ABSTRACT
Light sensing in chemotrophic bacteria has been relatively recently ascertained. In the human pathogen Acinetobacter baumannii, light modulates motility, biofilm formation, and virulence through the blue-light-sensing-using flavin (BLUF) photoreceptor BlsA. In addition, light can induce a reduction in susceptibility to certain antibiotics, such as minocycline and tigecycline, in a photoreceptor-independent manner. In this work, we identified new traits whose expression levels are modulated by light in this pathogen, which comprise not only important determinants related to pathogenicity and antibiotic resistance but also metabolic pathways, which represents a novel concept for chemotrophic bacteria. Indeed, the phenylacetic acid catabolic pathway and trehalose biosynthesis were modulated by light, responses that completely depend on BlsA. We further show that tolerance to some antibiotics and modulation of antioxidant enzyme levels are also influenced by light, likely contributing to bacterial persistence in adverse environments. Also, we present evidence indicating that surfactant production is modulated by light. Finally, the expression of whole pathways and gene clusters, such as genes involved in lipid metabolism and genes encoding components of the type VI secretion system, as well as efflux pumps related to antibiotic resistance, was differentially induced by light. Overall, our results indicate that light modulates global features of the A. baumannii lifestyle.
IMPORTANCE The discovery that nonphototrophic bacteria respond to light constituted a novel concept in microbiology. In this context, we demonstrated that light could modulate aspects related to bacterial virulence, persistence, and resistance to antibiotics in the human pathogen Acinetobacter baumannii. In this work, we present the novel finding that light directly regulates metabolism in this chemotrophic bacterium. Insights into the mechanism show the involvement of the photoreceptor BlsA. In addition, tolerance to antibiotics and catalase levels are also influenced by light, likely contributing to bacterial persistence in adverse environments, as is the expression of the type VI secretion system and efflux pumps. Overall, a profound influence of light on the lifestyle of A. baumannii is suggested to occur.
KEYWORDS: Acinetobacter baumannii, light, metabolism
INTRODUCTION
Acinetobacter baumannii is a threatening human pathogen considered to be the paradigm of multidrug resistance (MDR). A key component in its success as a pathogen is not only its outstanding capability to acquire resistance but also its peculiar ability to reside and persist in the hospital environment, which contributes to colonization, infection, and dissemination of the MDR clones (1–3). This situation, in addition to the scarce release of new antibiotics in the last years (4, 5), results in limited therapeutic options left to control infections caused by them (1, 6). Alternatives to control the microorganisms are urgently required and currently rely on the reestablishment and usage optimization of old antimicrobials and combinatorial regimes. Gaining detailed knowledge on the bacterial pathophysiology could provide other resources for control strategies. We have recently recognized a new aspect of A. baumannii physiology: its ability to perceive and respond to light-modulating aspects related to persistence in the environment, virulence against Candida albicans (7), and resistance to antibiotics (8). Indeed, we have previously shown that blue light inhibited motility and the formation of biofilms and pellicles in A. baumannii and enhanced the ability of the bacteria to kill the filamentous form of the eukaryotic fungus Candida albicans at 24°C (7). By means of biophysics and genetic studies, we have shown that these responses to light depend on BlsA, the only “traditional” photoreceptor encoded in its genome (7). These responses occurred at environmental temperatures, such as 24°C, but not at 37°C, a condition under which blsA expression is significantly reduced (7). It is therefore clear that light could have a direct effect on persistence of the bacteria in the environment, for example, by directly modulating virulence and/or competition against other microorganisms sharing a habitat. On the contrary, light may not play a role in systemic infections in humans but rather might be relevant in the pathogenesis of surface-exposed wound infections, considering the exposure of the bacteria to light and the relatively lower temperatures of skin wounds (7).
The response to light was found not to be restricted to A. baumannii but was widespread within the genus Acinetobacter, in many species of which photoregulation occurred both at 24 and 37°C, probably as a result of the presence of additional photoreceptors (9). Finally, we have presented evidence indicating that light modulates susceptibility to minocycline and tigecycline antibiotics in Acinetobacter, as well as other clinically relevant pathogens, by a BlsA-independent mechanism that most likely involves induction of the expression of resistance genes by singlet oxygen (8, 10).
