ABSTRACT
Gram-negative bacteria acquire ferric siderophores through TonB-dependent outer membrane transporters (TBDT). By fluorescence spectroscopic hgh-throughput screening (FLHTS), we identified inhibitors of TonB-dependent ferric enterobactin (FeEnt) uptake through Escherichia coli FepA (EcoFepA). Among 165 inhibitors found in a primary screen of 17,441 compounds, we evaluated 20 in secondary tests: TonB-dependent ferric siderophore uptake and colicin killing and proton motive force-dependent lactose transport. Six of 20 primary hits inhibited TonB-dependent activity in all tests. Comparison of their effects on [59Fe]Ent and [14C]lactose accumulation suggested several as proton ionophores, but two chemicals, ebselen and ST0082990, are likely not proton ionophores and may inhibit TonB-ExbBD. The facility of FLHTS against E. coli led us to adapt it to Acinetobacter baumannii. We identified its FepA ortholog (AbaFepA), deleted and cloned its structural gene, genetically engineered 8 Cys substitutions in its surface loops, labeled them with fluorescein, and made fluorescence spectroscopic observations of FeEnt uptake in A. baumannii. Several Cys substitutions in AbaFepA (S279C, T562C, and S665C) were readily fluoresceinated and then suitable as sensors of FeEnt transport. As in E. coli, the test monitored TonB-dependent FeEnt uptake by AbaFepA. In microtiter format with A. baumannii, FLHTS produced Z′ factors 0.6 to 0.8. These data validated the FLHTS strategy against even distantly related Gram-negative bacterial pathogens. Overall, it discovered agents that block TonB-dependent transport and showed the potential to find compounds that act against Gram-negative CRE (carbapenem-resistant Enterobacteriaceae)/ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens. Our results suggest that hundreds of such chemicals may exist in larger compound libraries.
IMPORTANCE Antibiotic resistance in Gram-negative bacteria has spurred efforts to find novel compounds against new targets. The CRE/ESKAPE pathogens are resistant bacteria that include Acinetobacter baumannii, a common cause of ventilator-associated pneumonia and sepsis. We performed fluorescence high-throughput screening (FLHTS) against Escherichia coli to find inhibitors of TonB-dependent iron transport, tested them against A. baumannii, and then adapted the FLHTS technology to allow direct screening against A. baumannii. This methodology is expandable to other drug-resistant Gram-negative pathogens. Compounds that block TonB action may interfere with iron acquisition from eukaryotic hosts and thereby constitute bacteriostatic antibiotics that prevent microbial colonization of human and animals. The FLHTS method may identify both species-specific and broad-spectrum agents against Gram-negative bacteria.
KEYWORDS: Acinetobacter baumannii, ESKAPE pathogen, FepA, TonB, antibiotic resistance, ferric enterobactin, fluorescence assays, high throughput, iron transport
INTRODUCTION
Iron is an essential metal in both pro- and eukaryotes, for respiration, DNA synthesis, intermediate metabolism, nitrite reduction, and reactive oxygen detoxification. Consequently, iron acquisition is a virulence determinant in the host-pathogen interaction. The sequestration of iron by eukaryotic proteins like transferrin and ferritin protects against iron-associated toxicity but also denies iron to invading pathogens, a defense mechanism termed nutritional immunity (1). Iron levels are tightly controlled at the organismal and cellular levels. During infection bacteria use complex, incompletely understood, energy- and TonB-dependent transporters (TBDT) to procure Fe3+ from host sources. Juxtaposed against mechanistic research, the specter of antimicrobial resistance calls for new compounds to combat Gram-negative bacteria, and their iron acquisition systems are potential targets for antimicrobial drug development.
Siderophores are a potent part of the microbial iron acquisition arsenal that complex Fe3+ with higher affinity than eukaryotic iron-binding proteins and thereby outcompete the host. Enterobactin is a catecholate siderophore of the Enterobacteriaceae that binds Fe3+ with the highest known affinity (Kd [dissociation constant] = 10−52 M [2]). Consequently, it takes the iron from host proteins (3, 4, 14). Ferric enterobactin (FeEnt) subsequently enters the cell through FepA, an outer membrane (OM) protein comprised of a 22-strand porin β-barrel (6), within which resides its N-terminal 150 residues, which regulate transport of FeEnt (5). FepA-mediated FeEnt transport requires the inner membrane (IM)-anchored protein TonB (7); hence, FepA is a TBDT (8–10). Gram-negative cells produce many TBDT for iron uptake (11): Escherichia coli K-12 encodes 7 (12) and wild species produce more (13) that internalize iron or strip it from eukaryotic proteins (11). The C terminus of TonB physically binds to a conserved “TonB-box” peptide sequence at the N termini of TBDT like FepA (15–19), which causes conformational changes that open TBDT channels for iron transport to the periplasm. The actions and motion of TonB require the proton motive force (PMF) across the IM. The PMF underlies the biochemical activities of TonB through the accessory proteins ExbB and ExbD (20–23). Furthermore, the interaction between TonB and peptidoglycan (PG) may affect the overall architecture and activities of TonB-ExbBD (24). TonB-dependent iron acquisition systems are indispensable to survival of bacterial pathogens in the host (25, 26). Bacteria have redundant iron (III) uptake systems, but all Fe3+ uptake through all Gram-negative bacterial OM transporters, including those of CRE (carbapenem-resistant Enterobacteriaceae)/ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens, requires facilitation by TonB. Inhibition of TonB causes iron deprivation, which slows bacterial growth and retards infection.
Fluorescence modification of site-directed Cys sulfhydryls in FepA enables spectroscopic observations of FeEnt transport in real time in live bacteria (27, 28). FeEnt binding quenches fluorescence, but as the bacteria internalize FeEnt and deplete it from solution, fluorescence recovers. By monitoring ligand binding-induced fluorescence quenching versus time, the method sensitively detects the uptake of even nanomolar concentrations of FeEnt. It also may discover compounds that prevent TonB-dependent, FepA-mediated FeEnt transport, because they interfere with fluorescence recovery in a titratable fashion.
Acinetobacter baumannii is an opportunistic Gram-negative ESKAPE pathogen that causes a range of infections in intensive care units worldwide; it is a leading cause of ventilator-associated pneumonia (29). Additionally, it persists in hospital environments (30, 31) by forming biofilms that survive on abiotic surfaces. Its high propensity for antibiotic resistance makes A. baumannii a priority threat that encompasses multidrug- and pan-drug-resistant strains (32, 33). The escalating number of infections by drug-resistant A. baumannii highlights the need for new antimicrobials against it. Like other bacterial pathogens, A. baumannii must survive in the iron-restricted environment of the host. During iron limitation A. baumannii undergoes Fur-regulated (34, 35) transcriptional changes that promote iron uptake. Under such iron-deficient conditions A. baumannii upregulates at least three siderophore biosynthetic and transport systems (35). Acinetobactin is a high-affinity, catecholate siderophore and a virulence factor (36–38) that is nearly ubiquitous among the currently sequenced A. baumannii strains. At least two other siderophore biosynthetic gene clusters exist in A. baumannii (34), one for the hydroxamates baumanoferrin A and B (39) and another for fimsbactin (40). Besides the secretion of these siderophores and uptake of their ferric complexes, A. baumannii produces transporters for iron complexes of other siderophores that it does not produce. FepA (AbaFepA) is one such Fur-regulated TBDT that is required for full virulence in a sepsis model of A. baumannii infection (41), and A. baumannii uses FeEnt as an iron source under low-iron conditions (41).
We used a fluorescence high-throughput screening (FLHTS) assay of FeEnt transport by FepA in living E. coli to identify inhibitors of TonB-dependent transport. The experiments revealed numerous compounds that inhibit TonB-dependent activity, as verified by their ability to interfere with FeEnt and ferrichrome (Fc) transport and colicin B (ColB) and Ia (ColIa) killing, without blocking PMF-dependent lactose uptake. We determined the MICs of these chemicals against both E. coli and A. baumannii. Besides characterizing the FeEnt transport system of A. baumannii, in these studies, we engineered Cys residues in AbaFepA and adapted FLHTS to observe FeEnt uptake by AbaFepA in A. baumannii. The experiments validated the assay against Gram-negative bacterial pathogens like A. baumannii. Compounds identified in this or larger FLHTS surveys against E. coli, A. baumannii, or other organisms may constitute new antimicrobial therapeutic agents against Gram-negative bacterial pathogenesis.
RESULTS
FLHTS for inhibitors of TonB-dependent FeEnt transport by E. coli FepA.
Considering TonB's essential role in iron acquisition, we sought to identify small molecules that inhibit TonB-dependent ferric siderophore transport in microtiter plates by the FLHTS approach, which spectroscopically measured FeEnt uptake by FepA in live E. coli (42). When bacteria harboring fluorescently labeled FepA were exposed to FeEnt, the ferric siderophore bound and quenched fluorescence intensity. However, as the cells transported FeEnt it became depleted from solution, FepA was vacated, and fluorescence rebounded. Control experiments in energy- and TonB-deficient bacteria (27, 28, 42) confirmed the interpretation of these observations. Thus, transport appeared in real time as sequential quenching and unquenching of fluorescence intensity. The extent and duration of quenching depended on the concentrations of bacteria and FeEnt, the temperature, and the incubation time. Variation of these parameters allowed customization of the assay. We previously optimized the test in 96-well plates, but to increase efficiency, we adapted it to 384-well format, where it similarly functioned: a control inhibitor, carbonyl cyanide m-chlorophenylhydrazone (CCCP), caused dose-dependent impairment of the fluorescence recovery (see Fig. S1A in the supplemental material). In addition, fluoresceinated E. coli cells that were cryopreserved at −70°C in 10% glycerol for up to a month were viable and transported FeEnt in the fluorescence assay (data not shown). When reconstituted, the stored cells had the same transport kinetics as freshly labeled cells, resulting in complete fluorescence recovery in 10 to 15 min in the microtiter format (data not shown). The viability of frozen, labeled cells enabled FLHTS experiments over multiple days with the same bacteria, allowing rapid, convenient FLHTS of large chemical libraries.
We performed FLHTS of 17,441 compounds in four chemical libraries at the University of Kansas High Throughput Screening Laboratory (KU-HTSL): the Microsource Library (2,000 compounds; 10 μM), FDA/Bioactives (5,233 compounds; 5 μM), University of Kansas Center of Excellence in Chemical Methodologies & Library Development (KU CMLD) Library (5,000 compounds; 10 μM), and TimTec Actiprobe 5K (5,000 compounds; 10 μM) (Table 1). Each test plate included the following controls: no bacteria (16 wells), fluoresceinated, otherwise untreated bacteria (8 wells), and fluoresceinated bacteria with 100 μM CCCP (8 wells). After initiating fluorescence readings at time zero, we dispensed FeEnt to 10 nM in all the wells and measured fluorescence 1 and 60 min thereafter. The three readings gave the initial fluorescence intensity, the extent of FeEnt-mediated quenching, and the extent of fluorescence recovery in response to FeEnt uptake. Across all plates the Z′ factor was 0.87 ± 0.02 (see Fig. S1B in the supplemental material). We selected 165 compounds that inhibited greater than or equal to 2.0 standard deviations (SD) from the median (Fig. S1C) for primary screen validation by dose-response assays that measured FeEnt uptake kinetics for 80 min in the presence of 0, 2.5, 5, 10, and 20 μM concentrations of each compound. From these data we categorized the compounds in 3 groups: those causing <20% inhibition, 20 to 30% inhibition, and >30% inhibition. We considered the 94 compounds with ≥20% inhibition as true primary hits, resulting in an overall hit rate of 0.54%. The primary screen yielded a reasonable number of candidate inhibitors to follow up with secondary screens.
TABLE 1.
Summary of primary screen results
| Library | Total no. of compounds | No. cherry-pickeda | No. with >30% inhibition |
No. with 20–30% inhibition |
No. with <20% inhibition |
Hit rateb (overall) |
|---|---|---|---|---|---|---|
| CMLD | 5,208 | 61 | 4 | 30 | 27 | 0.19 |
| TimTec Actiprobe 5K | 5,000 | 39 | 9 | 9 | 21 | 0.10 |
| FDA/Bioactives | 5,233 | 37 | 15 | 6 | 16 | 0.12 |
| Microsource Spectrum | 2,000 | 28 | 16 | 5 | 7 | 0.12 |
| Total | 17,441 | 165 | 44 | 50 | 71 | 0.54 |
Compounds were cherry-picked by selection of any compounds whose percent inhibition at read 3 was ≥2 SD from the median (15% cutoff).
