locations of FepA pBpa substitutions used for TonB-FepA photo-cross-linking analysis mapped onto the FepA crystal structure. Swiss-Pdb viewer (66) was used to create images of the FepA crystal structure (Protein Data Bank no. 1fep). (A) Positions of pBpa substitutions generated within the amino-terminal 51 residues of FepA. This region is accessible to the thiol-specific probe BMCC in vivo during transport of colicin B (29). The FepA beta barrel is colored black. The polypeptide backbone for cork domain residues is shown in space-filling format (yellow), with corresponding positions of diagnostic pBpa substitutions in red. The periplasm corresponds to the area underneath the beta barrel. (B) Positions of pBpa substitutions generated within FepA residues 63 to 145. The FepA beta barrel and cork polypeptide backbone is presented in the same manner as in panel A, but the polypeptide backbone residues are now colored blue to reflect the observation that many of these side chains are sequestered from BMCC when substituted with cysteine (29). Positions 85, 91, 92, and 133 were not visible (NV) when presented as shown in panel B, but approximate positions are labeled. (C) Alignment of TonB box sequences of FepA, FhuA, FecA, and BtuB. The numbers correspond to the primary amino acid sequence of the mature protein.
Based on site-directed spin labeling in vitro, it is proposed that binding of a ligand on the external surface of an OM transporter causes the otherwise-sequestered TonB box to extend into the periplasmic space independently of TonB. This newly available TonB box is then recognized by TonB, which causes release of the ligand into the periplasmic space (21). It is also hypothesized that pulling by TonB region Q160, bound to the TonB box, with downward force perpendicular to the beta sheets could be sufficient to completely remove the cork from the barrel and allow transport (15). In support of this idea, molecular dynamics simulations have modeled TonB pulling down on the cork domain by applying a force perpendicular to the cork domain. This perpendicular force then unwinds the cork such that the amino-terminal portions of the cork become exposed to the periplasm, revealing a channel just large enough for transport ligands to pass through (12, 15). After ligand transport, the cork may spontaneously refold back into the barrel. Consistent with this, individually expressed cork and barrel domains are, as predicted (22), able to reconstitute a functional FhuA transporter in vivo (23). Also, a partially denatured purified BtuB cork can spontaneously renature to its original condition upon removal of denaturant (24).
There are species of Gram-negative bacteria that encode multiple TonB proteins as well as many TonB-dependent transporters. Xanthomonas campestris pv. campestris has eight predicted TonB homologues with 48 predicted TonB-dependent transporters (7). It seems unlikely that the 5- to 7-residue TonB boxes of the transporters carry sufficient information to allow the correct TonB to locate and bind to the correct transporter.
Consistent with that idea, in vivo studies suggest that the mechanism of energy transduction and TonB-dependent transport is more complicated than just the residues surrounding TonB Q160 pulling on the TonB box handle: individual Cys substitutions in TonB residues 158 to 163 do not inactivate TonB, a deletion of TonB residues 158 to 162 does not inactivate TonB, inactivating mutations in the BtuB TonB box (L8P and V10P) do not prevent TonB binding to TonB box residues, and binding of two different ligands to BtuB elicits different degrees of exposure of the TonB box (4, 25–28). Furthermore, both in vivo and in vitro studies have detected significant movement of only roughly the amino-terminal one-third of the cork residues that are accessible from the periplasm. While removing one-third of the cork residues from the barrel would leave an opening sufficient for passage of transport ligands such as siderophores and vitamin B12, it may not be sufficient for larger ligands such as the 55-kDa protein toxin colicin B (24, 29–32).
While TonB interactions with the OM transporters have been observed in vitro and inferred in silico, they have not been totally congruent with subsequent in vivo results. The BtuB- and FhuA-TonB cocrystal structures suggest that TonB R158 and R166 are, respectively, important in formation of salt bridges with the transporters. The molecular dynamics simulations based on the BtuB-TonB cocrystal structure also predict many transient and some stable salt bridges during the equilibrium simulation (12). However, comparison of 4 transporter sequence alignments shows that none of the BtuB participants in the salt bridges are conserved among transporters (9, 15). Furthermore, simultaneous mutation of three TonB residues (TonB R158A, R166A, and R171A) proposed to participate in the salt bridges still allows for 75% of TonB activity, indicating that salt bridges through these Arg residues are not essential to the transport mechanism (26).
In addition to binding to TonB-dependent transporters, the TonB carboxy-terminal domain exhibits “conformational plasticity” by binding to a diverse array of nonfunctional targets such as OmpA and Lpp, to peptides corresponding to extracellular loops of the OM transporter FhuA, and to hen egg white lysozyme (33–35). Consistent with that, the carboxy terminus also contains significant regions of predicted intrinsic disorder (36). Our hypothesis is that the TonB carboxy-terminal domain can adopt several conformations in order to bind to structurally diverse outer membrane targets, but only the correct conformations elicited by prior interaction with the ExbD periplasmic domain will bring about correct contacts with transporters that lead to energy transduction (6). Our data suggest that other, yet-to-be-discovered important transporter-TonB interactions exist (29). This idea was also proposed years ago by Kadner and colleagues based on the low degree of structural discrimination to be found in TonB boxes (16).
In this study, we used FepA as a model TonB-dependent transporter and site-specific photo-cross-linking to identify residues both within and outside the FepA TonB box that made contact with TonB in vivo. Our strategy was to engineer amber codons that allowed introduction of a nonnative photo-cross-linkable amino acid, p-benzoylphenylalanine (pBpa) at those sites (37). In the presence of UV light, cross-links between FepA and TonB could be formed. This technique allowed us to choose the sites in FepA to examine while leaving the target sites in TonB unconstrained. In this study, we showed for the first time that the core TonB box is a region through which the transporter FepA, like others, contacts TonB. Several novel contacts in addition to the TonB box were discovered on the periplasmic surface of FepA. Some of the contacts occurred more strongly in the absence of transport ligand, consistent with the idea that TonB samples sites on transporters in the process of identifying those with ligand bound, as previously suggested (35). We also identified interactions that occurred without a requirement for the TonB box, consistent with our earlier finding that TonB is required to bind FepA at unknown sites in order to cause exposure of the FepA TonB box (29). Taken together, our results suggest that current models, where ligand binding to the transporter is a prerequisite for interaction with TonB and where TonB interacts only with the transporter TonB box, need to be revised.
RESULTS
Rationale for FepA sites of in vivo photo-cross-linking.
To examine in vivo interactions between FepA and TonB, individual amber codons (UAG) were engineered in the FepA cork domain (Fig. 1A and B). Unnatural amino acids such as p-benzoyl-l-phenylalanine (pBpa) can be incorporated into proteins of interest through amber codon suppression using a plasmid-encoded orthogonal suppressor tRNA and tRNA synthetase (37, 38). Upon excitation with near-UV light at 365 nm, pBpa cross-links to aliphatic side chains within 3.0 Å of its benzophenone group (39). Thus, to trap an interaction between two proteins using pBpa-mediated photo-cross-linking, only one of the two proteins needs to be replaced with pBpa. We chose to insert the pBpa in the FepA protein to identify residues of the transporter that interacted with TonB.
Engineered Cys residues in the BtuB TonB box D6, T7, L8,V9, and V10 are sites of interaction with engineered Cys residues in the region of TonB Q160 (4). To characterize interactions of the FepA TonB box with TonB, consecutive amber codons were engineered in the FepA TonB box from residue 12 to 16 (in the mature FepA sequence). We also examined the possibility of novel interactions between 21 other sites in the FepA cork and TonB.
Based on its crystal structure, the FepA cork domain contains a four-strand β sheet (denoted here with approximate boundaries as β1 [residues 25 to 31], β2 [residues 82 to 87], β3 [residues 120 to 126], and β4 [residues 140 to 146]) that is a common feature among transporters (14, 15, 40). To test the interaction of FepA strand β1, the strand that is most accessible to the periplasm in the FepA crystal structure, with TonB, nonsense codons were generated at FepA codons 28 and 29 as well as 32 and 33 just beyond the β1 boundary (Fig. 1A). Amber codons were also generated at codons 82 and 85, where the respective residues are located on opposite sides of strand β2. In addition, residue L85 is part of a conserved LIDG motif of unknown function (15, 40). To determine if strands β3 and β4, which contain some periplasmically exposed residues, also interact with TonB, nonsense codons were also engineered in place of E120 and V124 on strand β3 and V142 and I145 on strand β4 (Fig. 1B).