In this work, we have identified other novel bacterial physiological processes modulated by light related to bacterial physiology and virulence, revealed by a whole transcriptome approach, followed by molecular biology, biochemical, and physiological validations. Indeed, we have recognized that light directly modulates metabolic pathways, such as the phenylacetic acid (PAA) catabolic pathway and biosynthesis of trehalose, which are involved in the pathogenesis of A. baumannii (11) and Xanthomonas citri (12), respectively. By means of genetic, physiological, and biochemical studies, we demonstrate that these responses depend on BlsA. In contrast, we show that BlsA performs a fine tuning of catalase activity levels in response to light. We further provide evidence indicating that light produces increases in surfactant production and tolerance to fluoroquinolones, which could directly contribute to persistence under adverse conditions. Finally, we show that light modulates the expression of the type VI secretion system (T6SS), lipid metabolism cluster, and efflux pump genes. The overall evidence indicates that light signaling is a key stimulus in the lifestyle of the human pathogen A. baumannii.
RESULTS
A. baumannii transcriptome adjustments in response to light.
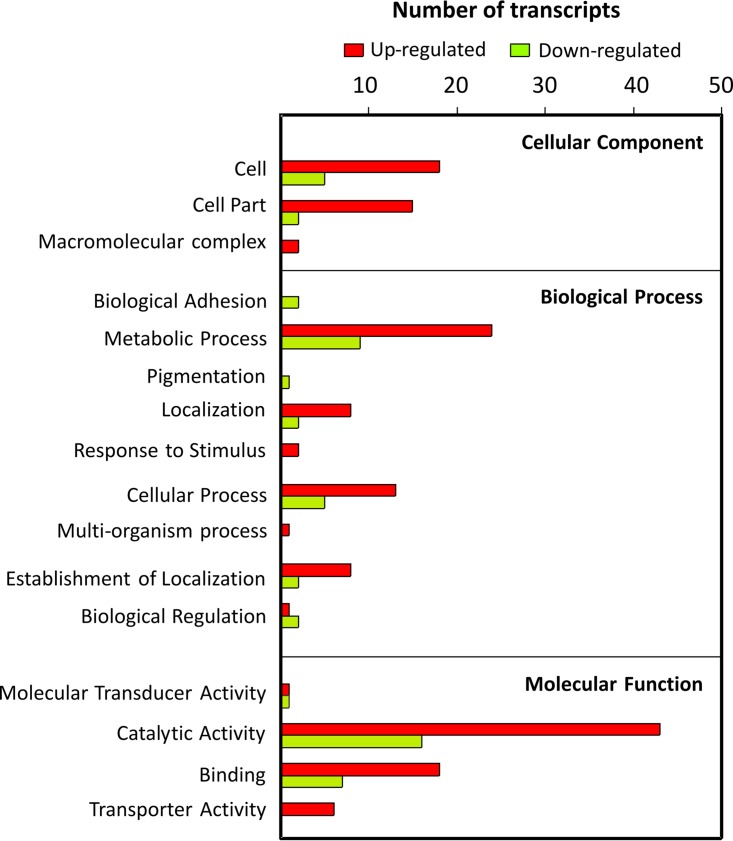
The RNA purified from A. baumannii ATCC 19606 cultures grown at 24°C under blue light or in the dark was analyzed by RNA sequencing (RNA-seq) to determine the gene expression profiles. We found 225 genes to be differentially expressed (see Table S1 in the supplemental material) (fold change, >2; P < 0.001) under light and dark conditions. One hundred ninety-three genes were upregulated by light, while 32 genes were downregulated, and the genes were dispersed across a wide variety of functions, according to gene ontology terms (Fig. 1). It is noteworthy that the majority of the up- and downregulated genes fell into the catalytic activity category, followed by metabolic process categories, indicating a significant change in the metabolism of this bacterium in response to light (Table S1 and Fig. 1).
FIG 1.
Graph representing the number of genes differentially expressed under light and dark conditions for A. baumannii ATCC 19606, organized per COG category. Green and red bars represent genes repressed or induced by light, respectively.
Complete pathways are modulated by light.