The hit rate represents the number of compounds with >20% inhibition out of the total number of compounds.
Secondary screening of primary-hit compounds.
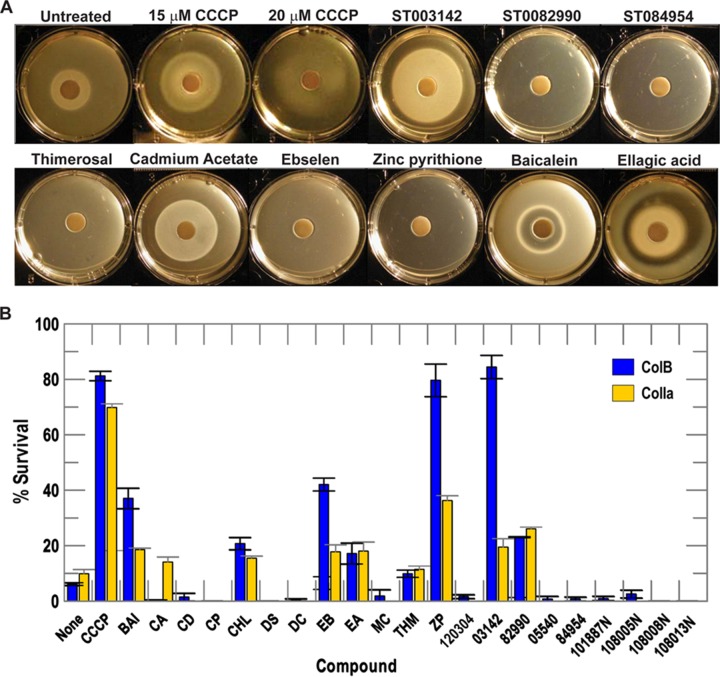
Among 44 compounds that inhibited >30%, we purchased 20 chemicals for secondary screening, based on their availability and structure (Fig. 1). The secondary screening assays monitored TonB-dependent FeEnt and Fc uptake in siderophore nutrition tests by FepA and FhuA, respectively (Fig. 2A; Table 2) (43), as well as TonB-dependent ColB and ColIa killing, through FepA and Cir, respectively (Table 2).
FIG 1.
Candidate inhibitors of TonB action identified by primary screening. Among the 44 compounds that inhibited fluorescence >30% in the primary FLHTS screening, we selected 20 compounds for further testing and obtained them from suppliers as fresh powders. These chemicals caused dose-dependent inhibition of fluorescence recovery; we did not consider compounds that reduced initial fluorescence and/or the extent of initial quenching. We also tested compound 120304 from reference 71, but it had no effects in our primary or secondary tests of TonB action (see also Table 2).
FIG 2.
Secondary screening by TonB-dependent siderophore nutrition assays and colicin killing tests. (A) We tested the 20 selected primary hits for interference with siderophore nutrition assays with FeEnt and Fc; the negative and positive controls were untreated bacteria and bacteria treated with CCCP, respectively. We observed several types of inhibition by the compounds: larger halos (that imply a lower rate of iron uptake [10, 44, 45]; illustrated by the control, CCCP, at 15 and 10 μM), distorted, aberrant halos, and the complete absence of a halo. Inhibition was identified by a loss of halo or an increase in the diameter of the halo around the ferric siderophore disc (Table 2). (B) We performed ColB and ColIa killing tests in the presence of the 20 selected compounds, with the same controls. The graph depicts the percent survival of bacteria in the presence of ColB or ColIa in the absence and presence of inhibitory compounds (see also Table 2). We performed each experiment 2 or 3 times and averaged the percent survival of colicin killing in the absence and presence of each compound. Error bars represent the standard deviations of the mean values. Compound abbreviations: CCCP, carbonyl cyanide m-chlorophenylhydrazone; Bai, baicalein; CA, cadmium acetate; CD, carbidopa; CP, carboplatin; CHL, p-chloranil; DS, dideoxyscleroin; DC, dequalinium chloride; EB, ebselen; EA, ellagic acid; MC, methylcatechol; THM, thimerosal; ZP, zinc pyrithione; 120304, lead compound from reference 71. See Fig. 1 and Table 2 for more information about the enumerated compounds.
TABLE 2.
Secondary screening of top 20 candidate compounds against E. coli and A. baumanniia
| Treatment |
E. coli |
A. baumannii |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| FeEnt (cm)b | Fc (cm)b | ColB hits/cellc | ColIa hits/cellc | [59Fe]Ent uptaked |
[14C]lactose uptakee |
MIC (μM)f | FeEnt (cm)g | MIC (μM)f | |
| None | 1.5 ± 0.1 | 1.5 ± 0.1 | 1.0 | 1.0 | 1.0 | 1.0 | NA | 1.2 ± 0.1 | NA |
| CCCP | 2.0 ± 0.1 | 1.9 ± 0.1 | 0.1 ± 0.0 | 0.2 ± 0.0 | 0.2 | 0.2 | NT | 1.5 ± 0.1 | NT |
| Baicalein | 1.7 ± 0.1 | 1.7 ± 0.1 | 0.3 ± 0.1 | 0.7 ± 0.0 | 1.0 | 1.0 | — | 1.2 ± 0.1 | — |
| Cadmium acetate | 2.2 ± 0.1 | 2.1 ± 0.4 | 1.1 ± 0.2 | 0.8 ± 0.1 | NT | NT | 512 | 0 | 256 |
| Carbidopa | 1.6 ± 0.1 | 1.6 ± 0.1 | 1.4 ± 0.5 | NT | NT | NT | — | 1.1 ± 0.0 | — |
| Carboplatin | 1.5 ± 0.0 | 1.6 ± 0.0 | 1.1 ± 0.1 | NT | NT | NT | — | 1.1 ± 0.1 | — |
| p-Chloranil | 1.5 ± 0.2 | 1.6 ± 0.1 | 0.7 ± 0.1 | 0.8 ± 0.0 | NT | NT | — | 1.1 ± 0.0 | — |
| Dideoxyscleroin | 1.6 ± 0.1 | 1.7 ± 0.2 | 1.1 ± 0.2 | NT | NT | NT | — | 1.2 ± 0.0 | — |
| Dequalinium-Cl | 1.4 ± 0.1 | 1.6 ± 0.1 | 1.1 ± 0.1 | NT | NT | NT | — | 1.1 ± 0.0 | — |
| Ebselen | 0h | 0h | 0.6 ± 0.1 | 0.8 ± 0.1 | 0.18 | 0.68 | 128 | 0 | 32 |
| Ellagic acid | 1.6 ± 0.1 | 1.4 ± 0.2 | 0.6 ± 0.1 | 0.7 ± 0.1 | NT | NT | — | 1.1 ± 0.0 | 256 |
| 3-Methoxycatechol | 1.5 ± 0.1 | 1.6 ± 0.5 | 0.9 ± 0.3 | NT | NT | NT | — | 1.1 ± 0.0 | — |
| Thimerosal | 0h | 0h | 1.1 ± 0.1 | 1.1 ± 0.1 | 0.2 | 0.2 | 2 | 0 | 1 |
| Zinc pyrithione | 0h | 0h | 0.0 ± 0.0 | 0.5 ± 0.0 | 0.2 | 0.2 | 4 | 0 | 16 |
| 120304 | 1.6 ± 0.1 | 1.6 ± 0.1 | 1.0 ± 0.2 | NT | NT | NT | — | 1.2 ± 0.1 | — |
| ST003142 | 2.7 ± 0.3 | 2.1 ± 0.0 | 0.0 ± 0.0 | 0.7 ± 0.1 | 1.0 | 1.0 | 256 | 1.7 ± 0.4 | 512 |
| ST005540 | 1.6 ± 0.2 | 1.7 ± 0.2 | 1.2 ± 0.2 | 1.2 ± 0.1 | NT | NT | — | 1.1 ± 0.0 | — |
| ST082990 | 0h | 0h | 0.6 ± 0.0 | 0.7 ± 0.0 | 0.44 | 0.74 | 128 | 0 | 16 |
| ST084954 | 0h | 0h | 1.3 ± 0.3 | 1.1 ± 0.1 | 1.0 | 1.0 | 32 | 1.4 ± 0.1 | 256 |
| KUC101887N | 1.4 ± 0.1 | 1.5 ± 0.1 | 1.1 ± 0.3 | NT | NT | NT | — | 1.1 ± 0.1 | — |
| KUC108008N | 1.5 ± 0.0 | 1.6 ± 0.0 | 1.3 ± 0.3 | NT | NT | NT | — | 1.1 ± 0.0 | — |
| KUC108005N | 1.5 ± 0.1 | 1.6 ± 0.1 | 1.0 ± 0.0 | NT | NT | NT | — | 1.1 ± 0.0 | — |
| KUC108013N | 1.5 ± 0.1 | 1.6 ± 0.1 | 1.3 ± 0.3 | NT | NT | NT | — | 1.1 ± 0.0 | — |
NA, not applicable; NT, not tested. —, MIC exceeded 512 μM.
Siderophore nutrition test using MG1655 with 50 μM FeEnt or Fc and a 100 μM concentration of a compound of interest. The data for CCCP derive from siderophore nutrition tests with the proton ionophore included at 15 μM.
Relative inhibition of ColB or ColIa measured by quantitative determination of colicin titer against MG1655 in the absence or presence of the inhibitors, according to S/S0 = e−k (79), with the titer against untreated bacteria set at 1.0. Addition of the compounds alone (diluted from a DMSO stock) caused ≤5% reduction in bacterial viability. The data for CCCP derive from siderophore nutrition tests with CCCP included at 15 μM.
Uptake of [59Fe]Ent, relative to E. coli MG1655. Data for CCCP derive from experiments with CCCP included at 15 μM.
Uptake of [14C]lactose, relative to E. coli MG1655. Data for CCCP derive from experiments with CCCP included at 15 μM.
MIC determined by titration of compounds against MG1655 or 17978 in LB. The noted concentration represents the point at which we observed growth inhibition in a series of serial 2-fold dilutions from 512 to 0.5 μM.
Siderophore nutrition test using 17978 with 50 μM FeEnt and a 100 μM concentration of a compound of interest.
Some compounds completely blocked bacterial growth at 100 μM, but at lower concentrations caused enlarged growth halos in siderophore nutrition tests (Fig. S2).
In siderophore nutrition tests, we included test compounds (at 100 μM) or the control, CCCP (at 15 μM or 20 μM), with the bacteria prior to plating the cells and applying the ferric siderophore to a paper disc on the agar surface. Iron utilization resulted in a growth halo around the disc (43). A decrease in the rate of iron uptake caused progressively larger and fainter growth halos in this test, until complete loss of iron uptake fully eliminates the halo (44, 45) (see CCCP in Fig. 2A). Compounds that inhibited TonB-dependent iron transport affected the size and morphology of the growth halos. We considered chemicals that caused >10% increase in halo diameter, or prevented growth halo formation, as valid inhibitors in this test (Table 2; Fig. 2A). Baicalein and ellagic acid conferred unique halos; we classified them as inhibitors, although the underlying causes of the atypical halos they created are unknown. Finally, at 100 μM, five candidate compounds entirely eliminated halo formation, and we retested them using lower concentrations in the FeEnt nutrition test. Their inclusion at lower concentrations resulted in dose-dependent increases in halo diameter (Fig. S2), demonstrating that they act by retarding siderophore utilization, rather than by general toxicity.
Various colicins, including ColB and ColIa, require TonB for OM translocation (46). We measured the ability of subtoxic concentrations of the 20 primary hit compounds to block killing of E. coli by ColB and ColIa, through their cognate, TonB-dependent receptors FepA and Cir, respectively. We considered compounds that increased survival in the presence of colicin >10% as inhibitors of TonB action (Table 2; Fig. 2B).