Additional nonsense codons were created at sites that we had previously characterized as being accessible to or sequestered from the periplasm based on thiol-specific labeling of engineered Cys substitutions (Fig. 1A and B). Because FepA substitutions A42C, S46C, and T51C are labeled by 1-biotinamido-4-[4′-(maleimidomethyl)cyclohexanecarboxamido]butane (BMCC) in a TonB-dependent manner (29), it was possible that these sites, when replaced by pBpa, would cross-link to TonB by virtue of being accessible to the periplasmic space during transport. In contrast, FepA cork substitutions V91C, S92C, S112C, V124, A131C, and V142C are sequestered from labeling by BMCC and thus either were not accessible to the periplasm under any circumstance or were shielded from the label by an unknown protein (29).
We also chose certain positions based on their location in the FepA crystal structure or sequence conservation (14, 15, 40). Highly conserved cork domain side chains R75 and R126 are sequestered from the periplasm in the crystal structure and form part of the conserved “lock region” of salt bridges that aligns the cork to the barrel wall (41). In FhuA, mutagenesis studies show that the salt bridges are not required for function (42). Mutations at these residues interfere with ligand transport but not binding, suggesting that they play a role in transport and potentially in contacting TonB (41, 43). Y133 is also a highly conserved side chain in TonB-dependent transporters and is positioned above the TonB box in the sequestered portion of the cork domain (14, 15). FepA S63, located at the apex of the cork domain, was engineered to test if the cork domain fully exits the barrel to contact TonB during transport. We chose not to mutate the numerous highly conserved Gly residues because that would likely introduce significant structural perturbations.
The majority of FepA pBpa substitutions retain activity.
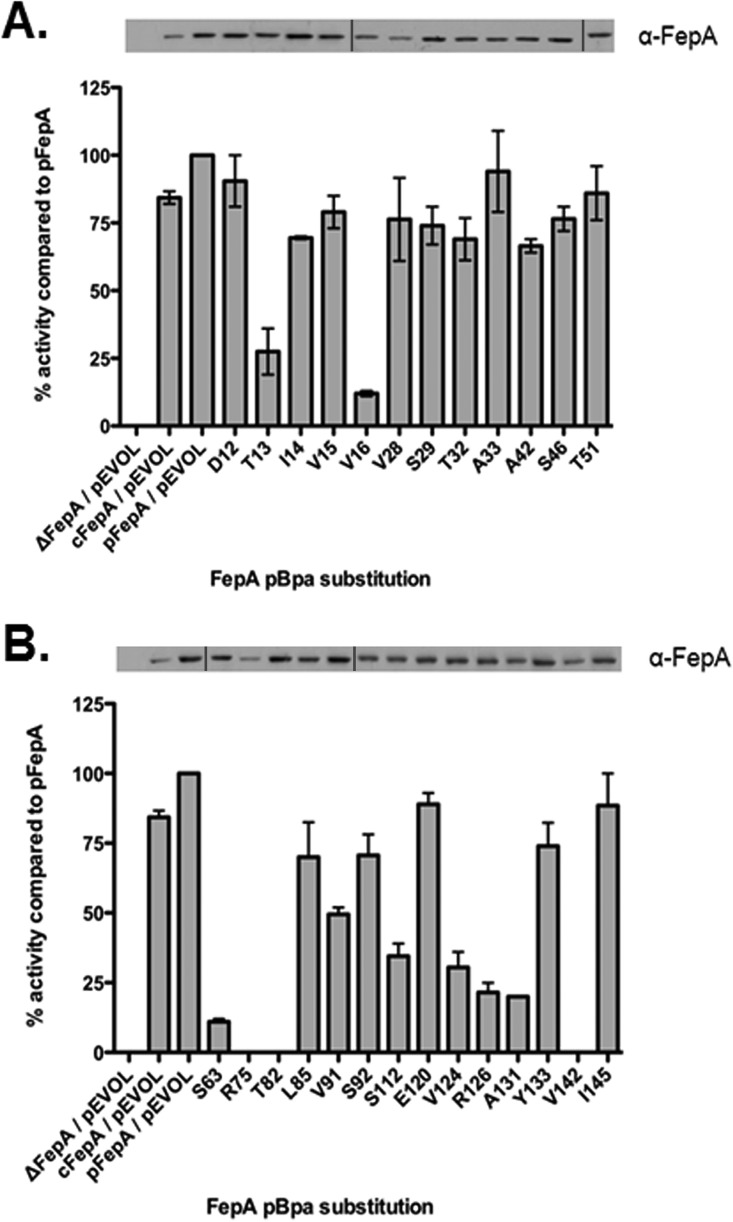
Mutagenic analyses of the amino-terminal TonB boxes of transporters show them to be generally tolerant to substitutions, except for Pro and Gly substitutions, which are structurally disruptive (42). Here, the pattern of inactivity within the FepA TonB box pBpa substitutions differed from what has been observed previously for cysteine substitutions in either BtuB or FhuA, where positions 3 and 5 (Fig. 1C) are those susceptible to inactivation (4, 16, 44). FepA T13pBpa, a position that is not generally very sensitive to substitution, was only about 25% active, whereas I14pBpA, a site where “classical” TonB box mutations occur, was ∼70% active here (45). It may be that the unnatural amino acid pBpa was having an unusual effect in the TonB box. As is commonly seen at the fifth position in the TonB box, FepA V16pBpa was largely inactive. Wild-type, His-tagged FepA expressed from pKP1693 supported full 55Fe-enterochelin uptake compared to chromosomal FepA and served as the standard for 100% uptake.
The amino-terminal residues from position 28 to 51 proved to be the most impervious to pBpa substitution and supported 55Fe-enterochelin transport at between 65 and 100% of the pFepA (pKP1693) initial uptake rate, indicating that they retained essentially wild-type function (Fig. 2A). However, several substitutions within FepA residues 63 to 145 (Fig. 2B) supported significantly decreased 55Fe-enterochelin uptake rates. The region of the FepA cork from S63 to I145 was fairly sensitive to pBpa substitutions, with over half unable to support transport of 55Fe-enterochelin at greater than ∼35% of wild-type rates. In fact, R75pBpa, T82pBpa, and V142pBpa were completely unable to transport 55Fe-enterochelin (Fig. 2B). This result raised the possibility that they were important regions of contact with TonB or that the bulky pBpa substitutions disrupted the structure of the FepA cork domain. Lack of function was not due to inefficient incorporation of pBpa, since each of the FepA pBpa derivatives analyzed was expressed to a level similar to that for the pFepA (pKP1693) control (Fig. 2B). The less active pBpa substitutions were all somewhat sensitive to colicin B and thus somewhat active, with V16pBpa supporting the least activity (see Table S2 in the supplemental material). Unlike Fe transport, the colicin B sensitivity assay discriminates among low levels of TonB activity (46).
FIG 2.
55Fe-enterochelin transport rates for FepA pBpa substitutions. (A) Initial rates of transport and FepA expression levels for D12pBpa-T51pBpa; (B) initial rates of transport and FepA expression levels for S63pBpa-I145pBpa. Substitutions were expressed in strain KP1490 (W3110 ΔfepA aroB) to approximately the same levels as uninduced plasmid-encoded wild-type FepA [KP1490/pFepA (pKP1693)/pEVOL-BpF]. Strain KP1270 (W3110 aroB) transformed with pEVOL-BpF (cFepA/pEVOL) and ΔFepA/pEVOL served as uptake controls. Initial rates of 55Fe-enterochelin transport were determined as described in Materials and Methods. At an A550 of 0.20 (early exponential phase), all samples were treated with 0.4 mM pBpa, and expression of pBpa substitutions was induced by the addition of sodium propionate. Expression of T13pBpa, V16pBpa, and R75pBpa required 3 mM sodium propionate, and all other substitutions were induced with 2 mM sodium propionate. Samples were assayed in triplicate, and linear regression was used to determine initial enterochelin uptake rates. Initial ferric enterochelin transport rates were determined as counts per minute/0.05 A550 ml cells/minute. The resulting slopes were normalized to the rate observed for pFepA/pEVOL. Average normalized activities from at least two independent experiments are shown. As indicated by the lines, this figure shows a composite of results from separate experiments due to the number of triplicate samples that can be analyzed in a single experiment. For this figure, exposures were chosen based on comparison to controls of chromosomally encoded FepA included in each separate immunoblot.
FepA TonB box pBpA residues cross-link differentially to TonB.
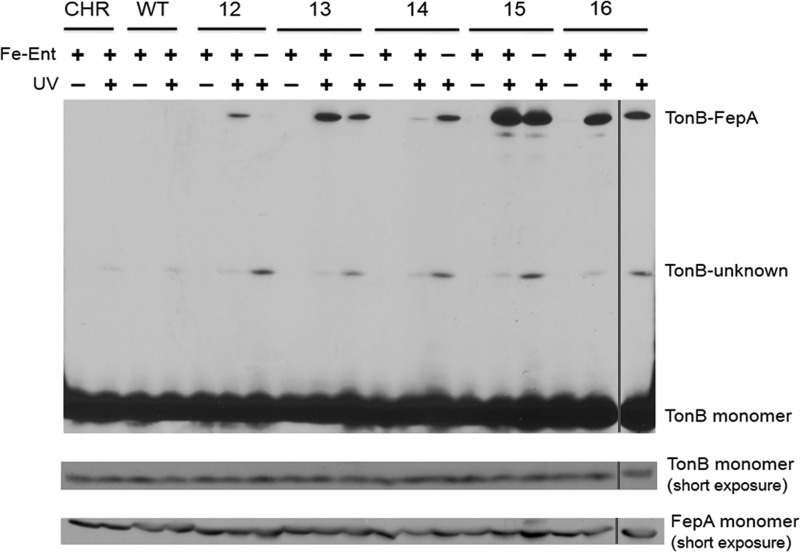
To better understand the activities of the TonB box pBpa derivatives, we photo-cross-linked them to unknown sites in TonB and analyzed the results by anti-TonB immunoblotting. Each FepA pBpa substitution in the TonB box could form a UV-dependent, ∼117 kDa complex with TonB (Fig. 3), consistent with the predicted molecular mass of a FepA (80 kDa)-TonB (37 kDa) heterodimer. Relative to the other substitutions, FepA V15pBpa cross-linked most strongly to TonB and also supported ∼80% of the wild-type enterochelin transport rate (Fig. 2A and 3), so it was chosen to be a cross-linking standard for the remaining experiments. The TonB-FepA complex was only faintly detected with anti-FepA antibodies, likely because only a fraction of the total FepA was physically interacting with TonB (data not shown). Chromosomal and plasmid-encoded wild-type FepA, which lacked pBpa substitutions, did not photo-cross-link to TonB.
FIG 3.
pBpa substitutions in the FepA TonB box cross-link to TonB in vivo. FepA pBpa substitutions were expressed in the enterochelin-producing strain KP1489 (Fe-Ent +) or the enterochelin-deficient strain KP1490 (Fe-Ent −). Substitutions were induced with the same concentrations of sodium propionate in both KP1489 and KP1490 as follows: D12pBpa, 2 mM; T13pBpa, 3 mM; I14pBpa, 2 mM; V15pBpa, 2 mM; and V16pBpa, 3 mM. Strain W3110/pPro24/pEVOL expresses FepA from the chromosome (CHR). Plasmid-encoded wild-type FepA (WT) is in KP1489/pKP1693/pEVOL and was not expressed with sodium propionate. Photo-cross-linking was performed as described in Materials and Methods, and samples were either exposed to UV (+) or not (−). Proteins were precipitated with TCA, and TonB-FepA cross-links were visualized on immunoblots of 9% SDS-polyacrylamide gels with anti-TonB antibodies. Membranes were reprobed with anti-FepA antibodies to determine steady-state levels of FepA. Positions of the FepA pBpa substitutions are indicated at the top. Positions of the TonB and FepA monomer, the TonB-FepA photo-cross-link, and the TonB-unknown protein photo-cross-link are indicated on the right. The rightmost lane (separated by lines) derives from the same experiment as the rest but, due to space considerations, was electrophoresed on a separate gel for the same length of time, transferred with the other gel in the same transfer tank, and blotted under equivalent conditions.
Since TonB is the limiting protein in the TonB system by a wide margin, it must engage and disengage from OM transporters during an energy transduction cycle (47). In vivo, the ability of TonB to engage productively with the transporters FepA, BtuB, and FhuA requires their cognate transport ligands (4, 5, 13). For FepA in E. coli K-12, the energy transduction cycle can be divided into events that occur prior to productive transporter interaction and those which follow by performing assays in an aroB strain (13). This strain lacks the ability to synthesize the siderophore enterochelin (also known as enterobactin) or any of its biosynthetic intermediates. Because it also lacks the ability to synthesize aromatic amino acids, these are supplied exogenously in the medium. If an interaction was prevented by the absence of enterochelin (aroB strain), then enterochelin binding was necessary for TonB to interact at that site on FepA.
In strain KP1490 (W3110 ΔfepA aroB) cross-links formed through D12pBpa, T13pBpa, V15pBpa, and V16pBpa were diminished to a modest degree relative to those in the aroB+ strain (Fig. 3). This result was expected since TonB interacts more efficiently with FepA in the presence of enterochelin (13). Surprisingly, FepA I14pBpa was able to cross-link more strongly in the absence of enterochelin than when enterochelin was present.
A UV-dependent, pBpa-dependent cross-link migrating at ∼55 kDa was detected by the monoclonal anti-TonB immunoblots for all strains examined, including those that did not contain a FepA pBpa substitution (Fig. 3 to 5; see Fig. S1 and S2 in the supplemental material). The 55-kDa complex was almost certainly formed between TonB (with its apparent mass of ∼36 kDa [48]) and an unidentified protein (of ∼19 kDa or less) that had pBpa incorporated at its native terminal amber codon and whose translation terminated at a subsequent unknown, nonamber codon. Because the tonB coding sequence terminates in an ochre codon (UAA), it was not the source of the cross-link. The absence of enterochelin enhanced formation of this complex, suggesting that this unidentified protein is more likely to photo-cross-link to TonB when TonB is not directly energizing transport. The identity of this complex remains unknown, but it does not contain either ExbB or ExbD since they lack amber codons. The complex running just beneath the TonB-FepA complex (Fig. 4A) is most likely FepA with a degradation fragment of TonB, as seen previously with other TonB-transporter cross-links (4, 5).
FIG 5.
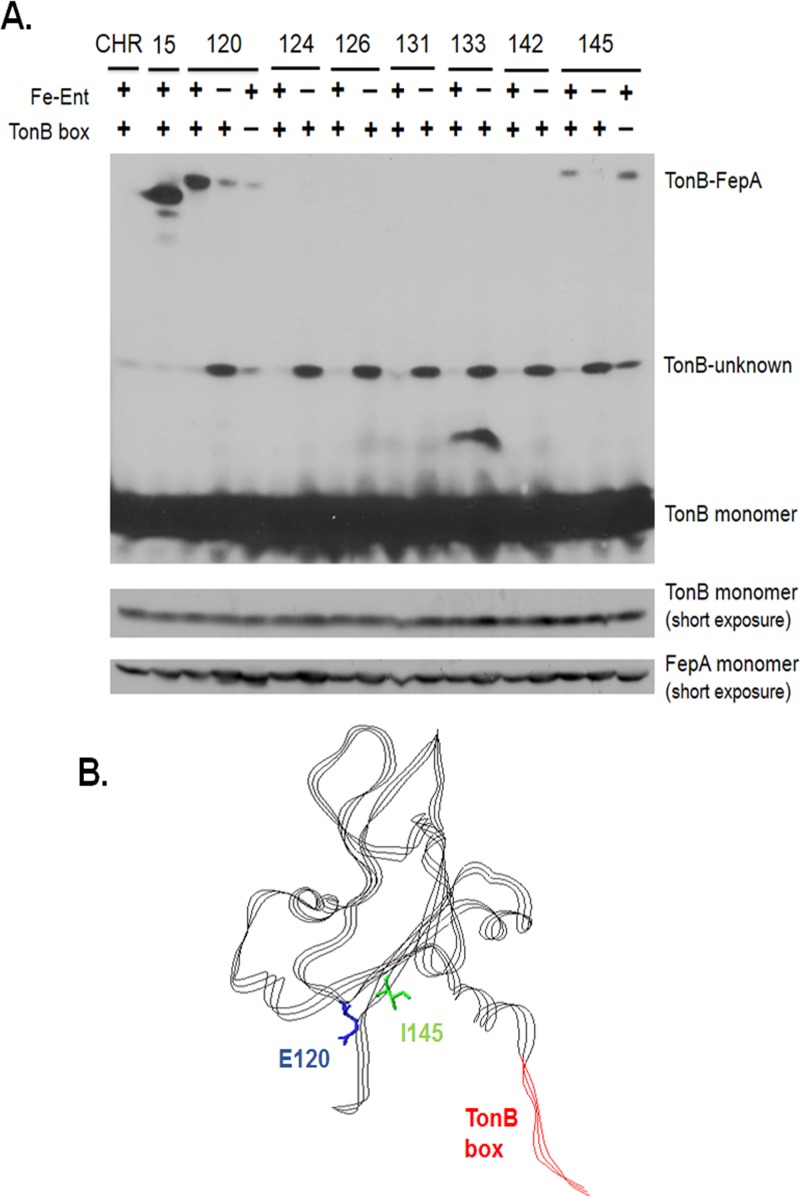
FepA E120pBpa and I145pBpa cross-link to TonB in vivo. Photo-cross-linking was performed as described in Materials and Methods. (A) Strains expressing pBpa substitutions were analyzed for photo-cross-link formation as described in Materials and Methods. Sodium propionate was used to express the FepA pBpa substitutions as follows: 2 mM for all substitutions in the presence or absence of enterochelin as long as the TonB box was present and 4 mM when the TonB box was deleted. As before, proteins were precipitated with TCA, and TonB-FepA cross-links and steady-state levels of TonB and FepA were visualized on immunoblots with anti-TonB and anti-FepA antibodies. Positions of monomeric TonB and FepA as well as cross-linked species are indicated. (B) Positions of cork domain side chains E120 and I145 in the crystal structure of FepA (Protein Data Bank no. 1fep). E120 is colored blue and is located on the cork β3 strand, and I145 is colored green and is located on the cork β4 strand. The side chains of both E120 and I145 are oriented toward the periplasm. The TonB box is colored red. The image was generated with Swiss-Pdb viewer (66).