Overall, the six compounds that inhibited in all four secondary screens of TonB dependence (FeEnt utilization, Fc utilization, ColB killing, and ColIa killing) were baicalein, ST003142, thimerosal, zinc pyrithione, ebselen, and ST0082990. Other compounds, including ST084954, ST005540, cadmium acetate, and ellagic acid, were inhibitory in two or three of the four secondary screens (Table 2).
[59Fe]Ent and [14C]lactose uptake measurements.
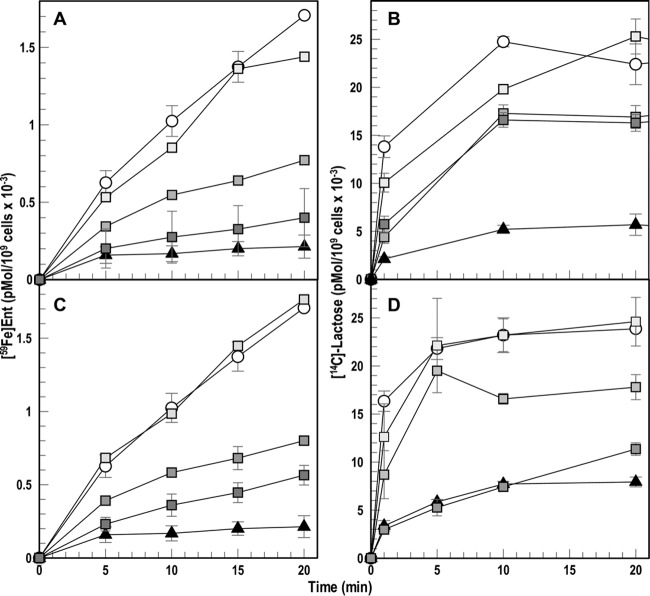
The electrochemical proton gradient across the IM energizes TonB action in OM transport, so we assessed the 6 chemicals that broadly inhibited the activities of TonB to identify (and exclude) probable proton ionophores. We did so by comparing their ability to impair the uptake of [59Fe]Ent and [14C]lactose, which both require PMF for transport through the OM and IM, respectively. Despite their effects on ferric siderophore uptake in nutrition tests, and the reduction of colicin lethality that they caused, neither baicalein nor ST003142 decreased accumulation of [59Fe]Ent or [14C]lactose, at concentrations as high as 0.5 mM (Table 2). Thimerosal and zinc pyrithione, on the other hand, were potent inhibitors of both [59Fe]Ent and [14C]lactose uptake, with roughly the same concentration dependence, in about the same manner as CCCP (Table 2). Lastly, ebselen and ST0082990 behaved differently (Fig. 3). First, relative to CCCP, which decreased [59Fe]Ent uptake 80 to 90% at 25 μM, both ebselen and ST0082990 caused comparable (but still less) inhibition only at a much higher concentration: 250 μM. Likewise, whereas CCCP at 25 μM blocked 80% of [14C]lactose transport, ebselen at 250 μM only decreased it ∼30%. Second, ebselen and ST0082990 inhibited [59Fe]Ent transport considerably more than they impaired [14C]lactose uptake. At 250 μM ebselen reduced [59Fe]Ent uptake 82%, but it only reduced [14C]lactose uptake 32%; 100 μM ST0082990 inhibited FeEnt uptake 66% but only reduced [14C]lactose uptake 26%. Hence, in this proof-of concept screen, 2 (10%) of the 20 primary hits that we fully analyzed (0.05% of the complete library) reduced iron transport and impaired colicin killing without abrogating PMF-dependent lactose transport. They differed from CCCP in both potency and scope of activity, suggesting that neither acts by PMF depletion. These two chemicals, ebselen and ST003142, remain potential candidate inhibitors of TonB-ExbBD.
FIG 3.
Inhibition of [59Fe]Ent and [14C]lactose uptake. E. coli MG1655 was grown in MOPS minimal medium until late exponential phase and independently assayed for its ability to accumulate [59Fe]Ent (A and C) and [14C]lactose (B and D) in the presence of either ebselen (A and B) or ST0082990 (C and D). We tested MG1655 in the absence of any inhibitors (circles), in the presence of CCCP at 25 μM (triangles), in the presence of ebselen at 25 (light gray squares), 200 (medium gray squares), and 250 (dark gray squares) μM, or in the presence of ST0082990 at 25 (light gray squares), 100 (medium gray squares), and 250 (dark gray squares) μM.
MIC tests.
To evaluate the bactericidal activity of the 20 chemicals, we grew E. coli MG1655 in the presence of a 2-fold dilution series of each compound in Luria broth (LB) and calculated the effective MIC up to 512 μM (Table 2). We also performed MIC studies in morpholinepropanesulfonic acid (MOPS) minimal medium under conditions that require active iron transport. The results were approximately the same in both media for the 20 compounds that we subjected to secondary screens. Cadmium acetate, ebselen, ST003142, ST0082990, ST0084954, thimerosal, and zinc pyrithione, which all inhibited of TonB-mediated physiology in some way, had MICs of ≤512 μM. Baicalein was not inhibitory to bacterial growth at concentrations as high as 512 μM, but ebselen and ST0082990, the most promising hits, both showed MICs of <128 μM.
We measured FeEnt nutrition and MIC in LB for the compounds of interest against the ESKAPE pathogen A. baumannii. With the exception of baicalein, all of the chemicals that inhibited FeEnt nutrition of E. coli also inhibited nutrition of A. baumannii (Table 2), and the compounds that inhibited E. coli growth in LB likewise inhibited A. baumannii, with some differences in sensitivity (Table 2). Relative to E. coli, certain compounds had higher or lower MICs against A. baumannii. Most notably, ebselen and ST0082990 were more active against A. baumannii (MICs of 32 μM and 16 μM, respectively), despite the 100-fold-lower OM permeability of the ESKAPE organism. These results suggested that a similar FLHTS screen against A. baumannii may identify compounds that inhibit its TonB-dependent transport processes.
Identification of the FeEnt transporter in A. baumannii.
The A. baumannii chromosome contains a fepA ortholog from A1S_0980 and A1S_0981, in a region that was incorrectly annotated as split by a stop codon (47). However, PCR analysis of genomic DNA from ATCC 17978 at the noted locus juncture revealed the error: the annotated stop codon was absent. Therefore, A1S_0980 and A1S_0981 represent a single open reading frame (ORF) (data not shown) (41). We precisely deleted A1S_0980 and A1S_0981 from the chromosome and tested the resulting ΔfepA strain for its ability to utilize FeEnt.
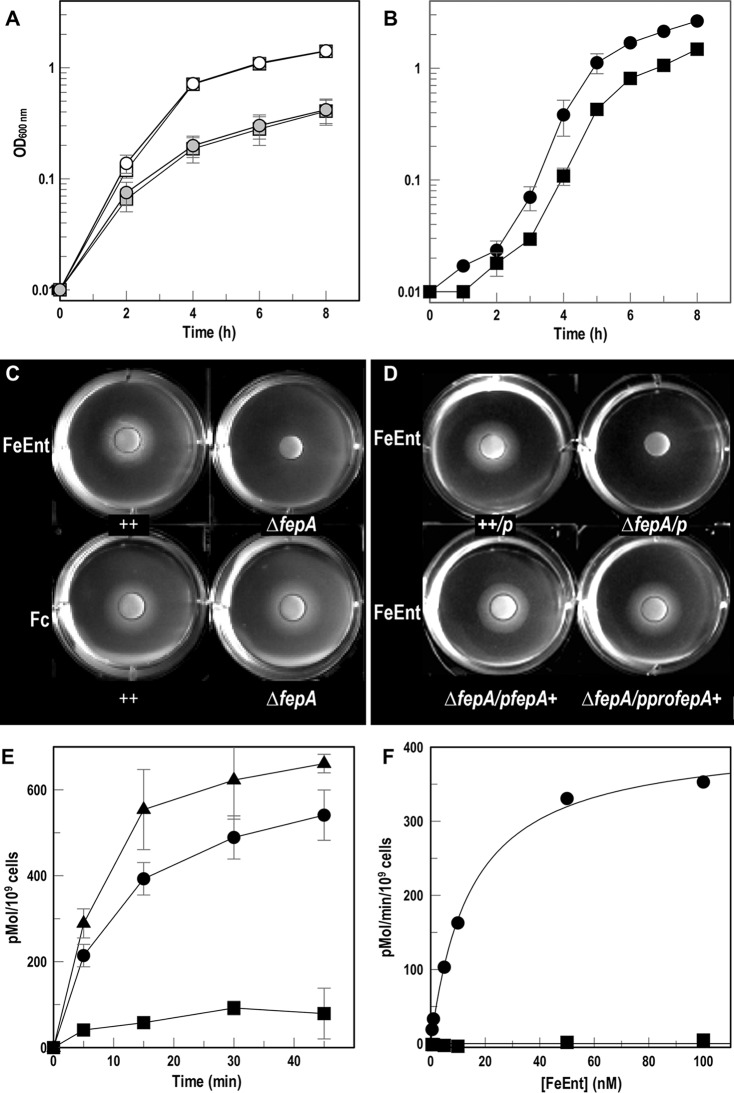
We discovered that like E. coli, A. baumannii cannot utilize iron from ferrichrome A (43), which allowed us to use apoferrichrome A (apoFcA) as an iron sequestering agent. We grew wild-type A. baumannii 17978 and its ΔfepA mutant in MOPS minimal medium plus or minus apoFcA (Fig. 4A). Under either condition, the ΔfepA derivative grew like the wild type, suggesting that other iron acquisition systems are sufficient for A. baumannii growth. However, unlike the wild type, the iron-starved ΔfepA strain did not utilize FeEnt as the sole iron source, indicating that locus A1S_0980-A1S_0981 encodes a FeEnt transporter (Fig. 4B). Siderophore nutrition tests confirmed that unlike wild-type A. baumannii, the ΔfepA derivative did not utilize FeEnt, whereas both strains normally utilized Fc (Fig. 4C). The expression of putative AbaFepA in trans under the control of its native promoter (pAbafepA) or an exogenous promoter (p2682proAbafepA) restored the ability to utilize FeEnt (Fig. 4D), confirming the identity of locus A1S_0980-AIS_0981 as the A. baumannii fepA gene.
FIG 4.
FeEnt transport by A. baumannii. (A) Growth of A. baumannii 17978 (circles) and its ΔfepA derivative (squares) in MOPS minimal medium without (open symbols) or with (gray symbols) 100 μM apoFcA (means from 3 experiments with standard deviations of the means as error bars) shows no growth defect from the ΔfepA mutation. (B) A. baumannii 17978 (●) and its ΔfepA derivative (■) were grown in LB, subcultured (1%) into MOPS minimal medium plus 100 μM apoFcA, and subcultured (1%) into MOPS minimal medium plus or minus 1 μM FeEnt (means and standard deviations from three experiments). The ΔfepA strain shows roughly half the growth of its parent. (C and D) Siderophore nutrition assays with A. baumannii 17978 and its ΔfepA mutant (C) or 17978/pWH1266 (++/p), ΔfepA/pWH1266 (ΔfepA/p), ΔfepA/pWH1266AbafepA+ (ΔfepA/pAbafepA+), and ΔfepA/p2682proAbafepA+ (ΔfepA/pproAbafepA+) (D). The bacteria were plated in top agar with 100 μM apoFcA, and discs containing FeEnt or Fc (10 μl of 50 μM) were placed on the agar surface. Both panels C and D are representative images from three separate experiments. (E) Accumulation of [59Fe]Ent over 45 min, with each data point performed in triplicate, by 17978/pWH1266 (●), ΔfepA/pWH1266 (■), and ΔfepA/p WH1266AbafepA+ (▲). The graph depicts the means and standard deviations of the means from two experiments. (F) Transport of 59Fe[Ent] by A. baumannii 17978 (●) or its ΔfepA mutant (■). The graph depicts the means and standard deviations of the means from triplicate measurements in two separate experiments.