FIG 4.
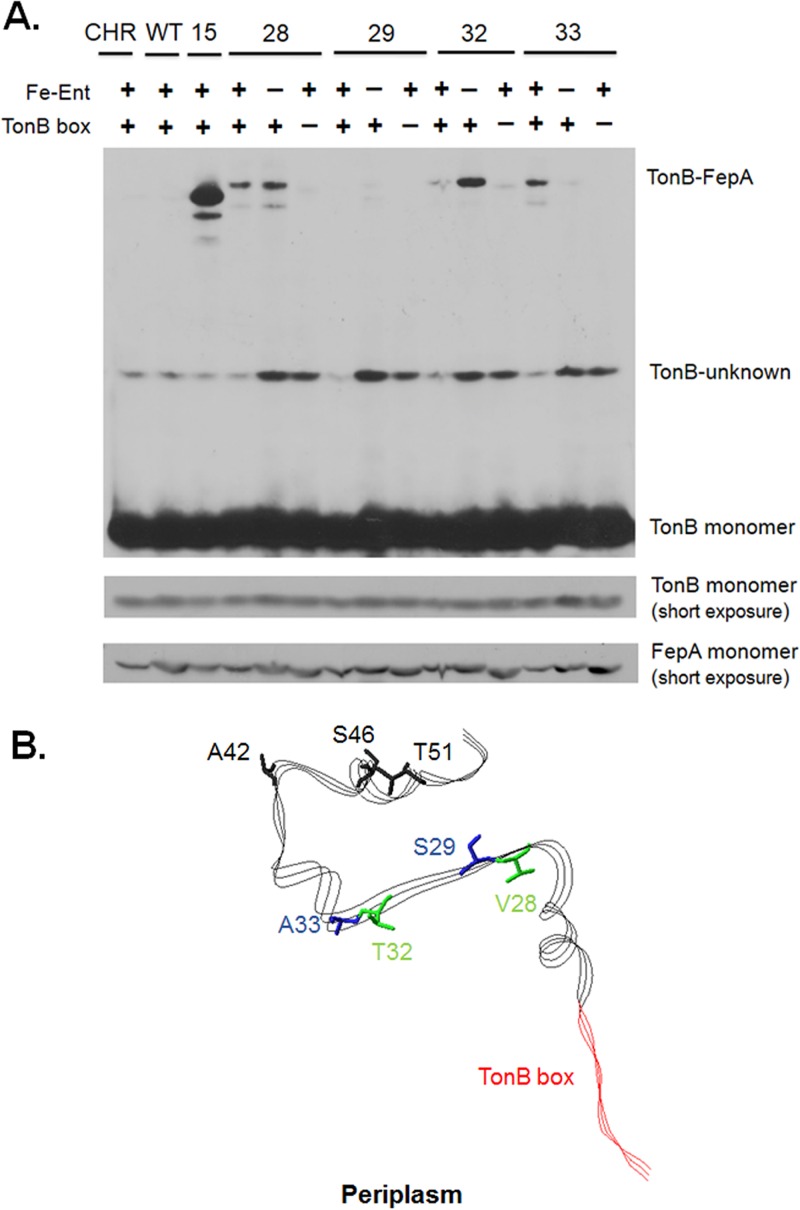
FepA pBpa substitutions in residues 28 to 33 cross-link to TonB in vivo. (A) Strains expressing FepA pBpa substitutions in the presence (+) or absence (−) of ferric enterochelin (Fe-Ent) or the TonB box were treated with UV (+) as described in Materials and Methods. During early exponential phase, pBpa was added to all strains at a final concentration of 0.4 mM. All pBpa substitutions expressed in the presence of a wild-type TonB box were induced with 2 mM sodium propionate, but substitutions combined with a TonB box deletion were induced with 4 mM sodium propionate. Cross-links were observed as higher-molecular-weight complexes on TonB immunoblots of 9% SDS-polyacrylamide gels. Steady-state levels of TonB and FepA (reprobe of TonB immunoblot) are shown underneath. Positions of photo-cross-linked complexes and TonB and FepA monomer are shown on the right, and the identity of each pBpa substitution is listed at the top. (B) Position of residues 28 to 33 in the crystal structure of FepA (Protein Data Bank no. 1fep) relative to the TonB box and residues A42, S46, and T51. This entire region of FepA has up to a 25-fold increase in exposure to the periplasm in the presence of colicin B and TonB (29). Residues 28 to 33 occupy a beta strand exposed to the periplasm. Side chains of V28 and T32 are colored green and are oriented toward the periplasm. Side chains S29 and A33 are colored blue and are oriented toward the interior of the cork. The TonB box is colored red, and side chains A42, S46, and T51 are colored black. The position of the periplasm is also indicated. The image was created with Swiss-Pdb viewer (66).
Identification of a potential ligand-dependent conformational switch following the β1 strand of the cork.
Under wild-type conditions and in the presence of the transport ligand colicin B, the amino-terminal 51 residues of FepA, including β1 residues S29C and A33C, are much more accessible to the periplasm than interior cork residues, as determined by in vivo BMCC labeling of these engineered Cys residues (29) (Fig. 1 and 4B). In this study of the pBpa substitutions in and near the β1 strand, V28pBpa, T32pBpa, and A33pBpa photo-cross-linked to TonB in the presence of enterochelin, while no cross-linking was observed for S29pBpa (Fig. 4A). The FepA β1 strand is the one most closely connected to the TonB box, a site of known TonB interaction (29).
Interestingly, in the absence of enterochelin, the behaviors of pBpa insertions at the neighboring residues 32 and 33 were completely opposite to one another. While FepA A33pBpa responded in expected fashion by exhibiting decreased cross-linking to TonB, formation of the T32pBpa-TonB complex was greatly enhanced in the absence of enterochelin (Fig. 4A). Thus, the residue 32 and 33 region of FepA appeared to be conformationally sensitive to the presence and absence of its transport ligand, reflected in the ability to photo-cross-link to TonB. Since both T32pBpa and A33pBpa FepAs are 70% or more active (Fig. 2), it seems likely that their photo-cross-linking behavior is similar to that of wild-type FepA and not the result of the pBpa substitution, although that cannot be ruled out entirely. The V28pBpa-TonB complex was only slightly enhanced in the absence of enterochelin. These results identify the first sites of TonB interaction with FepA seen outside the TonB box. FepA A42C, S46C, and T51C are the most highly periplasmically exposed residues upon colicin B addition. Nonetheless, as pBpa derivatives, they did not cross-link significantly to TonB, indicating that periplasmic exposure is not sufficient to cause photo-cross-linking to occur (Fig. S1).
FepA E120pBpa and I145pBpa in the cork β4 sheet make novel contacts with TonB.