To biochemically define FeEnt transport by AbaFepA, we measured both the accumulation (Fig. 4E) and concentration dependence (Fig. 4F) of [59Fe]Ent transport by A. baumannii 17978 and its derivatives. Whereas the wild type and the complementation strain accumulated 500 to 600 pM FeEnt per 109 cells in 45 min, uptake of FeEnt by the ΔfepA strain was much less, confirming that AbaFepA is its primary transporter. Kinetic measurements of [59Fe]Ent uptake over a range of concentrations revealed the following parameters for AbaFepA: Km = 14.6 nM and Vmax = 413.05 pmol/109 cells/min (Fig. 4F). Overall, these data showed both chromosomally encoded and plasmid-encoded AbaFepA confer utilization and transport of FeEnt as an iron source for A. baumannii.
Generation of Cys substitution mutants in A. baumannii FepA.
The genetic engineering of Cys substitutions in the surface loops of AbaFepA required knowledge of its tertiary structure. Because no crystal structure yet exists for AbaFepA, we used the “Modeler” function of Chimera to predict the architecture of AbaFepA from the crystal structure of its EcoFepA ortholog (Fig. 5A and B) (48). Guided by past Cys substitutions in the loops of EcoFepA (28, 49), we identified potential sites for fluoresceination in 7 loops of AbaFepA and used site-directed mutagenesis to convert the native residues to Cys: T223C, S279C, A326C, T383C, T482C, T562C, S665C, S712C (Fig. 5). After transforming the A. baumannii ΔfepA derivative with the plasmids carrying mutant AbafepA genes, we expressed the AbaFepA Cys mutants, labeled them with fluorescein maleimide (FM), and evaluated the extent of Cys-specific labeling by SDS-PAGE and fluorescence imaging (Fig. 6A). Wild-type AbaFepA was not significantly modified by FM, but all the AbaFepA Cys mutants were strongly fluoresceinated, much like EcoFepA-S271C. Each AbaFepA Cys mutant utilized FeEnt in siderophore nutrition like wild-type AbaFepA and EcoFepA-S271C (data not shown). To verify their functionality for FLHTS, we measured [59Fe]Ent uptake by each mutant before and after FM labeling. In total we generated 9 functional AbaFepA Cys mutants that had a range of FeEnt transport capabilities (Fig. 6B); 3 showed >30% of wild-type uptake levels even when fluoresceinated: S279C, T562C, and S665C mutants.
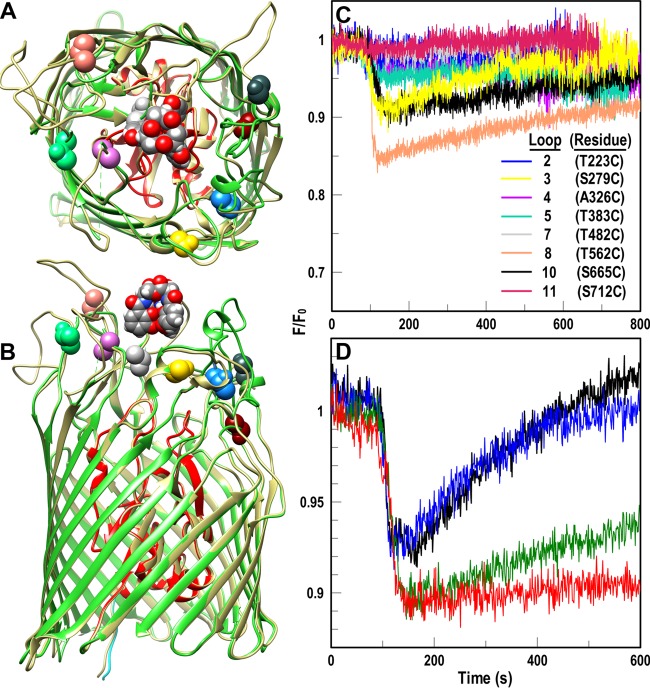
FIG 5.
Fluoresceination of Cys substitutions in AbaFepA facilitates measurement of FeEnt uptake by A. baumannii. (A and B) Cys mutagenesis of AbaFepA. The Modeller function of CHIMERA (48) predicted the tertiary structure of AbaFepA (dark beige) based on its 46% identity to EcoFepA (PDB file 1FEP; N domain, red; C domain, lime green). We selected eight amino acids in the surface loops of AbaFepA loops (colored and depicted in space-filling format) with Cys, from their similarity to residues in EcoFepA that when fluoresceinated gave substantial quenching during FeEnt binding (28). (C and D) Fluorescence spectroscopic measurement of FeEnt transport in A. baumannii. (C) After transforming mutant plasmids encoding AbaFepA Cys mutants into A. baumannii 17978 ΔfepA, we maximized expression of the AbaFepA Cys mutants by growth in iron-deficient MOPS medium and labeled the cells with FM. We monitored fluorescence voltage from FM-labeled ΔfepA/pAbafepA-Cys mutants in response to addition of 10 nM FeEnt at 100 s and normalized the data to the initial fluorescence of the bacteria (F/F0). Binding of FeEnt to AbaFepAS279C-FM, T562C-FM and S665C-FM quenched their fluorescence; subsequent FeEnt uptake by the cells depleted the ferric siderophore from solution, resulting in a gradual increase to initial fluorescence levels (recovery). The plotted data are the averages from three separate experiments. (D) Normalized fluorescence of FM-labeled 17978 ΔfepA/pAbafepAS279C following exposure to various concentrations of CCCP (black, no CCCP; blue, 2.5 μM CCCP; green, 5 μM CCCP; red, 10 μM CCCP). The plotted data are the averages from two separate experiments.
FIG 6.
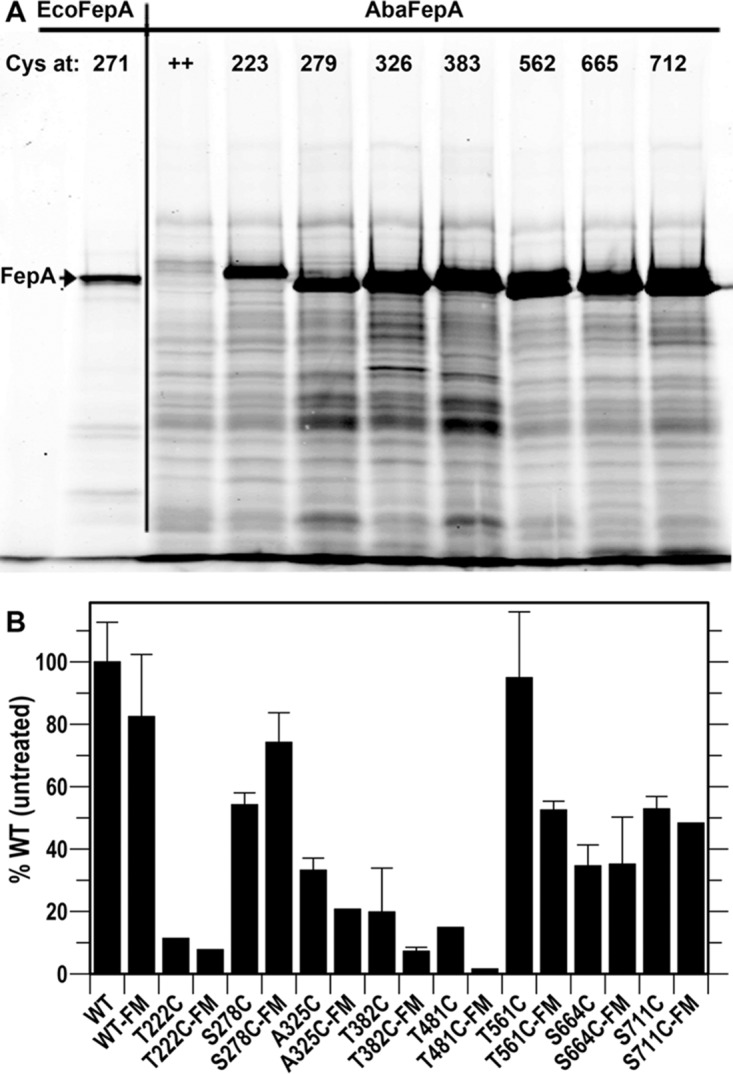
FeEnt uptake by fluoresceinated AbaFepA-Cys mutants. (A) FM labeling. A. baumannii 17978 ΔfepA harboring plasmids expressing AbaFepA Cys mutants were labeled with 5 μM FM and subjected to SDS-PAGE, followed by fluorescent imaging. We included E. coli OKN3/pITS23-S271C as a positive control. The imaged gel was representative of three experiments. (B) FeEnt transport Vmax screening. All unlabeled mutants normally utilized FeEnt in siderophore nutrition tests (data not shown), but quantitative measurements of [59Fe]Ent uptake showed that certain Cys substitutions (T223C, A326C, T383C, and S482C) impaired iron transport, especially when fluoresceinated. The graph depicts the transport of [59Fe]Ent by the mutant strains before and after FM labeling, relative to wild-type A. baumannii that was untreated but harboring the empty plasmid vector. S279C, T562C, and T665C mutants transported [59Fe]Ent with 38 to 75% of the efficiency of wild-type AbaFepA. Each bar derives from the average of 2 or 3 experiments, performed in triplicate.
Fluorescence observations of FeEnt transport with AbaFepA Cys mutants.
We assessed the ability of the individual Cys substitution derivatives of AbaFepA to monitor FeEnt uptake (Fig. 5C). The functionality of AbaFepA S279C, T562C, and S665C mutants, even when fluoresceinated, made them candidates for use in FLHTS. Both the fluorescence emissions of cells expressing them and fluorescence scans of SDS-PAGE gels of their cell lysates showed strong specific labeling of the mutants. Wild-type AbaFepA was only labeled at background levels that did not result in detectable fluorescence quenching or recovery (Fig. S3A and B). FM quantitatively labeled AbaFepA S279C, T562C, and S665C mutants (Fig. 6A), and FeEnt binding quenched the fluorescence of the cells 10% to 20%. AbaFepAS279C-FM, -T562C-FM, and -S665C-FM had, respectively, 75%, 53%, and 38% FeEnt uptake activity of wild-type, unmodified AbaFepA (Fig. 6B), resulting in fluorescence recovery in all three strains within 10 to 15 min (Fig. 5C). We determined the concentration dependence of FeEnt activity in the FLHTS system for AbaFepAS279C-FM (Fig. S3). Across a range of 5 to 40 nM FeEnt, 10 to 20 nM gave maximum quenching with complete recovery in 10 min. Furthermore, CCCP blocked FeEnt transport by AbaFepAS279C-FM (Fig. 5D), confirming the physiological relevance of the system. A. baumannii was ∼5-fold more sensitive than E. coli to CCCP, in that a 5 μM concentration of the proton ionophore abrogated FeEnt transport by bacteria harboring AbaFepAS279C-FM. Like E. coli, cryopreserved, fluoresceinated A. baumannii ΔfepA/pAbafepAS279C cells were viable in the assay (Fig. S3A). Finally, we compared the three AbaFepA Cys mutants that performed best in cuvettes (S279C, S665C, and T562C mutants) for FeEnt transport in 96-well plates (Fig. 7). In microtiter wells containing 5 × 107 cells/ml of FM-labeled A. baumannii ΔfepA/pAbafepAS279C or -T562C, 20 nM FeEnt quenched fluorescence 14% and 18%, respectively. The nearly complete recovery that occurred during subsequent iron transport (Fig. 7A and B) yielded Z′ factors of 0.6 to 0.8 after 8 to 15 min (Fig. 7C). These data showed the adaptability of FLHTS to A. baumannii, for discovery of inhibitors that block TonB-dependent transport in the ESKAPE pathogen.
FIG 7.