We examined pBpa substitutions between positions 63 and 145, most of which, when previously analyzed as Cys replacements, were labeled only to a slight extent with the thiol-specific reagent BMCC (V91C, S92C, S112C, V124C, A131C, and V142C) (29). Consistent with those results, the pBpa derivatives did not photo-cross-link to TonB, and this suggested that the cork does not fully denature during enterochelin transport since none of those residues are periplasmically exposed. We examined two additional residues that are periplasmically exposed and that cluster together at the ends of the β3 and β4 strands, E120pBpa and I145pBpa, both of which retained essentially full activity in 55Fe-enterochelin uptake assays (Fig. 2). Each of these FepA substitutions photo-cross-linked to TonB, even though they are not in the immediate vicinity of the TonB box (Fig. 5). The absence of enterochelin diminished the cross-linking efficiency of E120pBpa and I145pBpa and did not result in increased cross-linking for any of the other pBpa substitutions in the region from S63pBpa to V142pBpa (Fig. 5 and S2).
FepA T32pBpa and I145pBpa fulfill predictions for TonB box-independent interactions.
The TonB boxes of the transporters are essential for transport. However, our previous results predicted the existence of FepA residues outside its TonB box that not only interacted with TonB but did not require the TonB box for that interaction (29). To search for such residues, we photo-cross-linked pBpa substitutions in the range from position 28 to 51 where the TonB box had been deleted. Because E120pBpa and I145pBpa were the only substitutions within residues 63 to 145 that could interact with TonB, the effect of a TonB box deletion on photo-cross-linking was examined only for these two sites.
Two sites of TonB interaction fulfilled the condition of being TonB box independent. FepA T32pBpa, identified above as a potential site of transport ligand-induced signaling, cross-linked to TonB, albeit not strongly, whether or not the TonB box was present. Likewise, I145pBpa cross-linked, if anything slightly more strongly, to TonB in the absence of the TonB box (Fig. 5A).
The absence of the TonB box significantly diminished cross-linking of TonB to V28pBpa and A33pBpa (Fig. 4A) and E120pBpa (Fig. 5), suggesting that their interactions might have occurred following TonB contact with the TonB box. However, no TonB-FepA complexes were detected for the remaining substitutions in the absence of the TonB box (Fig. 4 and S1).
DISCUSSION
Our long-term goal is to understand the mechanism of TonB-dependent energy transduction, in part by identifying sites of and requirements for the complex protein-protein interactions among TonB system components FepA, TonB, ExbB, and ExbD that occur in vivo. Previously, we engineered Cys substitutions in each of two TonB system proteins to identify sites of interaction via disulfide cross-linking (49–54). Because the exact sites of interaction between the two proteins are unknown, many combinations of cysteine substitutions must be engineered in order to map the interaction interface. Even still, some important interactions could go undetected simply because the “right” sites were not tested. Here, we used in vivo photo-cross-linking to examine FepA TonB box interactions for the first time and to broaden the search for sites outside the TonB box where FepA interacts with TonB, since these were predicted to exist based on our previous work (29). By use of this technique, sites of photo-cross-linking were investigated within specific structural elements of FepA while leaving sites of TonB interaction intentionally undefined. There is a solved crystal structure for FepA (14), whereas a crystal structure for full-length TonB in its in vivo conformation has not been solved to date (25), possibly due in part to the predicted intrinsic disorder for the majority of TonB (36) and the dynamics of cyclic conversion between TonB homodimeric and TonB-ExbD heterodimeric carboxy termini (6).
As a result of these experiments, we identified interactions between the FepA TonB box and TonB for the first time and a new ligand-independent interaction between a TonB box residue, FepA I14, and TonB, and we discovered additional FepA-TonB interactions outside the TonB box. The FepA-TonB interaction is therefore a more dynamic one than has hitherto been suspected, with interactions occurring over the periplasmic face of FepA and both with and without the presence of the transport ligand enterochelin. FepA interacted more strongly through some pBpa substitutions than others. When the various FepA-pBpa substitutions were mostly active, largely ruling out effects of the pBpa itself, the detected interactions were likely to reflect native interactions between wild-type FepA and TonB. Reasons for lack of interaction could include proximity, dwell time, or both. The data from this characterization of FepA-TonB interactions that occur outside the TonB box can be summed up as follows based on the original FepA crystal structure (14) (Fig. 6).
FIG 6.
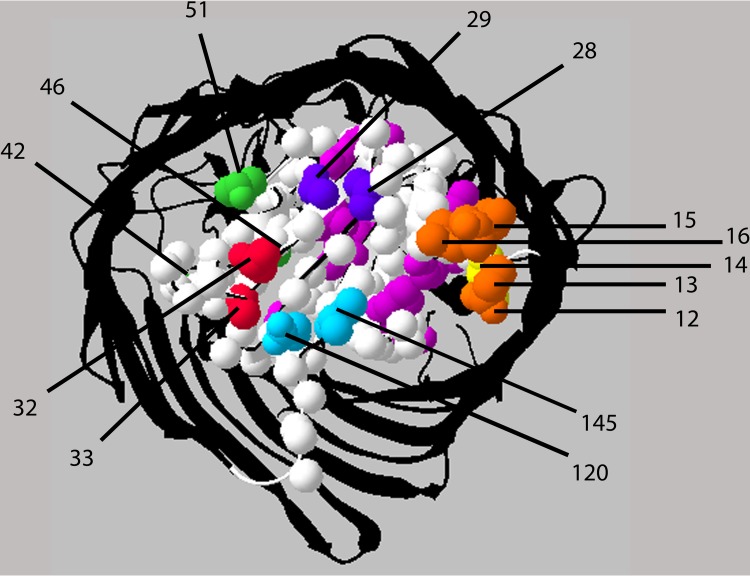
Summary of FepA pBpa Sites that photo-cross-link to TonB. The view is of the periplasmic face of the FepA cork, with the surrounding beta barrel in black (Protein Data Bank no. 1fep). Orange, FepA TonB box (residues D12, T13, I14, V15, and V16) with I14 in yellow. Purple, FepA residue V28, which photo-cross-links to TonB, and neighboring residue S29, which does not. Red, FepA residues T32 and A33, which are candidates for a conformational switch that is sensitive to the presence and absence of enterochelin. Green, FepA residues A42, S46, and T51, all of which become highly periplasmically exposed upon addition of the FepA ligand colicin B (29) but which, as pBpa substitutions, do not photo-cross-link significantly to TonB. A42 and S46 are not periplasmically accessible. Cyan, FepA residues E120 and I145, with E120 pBpa the stronger of the two interactions. FepA I145 pBpa interaction with TonB occurs prior to TonB box interaction with TonB but somehow still requires the presence of enterochelin. Magenta, all the other FepA positions tested (S63, R75, T82, L85, V91, S92, S112, V124, R126, A131, Y133, and V142). The image was generated with Swiss-Pdb viewer (66).
(i) There is specificity in the FepA-TonB interactions, since not all of the FepA pBpa substitutions on the periplasmically accessible face of the cork cross-linked to TonB. Notably, while S29 is periplasmically accessible in the FepA crystal structure and undergoes a 13-fold increase in accessibility when the transport ligand colicin B is present (29), FepA S29pBpa does not detectably photo-cross-link to TonB, whereas its neighboring residue, V28, photo-cross-links to TonB if the FepA TonB box is present.
(ii) Outside of the TonB box, the sites of FepA interaction with TonB are not highly conserved, and only one, T32pBpa, occurred in a highly conserved residue (9, 15). While FepA R75, R126, and Y133 are other highly conserved sites, as pBpa substitutions they did not photo-cross-link to TonB. That the remaining sites of interaction were not in conserved residues is consistent with the idea that TonB might be required to recognize the transporters through induced fit (36). This suggests the means of fine-tuning productive interactions among the several TonB-dependent transporters in E. coli K-12 and the single TonB that serves them. The fact of multiple sites for transporter interaction with TonB also suggests a means by which those bacteria with multiple sets of TonB proteins can discriminate among a large number of transporters.
(iii) FepA sites that photo-cross-link to TonB were distributed all over the periplasmic face of the cork but not within interior regions of the cork. It may be that we made unfortunate choices among interior residues to examine, but we find no evidence for TonB interaction with nonexposed regions of the FepA cork, consistent with previous results reported by ourselves and others (24, 29, 55). Although the side chains of R75, L85, V91, S112, R126, and A131 also project outward from the cork domain, they are not exposed to the periplasmic space in the crystal structure and do not detectably photo-cross-link to TonB when replaced with pBpa. Many of the pBpa substitutions at the interior cork residues supported less than 50% activity (R75, T82, S112, V124, R126, A131, and V142), suggesting prior to getting the photo-cross-linking results that they might be residues through which FepA needed to contact TonB. Since none of these residues photo-cross-linked to TonB as pBpa substitutions, it is more likely that they were required for correct packing of residues in that region. This was somewhat surprising since transporter structures have proven to be rather tolerant to mutation, but it is also possible that the rather bulky, aromatic pBpa side chain could have disrupted important interactions within the cork. For example, R126 corresponds to FhuA R133, which exhibited little effect of replacement with Ala or His (42).