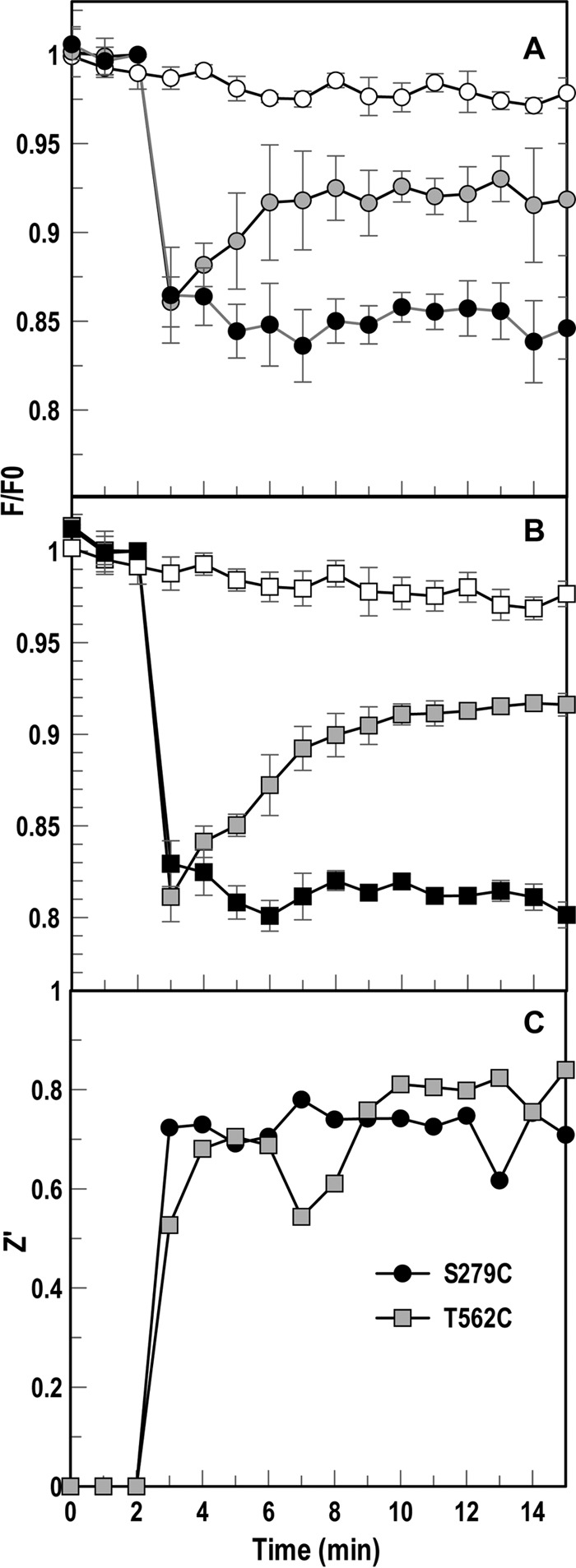
Adaptation of fluorescence assay to HTS format in A. baumannii. We tested 17978 ΔfepA expressing AbaFepAS279C-FM, T562C-FM, and S665C-FM (not shown) in the fluorescence assay in a 96-well microtiter plate format. The tracings depict normalized fluorescence of FM-labeled ΔfepA/pAbafepAS279C (A) or T562C mutant (B) following exposure to 10 μM CCCP (negative control; filled symbols) or an equivalent volume of DMSO; open symbols. Following three readings of initial fluorescence (F0), we injected 20 nM FeEnt in the absence (gray symbols) or presence (filled symbols) of CCCP and measured changes in the ensuing fluorescence (F) over time. We also included cells to which no FeEnt was added (positive control; open symbols). The graph averages data from two separate experiments. (C) Z′ factors for each mutant's controls were calculated at each read during experiment, averaging between 0.6 and 0.8 for the controls from 8 to 15 min.
DISCUSSION
In 2009, 1.7 million hospital-associated bacterial infections caused or contributed to 99,000 deaths. Gram-negative bacteria were responsible for two-thirds of the mortality, and 20% of the isolates were resistant to all known antibiotics (50). This large fraction of resistance derives in part from the selective permeability and antibiotic export capabilities of the prokaryotic cell envelope (51, 52). In 2011, a survey of 183 hospitals involving 11,282 patients revealed that 4% developed health care-associated bacterial infections (53). The CRE (carbapenem-resistant Enterobacteriaceae) and ESKAPE pathogens cause the majority of nosocomial infections as a result of their antibiotic resistance, and the clinical options for these bacteria are limited (54). Against this backdrop, drug and pharmaceutical companies lessened efforts to combat bacterial infections (55).
Microbes must acquire iron during colonization of humans and animals (25), and the ubiquitous TonB-dependent iron uptake systems of Gram-negative bacteria are virulence determinants (56, 57). The spectroscopic methodology we developed to study TonB-dependent transport, and customized for FLHTS, allows facile discovery of chemicals that may block or impair TonB action; these may become antibacterial drugs. Application of FLHTS in vivo obviates reconstitution of TonB-dependent transport in vitro, which has not yet been accomplished. It also offers the many advantages of live-cell assays: convenience, predictability, miniaturization, automation, and multiplexing (58). The findings reported herein create a template for comparable use of the iron transport systems of other bacteria: FLHTS against A. baumannii using FeEnt uptake produced Z′ values similar to those with E. coli. Its application to bacterial pathogens may lead to discovery of compounds that block specific iron transporters or broad-spectrum inhibitors of all Gram-negative bacterial TonB-dependent systems. Both classes may retard infections by Gram-negative ESKAPE organisms, and our findings with a small chemical library portend hundreds of relevant compounds in large chemical libraries.
Statistically, the FLHTS primary screen of 17,441 compounds against E. coli (42) in a 384-well format gave a sufficiently high mean Z′ factor (59) of 0.87 across all plates and an overall hit rate of 0.54%. We used CCCP as a positive control in these experiments because it depletes the PMF underlying TonB action in OM transport. However, it had limited value because it does not specifically target TonB. Many of the 165 compounds that impaired or retarded FeEnt uptake in the primary screen did not fit the desired inhibitor profile, illustrating that numerous types of compounds may inhibit iron uptake by mechanisms that do not involve TonB-ExbBD. Chemicals that spectrally overlap fluorescein or specifically or nonspecifically adsorb to FepA, for instance, may quench by energy transfer or ground-state complex formation, respectively. We excluded such compounds from further consideration because they quenched fluorescence in the 1st read, prior to the addition of FeEnt. Among 44 other chemicals that did fit the expected profile for inhibition of TonB action, we analyzed 20 and found 6 that affected TonB-dependent secondary screens; 5 more impaired activity in all 4 TonB-dependent transport in multiple ways, but not in every test we conducted. For E. coli, concomitant impairment of siderophore nutrition and colicin killing potentially implicates TonB as the drug target. We tested FLHTS primary hits against three different OM TBDT: FepA (FeEnt and ColB), FhuA (Fc), and Cir (ColIa). Simultaneous blockage of FeEnt and Fc uptake, or ColB and ColIa killing, eliminated the possibility of steric hindrance of ligand adsorption by an inhibitor, because TBDT ligand recognition is highly specific (8, 10, 45). Next, concomitant reduction of both ferric siderophore uptake and colicin killing excluded inhibition of an IM (iron) ABC transporter or of ATP hydrolysis as explanations, because colicin killing does not involve these processes. Still, these tests do not discriminate chemicals that directly inhibit TonB-ExbBD from compounds that affect related but different physiology, such as PMF. Proton ionophores are abundant in chemical libraries, and we used [14C]lactose uptake assays to identify them. The OM FeEnt transport system is similarly sensitive to PMF depletion as the IM lactose permease: the CCCP 50% inhibitory concentrations (IC50) were 5 to 15 μM for FeEnt uptake (this study) and 2 to 5 μM for lactose uptake (60). Comparable inhibition of [59Fe]Ent and [14C]lactose uptake suggested the actions of a proton ionophore. Conversely, [14C]lactose accumulation data provided the rationale for excluding PMF depletion as the underlying inhibition mechanism of ebselen and ST082900: both compounds were effective only at much higher concentrations (250 μM and 100 μM, respectively), and even at those high levels they were ineffective in blocking [14C]lactose uptake (ebselen, 32%; ST082990, 26%). Chemicals that passed through this filter of secondary tests may interfere with TonB biochemistry, including interactions between TonB and the TonB box of ligand-bound TBDT (16), between TonB and PG (24), or between TonB and ExbBD (61). Compounds of interest may also block TonB-ExbBD utilization of PMF or mechanistic motion by TonB (22). Even using TonB-deficient strains, authentic inhibitors of TonB action are difficult to confirm, because ΔtonB strains grow poorly under iron-deficient conditions. They form tiny colonies on plates and have long doubling times and low yield in low-iron liquid media (e.g., nutrient broth/plates and minimal media), confounding the antagonistic effects of chemicals with their already compromised growth. TolAQR partially compensates and complements the absence of TonB-ExbBD (62–64), so inhibitors of TonB may further curtail ΔtonB strains by blocking TolAQR. Lastly, the TonB C terminus binds PG in the sacculus (24), and chemicals that block this protein-PG interaction may interfere with PG metabolism and hence inhibit even ΔtonB strains.
Our small-scale FLHTS of 17,441 chemicals yielded 94 (0.54%) primary hits that inhibited fluorescence recovery ≥20%. Among the 20 primary hits we further tested, 2 (10%), ebselen and ST0082990, remained as candidates against TonB. These data extrapolate to an overall frequency of 0.054% potentially relevant TonB antagonists in a diverse compound collection, which predicts several hundred relevant, non-proton ionophore chemicals in a larger library (e.g., KU-HTSL encompasses 350,000 chemicals).
The screening results provided insight into the nature of chemicals that may affect TonB action. Numerous types of inhibition may occur, including the oligodynamic effects of organomercurials like thimerosal (65) that react with protein amino and sulfhydryl groups. TonB residue His20 is crucial to its physiological actions (66), which may explain finding thimerosal as an inhibitor of TonB-dependent FeEnt uptake. Zinc pyrithione, another potent primary hit, is a coordination complex proton ionophore (67–70), with bacteriostatic effects from disruption of membrane transport, whose actions resemble those of CCCP. A few compounds differentially affected TonB-dependent physiology: ST084954 and ST005540 inhibited ferric siderophore uptake but not colicin killing, whereas p-chloranil showed the opposite effect. The ability of these chemicals to differentiate the TonB-dependent activities of FepA may provide insight into the mechanisms of its interactions with its ligands. Others investigators searched for TonB inhibitors with an iron-dependent growth assay that used an E. coli tolC strain with altered cell envelope permeability (71): the inactivation of TolC causes greater retention of chemicals in the periplasm. We tested a lead compound from the study described in reference 71, compound 120304, but at 100 μM it was inactive in both the primary fluorescence FeEnt uptake test and other secondary screens of TonB activity. We did not define or characterize the permeability properties of compound 120304, but its mass (261 Da) implies rapid equilibration through general porin channels (72) into the bacterial periplasm. The tolC mutant employed by Yep et al. (71) decreases antibiotic export, potentially increasing the active concentration of compound 120304 in the periplasm, IM, and/or cytoplasm. This caveat may explain the activity of compound 120304 against the tolC derivative, but the FLHTS assay revealed that compound 120304 apparently does not affect TonB action in wild-type E. coli. It is noteworthy that secondary screens do not yet exist to identify inhibitors that directly target TonB-ExbBD. Nevertheless, the small-scale, proof-of-concept screen we completed identified 6 compounds that block TonB-mediated transport, and 2 of those may antagonize TonB itself. Further analyses of the remaining untested 165 primary hits will likely discover more, and FLHTS of larger libraries (∼350,000 compounds in the KU-HTSL) may find hundreds more.
A. baumannii ATCC 17978 encodes siderophore biosynthetic systems for acinetobactin (73), baumanoferrin (39), and fimsbactin (40), as well as Fur-regulated (34, 35) TBDT for ferric siderophores that it does not synthesize (e.g., FhuA-Fc, FhuE-Fe-rhodotorulate, and FepA-FeEnt). Hence, A. baumannii ORF A1S_0980-A1S_0980 encodes AbaFepA, the iron-regulated OM receptor for FeEnt, which efficiently transported the catecholate ferric siderophore. So, like other Gram-negative bacteria (74), A. baumannii responds to iron limitation by acquiring the ferric complexes in its environment. Enterobactin has the highest affinity for Fe3+ of any siderophore; the expression of AbaFepA allows A. baumannii to compete for iron in the presence of enterobactin-secreting organisms.
Using the 46% identity in primary structures between AbaFepA and EcoFepA in the Modeler function of CHIMERA (6, 48), we predicted the tertiary structure of AbaFepA from the crystal structure of EcoFepA (6). We then targeted sites in its surface loops and introduced single Cys residues in AbaFepA. Cys mutations in AbaFepA at equivalent sites to those in EcoFepA generally did not compromise FeEnt transport, even when AbaFepA was fluoresceinated. EcoFepA contains a disulfide-bonded Cys pair in L7 (6), and AbaFepA contains a homologous Cys pair in L7. Like wild-type EcoFepA, wild-type AbaFepA was not fluoresceinated by our procedures. FM specifically labeled only the engineered individual AbaFepA Cys substitutions, with minimal background labeling. Unfortunately, at present the only known assays of TonB action in A. baumannii are ferric siderophore and heme uptake; additional secondary tests for inhibition of TonB-ExbBD are needed. Nevertheless, our findings with A. baumannii validate the FLHTS strategy against Gram-negative bacteria that encode FepA and transport FeEnt. It is conceivable that other iron transport systems (for example, Fc-FhuA) are amenable to study with similar fluorescence approaches.
When applied to A. baumannii the fluorescence transport assay sensitively monitored uptake of FeEnt, as it does in E. coli. In A. baumannii, however, binding of FeEnt to AbaFepA-FM quenched fluorescence less than when bound to EcoFepA-FM. The maximum quenching of AbaFepA-FM (labeled at S279C or T562C) was ∼20%, whereas FeEnt binding to EcoFepA-FM (labeled at A698C) quenched ∼60% of the original fluorescence (28). FeEnt-induced quenching in EcoFepA-FM originates from conformational motion in the surface loops that relocates the fluorophore (27, 28). It is conceivable that our survey of AbaFepA surface loops did not identify an optimum site for Cys substitution and fluorophore attachment. Alternatively, the lesser fluorescence quenching in A. baumannii may derive from the full-length LPS O-antigen and/or polysaccharide capsule in its cell envelope, which create a more hydrophilic surface than that of rough E. coli K-12. Additional hydrophilic cell surface structures may decrease base fluorescence by collisional quenching, thereby limiting the extent of further quenching from conformational changes during FeEnt adsorption. We still observed dose-dependent quenching and recovery during FeEnt uptake in A. baumannii that yielded acceptable reproducibility and Z′ values in FLHTS.
In summary, this proof-of-concept FLHTS study against E. coli discovered compounds that antagonize the functions of TonB. FLHTS against an ESKAPE bacterium is a logical next step in its application to antibiotic discovery. The efficacy of the assay against A. baumannii creates a methodological template for identification of drugs against clinically important Gram-negative bacteria, as opposed to the laboratory E. coli strain used in our initial experiments. This capability is sorely needed in a world that faces an increasingly uncertain outcome of microbiological infection. We also expect that once found, bona fide inhibitors of TonB-ExbBD activity will provide new mechanistic insight into the biochemistry of TonB action.
MATERIALS AND METHODS
Bacterial strains and reagents.
The experiments involved E. coli strains MG1655 or BN1071 and their derivatives OKN1 (ΔtonB), OKN3 (ΔfepA), OKN7 (ΔfhuA), and OKN3/pITS23 (ΔfepA/pEcofepA; contains the wild-type EcofepA structural gene under the control of its natural, Fur-regulated promoter, cloned on the low-copy-number plasmid pHSG575 [75]) and its derivative OKN3/pFepAS271C (10, 76, 77). Experiments with A. baumannii were performed with strain ATCC 17978 or its ΔfepA derivative. Bacteria were cultured in Luria broth (LB) at 37°C with aeration. When appropriate, we added antibiotics at the following concentrations: ampicillin (Sigma), 100 μg/ml for E. coli and 500 μg/ml for A. baumannii; streptomycin (Sigma), 100 μg/ml; and chloramphenicol (Sigma), 20 μg/ml. For iron deprivation we used iron-deficient MOPS minimal medium (78). We prepared carbonyl cyanide-m-chlorophenylhydrazone (CCCP; Sigma) as a 10 mM stock in dimethyl sulfoxide (DMSO) and stored it at −20°C. For secondary screening, we purchased compounds as fresh solids (>95% purity) from commercial suppliers or the University of Kansas (KU CMLD), dissolved them in DMSO to 10 mM, and stored them at −20°C.
Generation of deletion mutants and complementation vectors in A. baumannii.
For generation of the A. baumannii ΔfepA derivative, we amplified approximately 1,000 bp of DNA in both the 5′ and 3′ flanking regions surrounding each open reading frame from A. baumannii genomic DNA. We subsequently amplified the pFLP2 suicide vector (79) and stitched the three PCR products together by the method of Gibson et al. (80), using the NEBuilder HiFi cloning kit (New England BioLabs), and verified the resulting construct by sequencing. After transforming this vector into A. baumannii by electroporation, we selected bacterial integrants on LB Amp500 agar and patched transformants onto LB Amp500 or LB with 10% sucrose: merodiploids were Ampr and sucrose sensitive. To resolve the integrated plasmid, we grew the merodiploid strains overnight in 3 ml of LB at 37°C with shaking, then serially diluted the cultures, plated them for single colonies on LB agar with 10% sucrose, and incubated them overnight at 37°C. We patched the transformants on both LB Amp500 and LB agar, incubated them at 37°C overnight, and then screened the Amps strains for the loss of fepA by PCR analysis.
For fepA complementation in A. baumannii, we amplified the complete A. baumannii fepA structural gene, including the native promoter, fepA gene, and terminator region from A. baumannii genomic DNA, incorporating BamHI and SalI restriction sites at the 5′ and 3′ ends, respectively. We also amplified the isolated fepA gene using the same strategy. Following digestion, we ligated these two PCR products into pWH1266 or p2682pro (81) to make pAbafepA and p2682proAbafepA, respectively. For each construct, we confirmed the sequences of the promoter, terminator, and structural gene (McLab, San Francisco, CA).
Site-directed mutagenesis to introduce Cys substitutions in AbaFepA.
We predicted the tertiary structure of AbaFepA using the Modeler algorithm of CHIMERA (48). The 46% identity of primary structure between AbaFepA and EcoFepA indicated a comparable overall fold for the former protein, based on the crystal structure of the latter (6). We used the model to select several residues in different predicted surface loops, which were generally structurally analogous to loop residues in EcoFepA that were successfully fluoresceinated (28, 49). We engineered the mutations in pAbafepA using a QuikChange II XL mutagenesis kit (Agilent, Santa Clara, CA). Specifically, with pAbafepA as the template, we generated site-directed Cys substitutions at positions T223, S279, A326, T383, T482, T562, S665, and S712 in AbaFepA. After verification by DNA sequence analysis, we transformed the constructs into the A. baumannii ΔfepA host and evaluated FeEnt transport of each strain by nutrition assays to verify complementation by the plasmid-mediated Cys mutant fepA genes.
Fluorescence labeling and spectroscopy.
For fluoresceination of FepA in live cells, after overnight growth in LB, we subcultured E. coli in MOPS minimal medium and A. baumannii in nutrient broth (NB) with shaking at 37°C until an optical density at 600 nm (OD600) of 1.0 to 1.5 was reached. We labeled the live bacteria with 5 μM fluorescein maleimide (ε493 nm = 81,500 M−1 at pH 8.0) in 50 mM NaHPO4, pH 6.7, for 15 min at 37°C, quenched the reaction with 1.3 mM β-mercaptoethanol, and pelleted, washed, and suspended the bacteria in phosphate-buffered saline (PBS) plus 0.4% glucose for E. coli (42), or 0.4% 50 mM sodium acetate for A. baumannii.
Spectroscopic assays were performed in an SLM/OLIS spectrofluorometer that has an SLM 8000 chassis (Aminco, Lake Forest, CA), upgraded to automatic control by an OLIS central processing unit, operating system, and analysis software (OLIS, Inc., Bogart, GA). The excitation and emission wavelengths were 490 and 520 nm, respectively. With the labeled cells diluted to 2.5 × 107/ml in 2 ml of the same buffer in a 3-ml quartz cuvette, we collected initial readings of fluorescence intensity, added FeEnt to the desired final concentration, and monitored changes in fluorescence emissions during its binding and transport.
For optimization in microtiter format, we used the Tecan GENios microplate reader (Tecan, Switzerland). We added frozen or fresh labeled cells to black, round-bottom 96-well (Corning, Lowell, MA) or 384-well (Greiner Bio-One, Monroe, NC) plates to a 190-μl or 95-μl total volume, respectively, of PBS plus 0.4% glucose or 50 mM acetate as for the SLM. After three reads of initial fluorescence at 535 nm, we added FeEnt at the designated concentration to each well by automatic injection. After shaking for 10 s, we recorded transport over a time course with reads every minute.
To determine the effect of cryopreservation on the labeled bacteria, we froze them at −70°C in 20% glycerol, thawed them on ice, and warmed them to 37°C prior to evaluation of transport in either format of the fluorescence assay. The cells remained viable and transported FeEnt with similar kinetics after cryostorage for at least 1 month by this method. For all fluorescence assays, we normalized raw fluorescence volt readings to account for variations in labeling efficacy and cell number, as instantaneous fluorescence (F) relative to initial fluorescence (F0). To determine the statistical effect size, we calculated Z′ factors for the positive and negative controls: Z′ = 1 − (3*σ+c + σ−c)/(|μ−c − μ+c|) (59). We plotted the raw and normalized values with GraFit 6.011.
Primary FLHTS assay.
The test strain in FLHTS, OKN3/pFepAS271C (entA ΔfepA/pEcofepAS271C), expresses FepAS271C under the control of its natural, Fur-regulated promoter from the low-copy-number plasmid pITS23, a derivative of pHSG575 (75). After growing and fluoresceinating E. coli OKN3/pFepAS271C as described above, we confirmed the extent of labeling and its ability to transport FeEnt by the spectroscopic assay. We aliquoted the labeled bacteria at 2 × 108 cells/ml, froze them at −80°C, and transported the frozen cells to University of Kansas HTS facility on dry ice. We next validated the FLHTS assay with frozen cells reconstituted in PBS, plus or minus CCCP, in the BioTek Synergy microplate reader (BioTek, Winooski, VT), prior to performing the HTS assay. We screened four libraries totaling 17,441 compounds in a 384-well plate format, with the bacteria at 2 × 107 cells/ml in PBS plus 0.4% glucose. Using the BioTek Synergy (gain = 90, excitation = 495 nm, and emission = 520 nm), we measured initial fluorescence (F1), quenched fluorescence 1 min after FeEnt addition (F2), and recovered fluorescence 60 to 80 min following addition of FeEnt and incubation at 37°C (F3). We included the same labeled cells plus FeEnt and 100 μM CCCP as controls on each plate.
Data analysis and dose-response curves from primary hits.
From the positive and negative controls, we calculated Z′ scores for each plate and plotted a scattergram of all compounds to determine the median and standard deviation (SD). We performed dose-response curves for compounds that inhibited fluorescence recovery more than 2 standard deviations from the mean of all compounds tested (165 compounds [Table 1]). We tested each compound at 0, 2.5, 5 10, and 20 μM in a kinetic format of the fluorescence assay described above, with fluorescence reads every minute for 80 min. The primary hit compounds fell into categories of percent inhibition: >30% inhibition, 20 to 30% inhibition, and <20% inhibition. We focused on 44 compounds with >30% inhibition and purchased 20 of them for secondary screening analysis.
Siderophore nutrition assays.
After overnight growth in LB and subculture in nutrient broth (NB) with appropriate antibiotics at 37°C for 5.5 h, we added 100-μl aliquots of E. coli and A. baumannii to NB top agar containing 100 μM apoFcA in 6-well plates (43, 82, 83). We applied 10 μl of 50 μM FeEnt or Fc to paper discs on the agar surface and incubated the plates overnight at 37°C. In secondary screens of primary hit compounds we added the chemicals of interest (diluted 100-fold from DMSO stock solutions to 100 μM) to the bacteria in top agar, plated the mixture, added discs with FeEnt or Fc, and incubated the assay overnight. Halos of growth around the discs revealed ferric siderophore utilization by the test bacteria; alterations in halo size or morphology or the complete absence of a halo indicated inhibition by the test compound. For some compounds we evaluated a range of concentrations from 1 to 75 μM in FeEnt nutrition tests. DMSO alone had no effect in the assay, and the positive control CCCP (at 15 or 20 μM) inhibited FeEnt uptake.
Colicin killing tests.