(iv) FepA S63 pBpa was specifically engineered to determine if the external regions of the cork came out of the barrel into the periplasm. It did not photo-cross-link to TonB, so either this loop in FepA does not interact with TonB at all or the loop interacts with TonB through residues other than S63.
(v) It is known that TonB does not transduce energy to ligand-free transporters, suggesting that TonB can tell when a ligand is bound (13, 55). In its simplest form, this might involve a ligand-dependent conformational change at the periplasmic face of the transporter. A pair of neighboring FepA residues were in the right place and exhibited behaviors suggesting that they could be candidates for such a signal. FepA T32pBpa and FepA A33pBpa exhibited opposite behaviors, with T32 showing greatly increased binding in the absence of enterochelin and A33 in its presence. Because these residues face in different directions, it might take only a twist in the FepA protein backbone to expose one or the other of them in response to binding or dissociation of ligand. The TonB energy transduction cycle can be divided into events that occur prior to ligand binding and those that occur following ligand binding, suggesting that unliganded FepA T32 binds to TonB before ligand-bound FepA T33 does (13, 55). FepA pBpa substitutions in V28, E120, and I145 (Fig. 6) also all cross-linked to TonB more efficiently in the presence of enterochelin, also possibly as a signal for ligand occupancy. Taken together, these results suggest that a more nuanced consideration of the TonB-dependent transport mechanism is warranted.
(vi) We found novel TonB box-independent interactions. Full-length TonB binds to transporters whether or not it is “energized,” as indicated by the ability of inactive TonB H20A to formaldehyde cross-link to FepA (49, 56). Likewise, purified inactive carboxy-terminal domains of TonB bind with various affinities to purified transporters but do not energize transport (18, 57, 58). Since both energized and unenergized TonB bind to transporters, the difference in the outcome of whether the ligand is transported across the outer membrane may rest with the nature of the contacts that are made by TonB. Furthermore, these novel interactions are predicted to exist since TonB is required for increased ligand-dependent exposure of the TonB box to the periplasm in vivo (29). Both FepA T32pBpa and I145pBpa represent these predicted interactions because they photo-cross-linked to TonB equally well in the presence and absence of the TonB box. Interactions of TonB with the FhuA barrel have also been suggested (59).
In summary, our in vivo data do not support the current model where the transporter TonB box becomes exposed and binds to TonB in response to the presence of transport ligand (21). This difference may reflect the difference between in vitro and in vivo studies. In this in vivo study, FepA TonB box residues T13, I14, V15, and V16 all photo-cross-linked to some extent with TonB in the absence of any transport ligand, as also seen previously for BtuB or FecA TonB box interactions in vivo with TonB (4, 5). Like in those prior studies, enhancement of the interaction was detectable, but generally modest, in the presence of the transport ligands (4, 5). Thus, the transporter is not waiting for a ligand to bind on its extracellular surface before TonB can bind to the TonB box. Consistent with that, we also found evidence for TonB interactions with FepA both before and following its interaction with the FepA TonB box.
We also found a tantalizing suggestion for the possibility of a site of interaction outside the TonB Q160 region. In this study, FepA I14pBpa, which has ∼70% activity, unexpectedly photo-cross-linked more strongly in the absence of enterochelin than in its presence, whereas the opposite is true for studies of the cognate TonB box residues of either BtuB or FecA. However, in those studies in vivo interactions were constrained and directed solely to the TonB Q160 region by the simultaneous use of Cys substitutions in the transporters and in TonB (4, 5). We speculate that since the region of TonB where FepA I14pBpa can cross-link is unconstrained, the complex might represent an interaction of the FepA TonB box with a site on TonB other than the Q160 region. It has been suggested that the transporter TonB box might interact with the β3 strand of the monomeric TonB structure (residues 221 to 230) determined by nuclear magnetic resonance (NMR) (60). Mey and Payne showed that for Vibrio cholerae TonB1, a single residue at the extreme carboxy terminus is sufficient to determine transporter specificity (61). In addition, they expanded the definition for transporter TonB boxes to show that the −1 residue plays an important role in relaxing transporter specificity for a specific TonB. We did not examine the role of that residue in our study.
In any case, based on our data, the mechanism of TonB-dependent transport is likely to be more complex than TonB simply pulling on the transporter TonB box that becomes exposed when ligand binds. The results of this study show that regions of FepA other than its TonB box are contact sites for TonB and suggest that it would be worthwhile to look for sites outside the region of TonB Q160 involved in FepA contact. A cysteine scan of the TonB carboxy terminus from residue 150 to 239 shows that 7 out of 90 residues give rise to idiosyncratic, assay-dependent decreases in TonB activity, suggesting that the 7 residues are involved in transporter recognition (25). We hypothesize that those residues might be involved in binding transporter residues outside the TonB box. Three of the residues, F202, W213, and Y215, lie within an amphipathic helix that could be involved in mediating TonB association with the transporters.
MATERIALS AND METHODS
Strains and plasmids.
All strains and plasmids used in this study are listed in Table S1 in the supplemental material. Strain KP1487 was constructed by replacing the W3110 fepA gene with a Kanr gene by using the λ red system. The Kanr gene was then excised from KP1487 with FLP recombinase encoded on plasmid pCP20 (62) to create KP1489.
To construct plasmid-encoded FepA derivatives, the wild-type fepA gene from pKP515 was amplified by PCR using primers with incorporated XbaI sites. Both the pPro24 vector and the fepA amplimer were then digested with XbaI and ligated together with T4 DNA ligase to generate pKP1302. A His6 tag was incorporated between residues 393 and 394 of the mature FepA protein in pKP1302 to create pKP1693. Amber codons were subsequently introduced into the fepA gene in pKP1693 by site-directed mutagenesis using primers with at least 12 bp on both sides of the introduced TAG sequence. TonB box deletion derivatives of pKP1693, lacking residues 12 to 16 of mature FepA, were constructed by extralong PCR using phosphorylated primers and ligation of PCR products with T4 DNA ligase. Sequences of all PCR constructs were verified at the Penn State Genomics Core Facility, University Park, PA.
55Fe-enterochelin transport.
Transport assays were performed in the enterochelin-deficient (aroB) strain KP1490 to eliminate competition between native enterochelin and 55Fe-enterochelin for binding to FepA. Plasmid-bearing strains expressing the FepA amber derivatives of pKP1693 and orthogonal pBpa tRNA/tRNA synthetase from pEVOL-BpF were grown in M9 minimal medium supplemented with 0.4% glycerol, 0.2% Casamino Acids, 40 μg/ml tryptophan, 4 μg/ml vitamin B1, 1 mM MgSO4, 0.5 mM CaCl2, and 1.85 μM FeCl3. Tyrosine and phenylalanine were also added to a final concentration of 40 μg/ml each to support growth of aroB strains, which are unable to synthesize aromatic amino acids. Ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) were added to maintain plasmids. Cells expressing chromosomal FepA (KP1270/pPro24/pEVOL), wild-type His-tagged FepA (KP1490/pKP1693/pEVOL), and no FepA (KP1490/pPro24/pEVOL) served as controls. Once strains reached an A550 of 0.20 (as determined on a Spectronic 20 spectrophotometer with a path length of 1.5 cm), pBpa (Bachem) was added to a final concentration of 0.4 mM, and sodium propionate was added to a final concentration of 2 to 3 mM to induce expression of FepA pBpa substitutions. Cells were harvested at an A550 of 0.45 to 0.50 and assayed for 55Fe-enterochelin uptake as described previously for 55Fe-ferrichrome transport (63). Aliquots of the same cultures were also collected by precipitation with trichloroacetic acid (TCA), and steady-state levels of FepA were visualized on immunoblots with polyclonal anti-FepA antibodies (47).
Spot titer assays.
Strain KP1490 was cotransformed with pEVOL-BpF and plasmids encoding FepA amber substitutions. Saturated overnight cultures were diluted 1:200 in T-broth supplemented with 34 μg/ml chloramphenicol and 100 μg/ml ampicillin. Once the culture reached an A550 of 0.20, pBpa was added to a final concentration of 0.4 mM. When the A550 reached 0.40, cells were spread on T-plates containing 0.4 mM pBpa, chloramphenicol, and ampicillin. Fivefold dilutions of colicin B were spotted onto the T-plates and incubated overnight at 37°C. Protein samples were collected by TCA precipitation, and FepA expression levels were visualized on immunoblots with FepA antiserum.