We used susceptibility to B-group colicins as additional tests of inhibition of TonB action. Colicins B and Ia kill E. coli by adsorbing to FepA and Cir, respectively, penetrating the OM in TonB-dependent fashion before forming a depolarizing channel in the IM (46). Hence, blockage of their killing action may reflect the inhibition of TonB. To assess the influence of primary hits, we determined the titer of each colicin (84, 85) in the absence and presence of the primary hit compound of interest. We cultured MG1655 overnight in 5 ml of LB, subcultured it at 1% in 5 ml of MOPS medium for 6 h, and adjusted the bacterial concentration to 1 × 104 CFU/ml by OD600 (confirmed by serial dilution and plating on LB agar). We incubated a subtoxic concentration (as determined by MIC assay) of compound with 100-μl aliquots of bacterial cells for 15 min at 37°C, then added a predetermined amount of colicin B or Ia to the mixture, and incubated it for an additional 15 min at 37°C before plating on LB agar. After incubation at 37°C for 16 h, we counted the colonies on the plates and determined the number of colicin hits per cell from S/S0 = e−k (85), where S is the number of colonies in the presence of colicin (cells that survive colicin exposure), S0 is the number of colonies in the absence of colicin, and k is the number of hits per cell. These experiments allowed quantitative comparisons of colicin killing in the absence and presence of test compounds. We evaluated each primary hit compound of interest in at least three separate experiments and averaged the three replicates as either percent killing (Table 2) or percent survival (Fig. 2B) using GraFit 6.011.
MIC assay.
E. coli MG1655 or A. baumannii ATCC 17978 was cultured overnight in LB and subcultured at 1% into LB in the absence or presence of the compound of interest. We tested each compound over a range of concentrations in a 2-fold dilution series: 0.25 to 512 μM. The MIC was the minimum final concentration that completely prevented bacterial growth; compounds that did not prevent growth at 512 μM were considered noninhibitory. We included untreated E. coli and E. coli exposed to the corresponding volumes of DMSO as controls. We tested each compound in 2 or 3 separate experiments.
Growth assay under iron-limiting conditions.
We cultured wild-type A. baumannii and its ΔfepA derivative overnight in LB, subcultured the bacteria at 1% in MOPS minimal medium plus or minus 100 μM apoFcA, and monitored growth over an 8-h time course. For growth with FeEnt as an iron source, we grew and subcultured the strains as described above, diluted them at 1% into MOPS minimal medium plus or minus 1 μM FeEnt, and evaluated them for growth over an 8-h time course.
[59Fe]Ent accumulations.
We measured accumulation (86) of freshly prepared, chromatotraphically purified (87) [59Fe]Ent (specific activity, ∼200 cpm/pmol). For E. coli we cultured the bacteria in LB, subcultured them at 1% in MOPS minimal medium plus 0.4% glucose at 37°C with shaking for 5.5 h, and maintained the cultures at 37°C during the uptake assay. To a 4-ml aliquot of bacterial culture in a water bath with gentle agitation, we added [59Fe]Ent to 1 μM at time zero. After 5, 15, 30, and 45 min at 37°C, we collected 100-μl aliquots of cells (in triplicate) on 0.45-mm nitrocellulose filters and washed them with 10 ml of 0.9% LiCl. For A. baumannii we cultured the bacteria in LB and subcultured them at 1% in MOPS minimal medium plus 50 mM sodium acetate at 37°C with shaking for 5.5 h. All other procedures were identical to those used for E. coli. After measuring the radioactivity on the filters in a Cobra Quantum gamma counter (PerkinElmer, Inc., Waltham, MA), we averaged the data from replicate experiments and plotted the results with GraFit 6.011.
[59Fe]Ent uptake kinetics in transport assays with A. baumannii.
We deposited bacteria, cultured as described above, in a 50-ml glass test tube in a 37°C water bath and without delay poured 10 ml of prewarmed MOPS minimal medium plus 20 mM sodium acetate containing various concentrations of [59Fe]Ent into the tube. After 5 s or 1 min 5 s, we collected the cells on 0.45-mm nitrocellulose filters, washed them with 10 ml of 0.9% LiCl, and determined their radioactivity in the gamma counter (86). We used the Enzyme Kinetics function of Grafit 6.011 to determine kinetic parameters from the initial rates of FeEnt uptake, which we determined at each substrate concentration from two independent measurements, each made in triplicate, at 5 s and 1 min 5 s. We subtracted the radioactivity associated with the cells at 5 s from the radioactivity associated with the cells at 1 min 5 s. We fitted curves by nonlinear regression analysis of the average of replicate experiments, which produced Km and Vmax values.
[14C]lactose accumulations.
To identify compounds that act as proton ionophores, we measured their effects on the accumulation of [14C]lactose. We cultured E. coli MG1655 in LB, subcultured the bacteria at 1% in MOPS minimal medium plus 0.4% glucose at 37°C with shaking for 3.5 h, added isopropyl-β-d-thiogalactopyranoside (IPTG) to 0.1 mM with shaking for 2 h more, and maintained the cultures at 37°C during the uptake assay. To a 4-ml aliquot of bacterial culture in a water bath with gentle agitation, we added [14C]lactose to 10 μM at time zero. After 5, 15, 30, and 45 min at 37°C, we collected 100-μl aliquots of cells (in triplicate) on 0.45-mm nitrocellulose filters and washed them with 10 ml of 0.9% LiCl. After measuring the radioactivity on the filters in a Cobra Quantum gamma counter (PerkinElmer, Inc., Waltham, MA), we averaged the data from replicate experiments and plotted the results with GraFit 6.011.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yan Shipelskiy for thoughtful discussions regarding the study.
This research was supported by grant R21AI115187 from the National Institutes of Health (NIH).
The content of this article does not necessarily represent the views of the NIH or NIAID and is solely the responsibility of the authors.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00889-16.
REFERENCES
- 1.Weinberg ED. 1975. Nutritional immunity. Host's attempt to withold iron from microbial invaders; JAMA 231:39–41. [DOI] [PubMed] [Google Scholar]
- 2.Harris WR, Carrano CJ, Raymond KN. 1979. Spectrophotometric determination of the proton-dependent stability constant of ferric enterobactin. J Am Chem Soc 101:2213–2214. doi: 10.1021/ja00502a053. [DOI] [Google Scholar]
- 3.Carrano CJ, Raymond KN. 1979. Ferric ion sequestering agents. 2. Kinetics and mechanism of iron removal from transferrin by enterobactin and synthetic tricatechols. J Am Chem Soc 101:5401–5404. [Google Scholar]
- 4.Tidmarsh GF, Klebba PE, Rosenberg LT. 1983. Rapid release of iron from ferritin by siderophores. J Inorg Biochem 18:161–168. doi: 10.1016/0162-0134(83)80019-1. [DOI] [PubMed] [Google Scholar]
- 5.Klebba PE. 2016. ROSET model of TonB action in Gram-negative bacterial iron acquisition. J Bacteriol 198:1013–1021. doi: 10.1128/jb.00823-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchanan SK, Smith BS, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat Struct Biol 6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 7.Evans JS, Levine BA, Trayer IP, Dorman CJ, Higgins CF. 1986. Sequence-imposed structural constraints in the TonB protein of E. coli. FEBS Lett 208:211–216. doi: 10.1016/0014-5793(86)81020-1. [DOI] [PubMed] [Google Scholar]
- 8.Scott DC, Cao Z, Qi Z, Bauler M, Igo JD, Newton SM, Klebba PE. 2001. Exchangeability of N termini in the ligand-gated porins of Escherichia coli. J Biol Chem 276:13025–13033. doi: 10.1074/jbc.M011282200. [DOI] [PubMed] [Google Scholar]
- 9.Schauer K, Rodionov DA, de Reuse H. 2008. New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem Sci 6:6. [DOI] [PubMed] [Google Scholar]
- 10.Newton SM, Igo JD, Scott DC, Klebba PE. 1999. Effect of loop deletions on the binding and transport of ferric enterobactin by FepA. Mol Microbiol 32:1153–1165. doi: 10.1046/j.1365-2958.1999.01424.x. [DOI] [PubMed] [Google Scholar]
- 11.Guerinot ML. 1994. Microbial iron transport. Annu Rev Microbiol 48:743–772. [DOI] [PubMed] [Google Scholar]
- 12.Saier MH., Jr 2000. Families of proteins forming transmembrane channels. J Membr Biol 175:165–180. doi: 10.1007/s002320001065. [DOI] [PubMed] [Google Scholar]
- 13.Andrade A, Agnol MD, Newton S, Martinez MB. 2000. The iron uptake mechanisms of enteroinvasive Escherichia coli. Braz J Microbiol 31:200–205. doi: 10.1590/S1517-83822000000300009. [DOI] [Google Scholar]
- 14.Konopka K, Bindereif A, Neilands JB. 1982. Aerobactin-mediated utilization of transferrin iron. Biochemistry 21:6503–6508. [DOI] [PubMed] [Google Scholar]
- 15.Chang C, Mooser A, Pluckthun A, Wlodawer A. 2001. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J Biol Chem 276:27535–27540. doi: 10.1074/jbc.M102778200. [DOI] [PubMed] [Google Scholar]
- 16.Chimento DP, Mohanty AK, Kadner RJ, Wiener MC. 2003. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat Struct Biol 10:394–401. doi: 10.1038/nsb914. [DOI] [PubMed] [Google Scholar]
- 17.Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW. 2006. Structure of TonB in complex with FhuA, E coli outer membrane receptor. Science 312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 18.Peacock RS, Weljie AM, Peter Howard S, Price FD, Vogel HJ. 2005. The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J Mol Biol 345:1185–1197. doi: 10.1016/j.jmb.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 19.Shultis DD, Purdy MD, Banchs CN, Wiener MC. 2006. Outer membrane active transport: structure of the BtuB:TonB complex. Science 312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 20.Celia H, Noinaj N, Zakharov SD, Bordignon E, Botos I, Santamaria M, Barnard TJ, Cramer WA, Lloubes R, Buchanan SK. 2016. Structural insight into the role of the Ton complex in energy transduction. Nature doi: 10.1038/nature19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgs PI, Larsen RA, Postle K. 2002. Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA. Mol Microbiol 44:271–281. doi: 10.1046/j.1365-2958.2002.02880.x. [DOI] [PubMed] [Google Scholar]
- 22.Jordan LD, Zhou Y, Smallwood CR, Lill Y, Ritchie K, Yip WT, Newton SM, Klebba PE. 2013. Energy-dependent motion of TonB in the Gram-negative bacterial inner membrane. Proc Natl Acad Sci U S A 110:11553–11558. doi: 10.1073/pnas.1304243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen RA, Chen GJ, Postle K. 2003. Performance of standard phenotypic assays for TonB activity, as evaluated by varying the level of functional, wild-type TonB. J Bacteriol 185:4699–4706. doi: 10.1128/JB.185.16.4699-4706.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaserer WA, Jiang X, Xiao Q, Scott DC, Bauler M, Copeland D, Newton SM, Klebba PE. 2008. Insight from TonB hybrid proteins into the mechanism of iron transport through the outer membrane. J Bacteriol 190:4001–4016. doi: 10.1128/JB.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pi H, Jones SA, Mercer LE, Meador JP, Caughron JE, Jordan L, Newton SM, Conway T, Klebba PE. 2012. Role of catecholate siderophores in gram-negative bacterial colonization of the mouse gut. PLoS One 7:e50020. doi: 10.1371/journal.pone.0050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsolis RM, Baumler AJ, Heffron F, Stojiljkovic I. 1996. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect Immun 64:4549–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao Z, Warfel P, Newton SM, Klebba PE. 2003. Spectroscopic observations of ferric enterobactin transport. J Biol Chem 278:1022–1028. doi: 10.1074/jbc.M210360200. [DOI] [PubMed] [Google Scholar]
- 28.Smallwood CR, Jordan L, Trinh V, Schuerch DW, Gala A, Hanson M, Shipelskiy Y, Majumdar A, Newton SM, Klebba PE. 2014. Concerted loop motion triggers induced fit of FepA to ferric enterobactin. J Gen Physiol 144:71–80. doi: 10.1085/jgp.201311159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doyle JS, Buising KL, Thursky KA, Worth LJ, Richards MJ. 2011. Epidemiology of infections acquired in intensive care units. Semin Respir Crit Care Med 32:115–138. doi: 10.1055/s-0031-1275525. [DOI] [PubMed] [Google Scholar]