In vivo photo-cross-linking.
Strain KP1489 or KP1490 expressing FepA amber derivatives of pKP1693 and orthogonal pBpa tRNA/tRNA synthetase from pEVOL-BpF (37) were grown in supplemented M9 as described for 55Fe-enterochelin transport assays along with 34 μg/ml chloramphenicol and 100 μg/ml ampicillin to maintain plasmids. Once cells reached an A550 of 0.20, pBpa was added to 0.4 mM to all cultures, and near-chromosomal expression of FepA pBpa substitutions was induced by adding sodium propionate to a final concentration of 2 to 4 mM. Cells were grown further to mid-exponential phase (A550 of 0.50) and 1.0 A550 ml cells (e.g., 2 ml of cells for an A550 of 0.50) were removed and resuspended in supplemented M9 lacking antibiotics, pBpa, and propionate. Cells were then incubated at 37°C with shaking for 20 min to allow cells to equilibrate and then transferred to a multiwell plate and immediately irradiated with 365-nm light at a distance of 2.5 cm for 30 min on ice, during which the photoactivatable pBpa located at various sites in FepA would cross-link to TonB. After irradiation, proteins were harvested by TCA precipitation, suspended in sample buffer (64) with freshly added 2-mercaptoethanol, and heated for 10 min at 95°C. TonB-FepA photo-cross-links and TonB steady-state levels were visualized on immunoblots of 9% SDS-polyacrylamide gels with the TonB monoclonal antibody 4F1 (65). Steady-state levels of FepA were visualized on immunoblots of 9% SDS-polyacrylamide gels with polyclonal FepA antibodies. Equal loading of gel lanes was confirmed by treating the immunoblots with Coomassie blue stain. Each of the FepA pBpa derivatives analyzed was expressed to roughly the same level as the pFepA (pKP1693) control without addition of inducer, indicating that the incorporation of pBpA was efficient.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ray Larsen for critical reading of the manuscript.
This work was supported by grants GM42146 and GM112710 to K.P.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00649-16.
REFERENCES
- 1.Andrews SC, Robinson AK, Rodriguez-Quinones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Krewulak KD, Vogel HJ. 2011. TonB or not TonB: is that the question? Biochem Cell Biol 89:87–97. doi: 10.1139/O10-141. [DOI] [PubMed] [Google Scholar]
- 3.Skare JT, Ahmer BMM, Seachord CL, Darveau RP, Postle K. 1993. Energy transduction between membranes—TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J Biol Chem 268:16302–16308. [PubMed] [Google Scholar]
- 4.Cadieux N, Kadner RJ. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc Natl Acad Sci U S A 96:10673–10678. doi: 10.1073/pnas.96.19.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogierman M, Braun V. 2003. Interactions between the outer membrane ferric citrate transporter FecA and TonB: studies of the FecA TonB box. J Bacteriol 185:1870–1885. doi: 10.1128/JB.185.6.1870-1885.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gresock MG, Kastead KA, Postle K. 2015. From homodimer to heterodimer and back: elucidating the TonB energy transduction cycle. J Bacteriol 197:3433–3445. doi: 10.1128/JB.00484-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schauer K, Rodionov DA, de Reuse H. 2008. New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem Sci 33:330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Choby JE, Skaar EP. 2016. Heme synthesis and acquisition in bacterial pathogens. J Mol Biol 428:3408–3428. doi: 10.1016/j.jmb.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fanucci GE, Cadieux N, Kadner RJ, Cafiso DS. 2003. Competing ligands stabilize alternate conformations of the energy coupling motif of a TonB-dependent outer membrane transporter. Proc Natl Acad Sci U S A 100:11382–11387. doi: 10.1073/pnas.1932486100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moeck GS, Tawa P, Xiang H, Ismail AA, Turnbull JL, Coulton JW. 1996. Ligand-induced conformational change in the ferrichrome-iron receptor of Escherichia coli K12. Mol Microbiol 22:459–471. doi: 10.1046/j.1365-2958.1996.00112.x. [DOI] [PubMed] [Google Scholar]
- 12.Gumbart J, Wiener MC, Tajkhorshid E. 2007. Mechanics of force propagation in TonB-dependent outer membrane transport. Biophys J 93:496–504. doi: 10.1529/biophysj.107.104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen RA, Thomas MG, Postle K. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol Microbiol 31:1809–1824. doi: 10.1046/j.1365-2958.1999.01317.x. [DOI] [PubMed] [Google Scholar]
- 14.Buchanan SK, Smith BS, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat Struct Biol 6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 15.Chimento DP, Kadner RJ, Wiener MC. 2005. Comparative structural analysis of TonB-dependent outer membrane transporters: implications for the transport cycle. Proteins 59:240–251. doi: 10.1002/prot.20416. [DOI] [PubMed] [Google Scholar]
- 16.Gudmundsdottir A, Bell PE, Lundrigan MD, Bradbeer C, Kadner RJ. 1989. Point mutations in a conserved region (TonB box) of the Escherichia coli outer membrane BtuB protein affect vitamin B12 transport. J Bacteriol 171:6526–6533. doi: 10.1128/jb.171.12.6526-6533.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell PE, Nau CD, Brown JT, Konisky J, Kadner RJ. 1990. Genetic suppression demonstrates direct interaction of TonB protein with outer membrane transport proteins in Escherichia coli. J Bacteriol 172:3826–3829. doi: 10.1128/jb.172.7.3826-3829.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freed DM, Lukasik SM, Sikora A, Mokdad A, Cafiso DS. 2013. Monomeric TonB and the Ton Box are required for the formation of a high-affinity transporter-TonB complex. Biochemistry 52:2638–2648. doi: 10.1021/bi3016108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shultis DD, Purdy MD, Banchs CN, Wiener MC. 2006. Outer membrane active transport: structure of the BtuB:TonB complex. Science 312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 20.Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW. 2006. Structure of TonB in complex with FhuA, E coli outer membrane receptor. Science 312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 21.Cafiso DS. 2014. Identifying and quantitating conformational exchange in membrane proteins using site-directed spin labeling. Acc Chem Res 47:3102–3109. doi: 10.1021/ar500228s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vakharia HL, Postle K. 2002. FepA with globular domain deletions lacks activity. J Bacteriol 184:5508–5512. doi: 10.1128/JB.184.19.5508-5512.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun M, Endriss F, Killmann H, Braun V. 2003. In vivo reconstitution of the FhuA transport protein of Escherichia coli K-12. J Bacteriol 185:5508–5518. doi: 10.1128/JB.185.18.5508-5518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flores Jimenez RH, Cafiso DS. 2012. The N-terminal domain of a TonB-dependent transporter undergoes a reversible stepwise denaturation. Biochemistry 51:3642–3650. doi: 10.1021/bi300118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Postle K, Kastead KA, Gresock MG, Ghosh J, Swayne CD. 2010. The TonB dimeric crystal structures do not exist in vivo. mBio 1:e00307–. doi: 10.1128/mBio.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vakharia-Rao H, Kastead KA, Savenkova MI, Bulathsinghala CM, Postle K. 2007. Deletion and substitution analysis of the Escherichia coli TonB Q160 region. J Bacteriol 189:4662–4670. doi: 10.1128/JB.00180-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cadieux N, Bradbeer C, Kadner RJ. 2000. Sequence changes in the ton box region of BtuB affect its transport activities and interaction with TonB protein. J Bacteriol 182:5954–5961. doi: 10.1128/JB.182.21.5954-5961.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cadieux N, Phan PG, Cafiso DS, Kadner RJ. 2003. Differential substrate-induced signaling through the TonB-dependent transporter BtuB. Proc Natl Acad Sci U S A 100:10688–10693. doi: 10.1073/pnas.1932538100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devanathan S, Postle K. 2007. Studies on colicin B translocation: FepA is gated by TonB. Mol Microbiol 65:441–453. doi: 10.1111/j.1365-2958.2007.05808.x. [DOI] [PubMed] [Google Scholar]