- 30.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 31.Vidal R, Dominguez M, Urrutia H, Bello H, Gonzalez G, Garcia A, Zemelman R. 1996. Biofilm formation by Acinetobacter baumannii. Microbios 86:49–58. [PubMed] [Google Scholar]
- 32.Durante-Mangoni E, Zarrilli R. 2011. Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol 6:407–422. doi: 10.2217/fmb.11.23. [DOI] [PubMed] [Google Scholar]
- 33.Doi Y, Murray GL, Peleg AY. 2015. Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin Respir Crit Care Med 36:85–98. doi: 10.1055/s-0034-1398388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. 2011. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 12:126. doi: 10.1186/1471-2164-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nwugo CC, Gaddy JA, Zimbler DL, Actis LA. 2011. Deciphering the iron response in Acinetobacter baumannii: a proteomics approach. J Proteomics 74:44–58. doi: 10.1016/j.jprot.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penwell WF, Arivett BA, Actis LA. 2012. The Acinetobacter baumannii entA gene located outside the acinetobactin cluster is critical for siderophore production, iron acquisition and virulence. PLoS One 7:e36493. doi: 10.1371/journal.pone.0036493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorsey CW, Beglin MS, Actis LA. 2003. Detection and analysis of iron uptake components expressed by Acinetobacter baumannii clinical isolates. J Clin Microbiol 41:4188–4193. doi: 10.1128/JCM.41.9.4188-4193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaddy JA, Arivett BA, McConnell MJ, Lopez-Rojas R, Pachon J, Actis LA. 2012. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun 80:1015–1024. doi: 10.1128/IAI.06279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penwell WF, DeGrace N, Tentarelli S, Gauthier L, Gilbert CM, Arivett BA, Miller AA, Durand-Reville TF, Joubran C, Actis LA. 2015. Discovery and characterization of new hydroxamate siderophores, baumannoferrin A and B, produced by Acinetobacter baumannii. Chembiochem doi: 10.1002/cbic.201500147. [DOI] [PubMed] [Google Scholar]
- 40.Proschak A, Lubuta P, Grun P, Lohr F, Wilharm G, De Berardinis V, Bode HB. 2013. Structure and biosynthesis of fimsbactins A-F, siderophores from Acinetobacter baumannii and Acinetobacter baylyi. Chembiochem 14:633–638. doi: 10.1002/cbic.201200764. [DOI] [PubMed] [Google Scholar]
- 41.Subashchandrabose S, Smith S, DeOrnellas V, Crepin S, Kole M, Zahdeh C, Mobley HL. 2016. Acinetobacter baumannii genes required for bacterial survival during bloodstream infection. mSphere 1:e00013-15. doi: 10.1128/mSphere.00013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanson M, Jordan LD, Shipelskiy Y, Newton SM, Klebba PE. 2016. High-throughput screening assay for inhibitors of TonB-dependent iron transport. J Biomol Screen 21:316–322. doi: 10.1177/1087057115613788. [DOI] [PubMed] [Google Scholar]
- 43.Wayne R, Frick K, Neilands JB. 1976. Siderophore protection against colicins M, B, V, and Ia in Escherichia coli. J Bacteriol 126:7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao Z, Qi Z, Sprencel C, Newton SM, Klebba PE. 2000. Aromatic components of two ferric enterobactin binding sites in Escherichia coli fepA. Mol Microbiol 37:1306–1317. doi: 10.1046/j.1365-2958.2000.02093.x. [DOI] [PubMed] [Google Scholar]
- 45.Annamalai R, Jin B, Cao Z, Newton SM, Klebba PE. 2004. Recognition of ferric catecholates by FepA. J Bacteriol 186:3578–3589. doi: 10.1128/JB.186.11.3578-3589.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao Z, Klebba PE. 2002. Mechanisms of colicin binding and transport through outer membrane porins. Biochimie 84:399–412. doi: 10.1016/S0300-9084(02)01455-4. [DOI] [PubMed] [Google Scholar]
- 47.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev 21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 49.Smallwood CR, Marco AG, Xiao Q, Trinh V, Newton SM, Klebba PE. 2009. Fluoresceination of FepA during colicin B killing: effects of temperature, toxin and TonB. Mol Microbiol 72:1171–1180. doi: 10.1111/j.1365-2958.2009.06715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elemam A, Rahimian J, Mandell W. 2009. Infection with panresistant Klebsiella pneumoniae: a report of 2 cases and a brief review of the literature. Clin Infect Dis 49:271–274. doi: 10.1086/600042. [DOI] [PubMed] [Google Scholar]
- 51.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu EW, McDermott G, Zgurskaya HI, Nikaido H, Koshland DE Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976–980. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
- 53.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Emerging Infections Program Healthcare-Associated I, Antimicrobial Use Prevalence Survey Team. 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J, Nation RL, Milne RW, Turnidge JD, Coulthard K. 2005. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int J Antimicrob Agents 25:11–25. doi: 10.1016/j.ijantimicag.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Pollack A. 27 February 2010. Deadly germs largely ignored by drug firms, p B4 New York Times, New York, NY. [Google Scholar]
- 56.Bullen J, Griffiths E, Rogers H, Ward G. 2000. Sepsis: the critical role of iron. Microbes Infect 2:409–415. doi: 10.1016/S1286-4579(00)00326-9. [DOI] [PubMed] [Google Scholar]
- 57.Bullen JJ. 1981. The significance of iron in infection. Rev Infect Dis 3:1127–1138. [DOI] [PubMed] [Google Scholar]
- 58.Michelini E, Cevenini L, Mezzanotte L, Coppa A, Roda A. 2010. Cell-based assays: fuelling drug discovery. Anal Bioanal Chem 398:227–238. doi: 10.1007/s00216-010-3933-z. [DOI] [PubMed] [Google Scholar]
- 59.Zhang JH, Chung TD, Oldenburg KR. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4:67–73. [DOI] [PubMed] [Google Scholar]
- 60.Kaback HR, Reeves JP, Short SA, Lombardi FJ. 1974. Mechanisms of active transport in isolated bacterial membrane vesicles. 18. The mechanism of action of carbonylcyanide m-chlorophenylhydrazone. Arch Biochem Biophys 160:215–222. [DOI] [PubMed] [Google Scholar]
- 61.Sverzhinsky A, Chung JW, Deme JC, Fabre L, Levey KT, Plesa M, Carter DM, Lypaczewski P, Coulton JW. 2015. Membrane protein complex ExbB4-ExbD1-TonB1 from Escherichia coli demonstrates conformational plasticity. J Bacteriol 197:1873–1885. doi: 10.1128/JB.00069-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Braun V. 1989. The structurally related exbB and tolQ genes are interchangeable in conferring TonB-dependent colicin, bacteriophage, and albomycin sensitivity. J Bacteriol 171:6387–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Braun V, Herrmann C. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol Microbiol 8:261–268. [DOI] [PubMed] [Google Scholar]
- 64.Pilsl H, Braun V. 1998. The Ton system can functionally replace the TolB protein in the uptake of mutated colicin U. FEMS Microbiol Lett 164:363–367. [DOI] [PubMed] [Google Scholar]
- 65.Nägeli KW. 1893. Über oligodynamische Erscheinungen in lebenden Zellen. Neue Denkschriften der allgemeinen Schweizerischen Gesellschaft für die gesamte Naturwissenschaft XXXIII. [Google Scholar]
- 66.Larsen RA, Deckert GE, Kastead KA, Devanathan S, Keller KL, Postle K. 2007. His(20) provides the sole functionally significant side chain in the essential TonB transmembrane domain. J Bacteriol 189:2825–2833. doi: 10.1128/JB.01925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chandler CJ, Segel IH. 1978. Mechanism of the antimicrobial action of pyrithione: effects on membrane transport, ATP levels, and protein synthesis. Antimicrob Agents Chemother 14:60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ermolayeva E, Sanders D. 1995. Mechanism of pyrithione-induced membrane depolarization in Neurospora crassa. Appl Environ Microbiol 61:3385–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reeder NL, Kaplan J, Xu J, Youngquist RS, Wallace J, Hu P, Juhlin KD, Schwartz JR, Grant RA, Fieno A, Nemeth S, Reichling T, Tiesman JP, Mills T, Steinke M, Wang SL, Saunders CW. 2011. Zinc pyrithione inhibits yeast growth through copper influx and inactivation of iron-sulfur proteins. Antimicrob Agents Chemother 55:5753–5760. doi: 10.1128/AAC.00724-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiong Q, Sun H, Li M. 2007. Zinc pyrithione-mediated activation of voltage-gated KCNQ potassium channels rescues epileptogenic mutants. Nat Chem Biol 3:287–296. doi: 10.1038/nchembio874. [DOI] [PubMed] [Google Scholar]
- 71.Yep A, McQuade T, Kirchoff P, Larsen M, Mobley HLT. 2014. Inhibitors of TonB function identified by a high-throughput screen for inhibitors of iron acquisition in uropathogenic Escherichia coli CFT073. mBio 5:e01089-13. doi: 10.1128/mBio.01089-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nikaido H, Rosenberg EY. 1981. Effect on solute size on diffusion rates through the transmembrane pores of the outer membrane of Escherichia coli. J Gen Physiol 77:121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mihara K, Tanabe T, Yamakawa Y, Funahashi T, Nakao H, Narimatsu S, Yamamoto S. 2004. Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiology 150:2587–2597. doi: 10.1099/mic.0.27141-0. [DOI] [PubMed] [Google Scholar]
- 74.Rutz JM, Abdullah T, Singh SP, Kalve VI, Klebba PE. 1991. Evolution of the ferric enterobactin receptor in gram-negative bacteria. J Bacteriol 173:5964–5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hashimoto-Gotoh T, Franklin FC, Nordheim A, Timmis KN. 1981. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization-defective pSC101-derived containment vectors. Gene 16:227–235. [DOI] [PubMed] [Google Scholar]
- 76.Klebba PE, McIntosh MA, Neilands JB. 1982. Kinetics of biosynthesis of iron-regulated membrane proteins in Escherichia coli. J Bacteriol 149:880–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma L, Kaserer W, Annamalai R, Scott DC, Jin B, Jiang X, Xiao Q, Maymani H, Massis LM, Ferreira LC, Newton SM, Klebba PE. 2007. Evidence of ball-and-chain transport of ferric enterobactin through FepA. J Biol Chem 282:397–406. doi: 10.1074/jbc.M605333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. [DOI] [PubMed] [Google Scholar]
- 80.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 81.Nairn BL, Lonergan ZR, Wang J, Braymer JJ, Zhang Y, Calcutt MW, Lisher JP, Gilston BA, Chazin WJ, de Crecy-Lagard V, Giedroc DP, Skaar EP. 2016. The response of Acinetobacter baumannii to zinc starvation. Cell Host Microbe 19:826–836. doi: 10.1016/j.chom.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luckey M, Pollack JR, Wayne R, Ames BN, Neilands JB. 1972. Iron uptake in Salmonella typhimurium: utilization of exogenous siderochromes as iron carriers. J Bacteriol 111:731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thulasiraman P, Newton SM, Xu J, Raymond KN, Mai C, Hall A, Montague MA, Klebba PE. 1998. Selectivity of ferric enterobactin binding and cooperativity of transport in gram-negative bacteria. J Bacteriol 180:6689–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guterman SK. 1971. Inhibition of colicin B by enterochelin. Biochem Biophys Res Commun 44:1149–1155. [DOI] [PubMed] [Google Scholar]
- 85.Guterman SK. 1973. Colicin B: mode of action and inhibition by enterochelin. J Bacteriol 114:1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Newton SM, Trinh V, Pi H, Klebba PE. 2010. Direct measurements of the outer membrane stage of ferric enterobactin transport: postuptake binding. J Biol Chem 285:17488–17497. doi: 10.1074/jbc.M109.100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wayne R, Neilands JB. 1975. Evidence for common binding sites for ferrichrome compounds and bacteriophage phi 80 in the cell envelope of Escherichia coli. J Bacteriol 121:497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.