- 30.Udho E, Jakes KS, Finkelstein A. 2012. TonB-dependent transporter FhuA in planar lipid bilayers: partial exit of its plug from the barrel. Biochemistry 51:6753–6759. doi: 10.1021/bi300493u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hilsenbeck JL, Park H, Chen G, Youn B, Postle K, Kang C. 2004. Crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution. Mol Microbiol 51:711–720. doi: 10.1111/j.1365-2958.2003.03884.x. [DOI] [PubMed] [Google Scholar]
- 32.Smallwood CR, Marco AG, Xiao Q, Trinh V, Newton SM, Klebba PE. 2009. Fluoresceination of FepA during colicin B killing: effects of temperature, toxin and TonB. Mol Microbiol 72:1171–1180. doi: 10.1111/j.1365-2958.2009.06715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higgs PI, Letain TE, Merriam KK, Burke NS, Park H, Kang C, Postle K. 2002. TonB interacts with nonreceptor proteins in the outer membrane of Escherichia coli. J Bacteriol 184:1640–1648. doi: 10.1128/JB.184.6.1640-1648.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter DM, Miousse IR, Gagnon JN, Martinez E, Clements A, Lee J, Hancock MA, Gagnon H, Pawelek PD, Coulton JW. 2006. Interactions between TonB from Escherichia coli and the periplasmic protein FhuD. J Biol Chem 281:35413–35424. doi: 10.1074/jbc.M607611200. [DOI] [PubMed] [Google Scholar]
- 35.Kaserer WA, Jiang X, Xiao Q, Scott DC, Bauler M, Copeland D, Newton SM, Klebba PE. 2008. Insight from TonB hybrid proteins into the mechanism of iron transport through the outer membrane. J Bacteriol 190:4001–4016. doi: 10.1128/JB.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsen RA, Deckert GE, Kastead KA, Devanathan S, Keller KL, Postle K. 2007. His20 provides the sole functionally significant side chain in the essential TonB transmembrane domain. J Bacteriol 189:2825–2833. doi: 10.1128/JB.01925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young TS, Ahmad I, Yin JA, Schultz PG. 2010. An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol 395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 38.Chin JW, Martin AB, King DS, Wang L, Schultz PG. 2002. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci U S A 99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farrell IS, Toroney R, Hazen JL, Mehl RA, Chin JW. 2005. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat Methods 2:377–384. doi: 10.1038/nmeth0505-377. [DOI] [PubMed] [Google Scholar]
- 40.Yue WW, Grizot S, Buchanan SK. 2003. Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J Mol Biol 332:353–368. doi: 10.1016/S0022-2836(03)00855-6. [DOI] [PubMed] [Google Scholar]
- 41.Chakraborty R, Lemke EA, Cao Z, Klebba PE, van der Helm D. 2003. Identification and mutational studies of conserved amino acids in the outer membrane receptor protein, FepA, which affect transport but not binding of ferric-enterobactin in Escherichia coli. Biometals 16:507–518. doi: 10.1023/A:1023485632520. [DOI] [PubMed] [Google Scholar]
- 42.Endriss F, Braun M, Killmann H, Braun V. 2003. Mutant analysis of the Escherichia coli FhuA protein reveals sites of FhuA activity. J Bacteriol 185:4683–4692. doi: 10.1128/JB.185.16.4683-4692.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnard TJ, Watson ME Jr, McIntosh MA. 2001. Mutations in the Escherichia coli receptor FepA reveal residues involved in ligand binding and transport. Mol Microbiol 41:527–536. doi: 10.1046/j.1365-2958.2001.02473.x. [DOI] [PubMed] [Google Scholar]
- 44.Schöffler H, Braun V. 1989. Transport across the outer membrane of Escherichia coli via the FhuA receptor is regulated by the TonB protein of the cytoplasmic membrane. Mol Gen Genet 217:378–383. doi: 10.1007/BF02464907. [DOI] [PubMed] [Google Scholar]
- 45.Larsen RA, Foster Hartnett D, McIntosh MA, Postle K. 1997. Regions of Escherichia coli TonB and FepA proteins essential for in vivo physical interactions. J Bacteriol 179:3213–3221. doi: 10.1128/jb.179.10.3213-3221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larsen RA, Chen GJ, Postle K. 2003. Performance of standard phenotypic assays for TonB activity, as evaluated by varying the level of functional, wild-type TonB. J Bacteriol 185:4699–4706. doi: 10.1128/JB.185.16.4699-4706.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Higgs PI, Larsen RA, Postle K. 2002. Quantitation of known components of the Escherichia coli TonB-dependent energy transduction system: TonB, ExbB, ExbD, and FepA. Mol Microbiol 44:271–281. doi: 10.1046/j.1365-2958.2002.02880.x. [DOI] [PubMed] [Google Scholar]
- 48.Postle K, Skare JT. 1988. Escherichia coli TonB protein is exported from the cytoplasm without proteolytic cleavage of its amino terminus. J Biol Chem 263:11000–11007. [PubMed] [Google Scholar]
- 49.Ghosh J, Postle K. 2005. Disulphide trapping of an in vivo energy-dependent conformation of Escherichia coli TonB protein. Mol Microbiol 55:276–288. doi: 10.1111/j.1365-2958.2004.04384.x. [DOI] [PubMed] [Google Scholar]
- 50.Jana B, Manning M, Postle K. 2011. Mutations in the ExbB cytoplasmic carboxy terminus prevent energy-dependent interaction between the TonB and ExbD periplasmic domains. J Bacteriol 193:5649–5657. doi: 10.1128/JB.05674-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ollis AA, Postle K. 2011. The same periplasmic ExbD residues mediate in vivo interactions between ExbD homodimers and ExbD-TonB heterodimers. J Bacteriol 193:6852–6863. doi: 10.1128/JB.06190-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ollis AA, Kumar A, Postle K. 2012. The ExbD periplasmic domain contains distinct functional regions for two stages in TonB energization. J Bacteriol 194:3069–3077. doi: 10.1128/JB.00015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ollis AA, Postle K. 2012. Identification of functionally important TonB-ExbD periplasmic domain interactions in vivo. J Bacteriol 194:3078–3087. doi: 10.1128/JB.00018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baker KR, Postle K. 2013. Mutations in Escherichia coli ExbB transmembrane domains identify scaffolding and signal transduction functions and exclude participation in a proton pathway. J Bacteriol 195:2898–2911. doi: 10.1128/JB.00017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Udho E, Jakes KS, Buchanan SK, James KJ, Jiang X, Klebba PE, Finkelstein A. 2009. Reconstitution of bacterial outer membrane TonB-dependent transporters in planar lipid bilayer membranes. Proc Natl Acad Sci U S A 106:21990–21995. doi: 10.1073/pnas.0910023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ollis AA, Manning M, Held KG, Postle K. 2009. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol Microbiol 73:466–481. doi: 10.1111/j.1365-2958.2009.06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khursigara CM, De Crescenzo G, Pawelek PD, Coulton JW. 2004. Enhanced binding of TonB to a ligand-loaded outer membrane receptor. Role of the oligomeric state of TonB in formation of a functional FhuA-TonB complex. J Biol Chem 279:7405–7412. [DOI] [PubMed] [Google Scholar]
- 58.Khursigara CM, De Crescenzo G, Pawelek PD, Coulton JW. 2005. Kinetic analyses reveal multiple steps in forming TonB-FhuA complexes from Escherichia coli. Biochemistry 44:3441–3453. doi: 10.1021/bi047882p. [DOI] [PubMed] [Google Scholar]
- 59.Killmann H, Herrmann C, Torun A, Jung G, Braun V. 2002. TonB of Escherichia coli activates FhuA through interaction with the beta-barrel. Microbiology 148:3497–3509. doi: 10.1099/00221287-148-11-3497. [DOI] [PubMed] [Google Scholar]
- 60.Peacock SR, Weljie AM, Peter Howard S, Price FD, Vogel HJ. 2005. The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J Mol Biol 345:1185–1197. doi: 10.1016/j.jmb.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 61.Mey AR, Payne SM. 2003. Analysis of residues determining specificity of Vibrio cholerae TonB1 for its receptors. J Bacteriol 185:1195–1207. doi: 10.1128/JB.185.4.1195-1207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Datsenko K, Wanner B. 2000. One-step inactivation of charomosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Postle K. 2007. TonB system, in vivo assays and characterization. Methods Enzymol 422:245–269. doi: 10.1016/S0076-6879(06)22012-3. [DOI] [PubMed] [Google Scholar]
- 64.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 65.Larsen RA, Myers PS, Skare JT, Seachord CL, Darveau RP, Postle K. 1996. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J Bacteriol 178:1363–1373. doi: 10.1128/jb.178.5.1363-1373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.