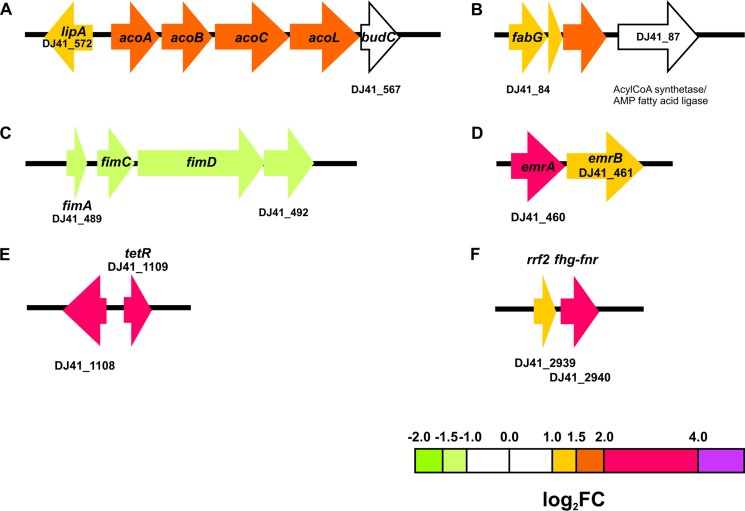
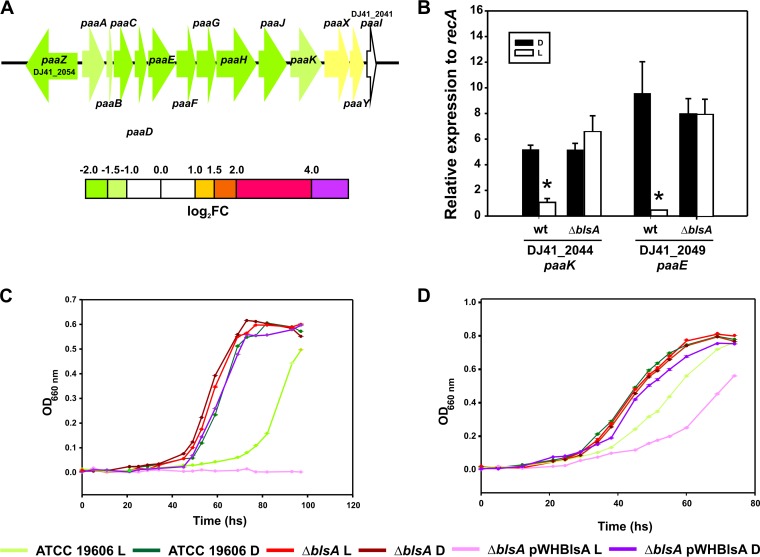
Analyses of differential expression data indicate that light modulates a discrete number of genes, many of which, interestingly, are arranged as gene clusters (see Fig. 2 to 4 and 6 to 9). In fact, complete gene pathways, but not the flanking genes, were found to be differentially expressed in the presence of light, strongly reinforcing the notion that these particular routes are modulated by this stimulus. Some of these correspond to crowded gene clusters, such as those coding for the phenylacetic acid catabolic pathway (11 genes) (see Fig. 3A), lipid metabolism (10 genes) (see Fig. 8B), and the whole type VI secretion system (T6SS; at least 18 contiguous genes) (see Fig. 9C). Moreover, pathways containing fewer genes, such as the acetoin utilization pathway (Fig. 2A) and a cluster of genes putatively involved in lipid metabolism, were induced (Fig. 2B), while a cluster of genes coding for fimbriae (Fig. 2C) were repressed by light. emrA and emrB, which code for components of an efflux pump of the resistance-nodulation-division (RND) and major facilitator superfamilies, as well as otsA and otsB, which are involved in trehalose biosynthesis, were also found to be modulated by light (Fig. 2D; see also Fig. 4A). Moreover, genes involved in iron recycling and its regulator (Fig. 2E), flavohemoglobin genes (DJ41_2939 and DJ41_2940) (Fig. 2F), a cluster harboring a catalase gene (see Fig. 6A), and dnpA, in the close proximity of blsA (see Fig. 7A), are also upregulated by light. Induction of the expression of emrA and tetR, as well as other representative genes of particular pathways (see below), by light was confirmed by reverse transcription-quantitative PCR (qRT-PCR) experiments (see Fig. S1).
FIG 2.
Schematic representation of some gene clusters differentially expressed under blue light. Each arrow represents an individual gene to scale. The colors of the arrows represent relative degrees of differential gene expression under light versus darkness based on the color scale at the bottom of the figure. The genomic organization of the different gene clusters was determined using databases for A. baumannii ATCC 19606 present in Ensembl Bacteria (47) and in the MAGE platform (48; http://www.genoscope.cns.fr/agc/microscope/home/). (A) Acetoin degradation pathway. Gene products are as follows: acoA, 2,6-dichlorophenol indophenol oxidoreductase subunit alpha; lipA, lipoyl synthase; acoB, 2,6-dichlorophenol indophenol oxidoreductase subunit beta; acoC, dihydrolipoamide acetyltransferase; acoL, dihydrolipoamide dehydrogenase; budC, diacetyl reductase [(S)-acetoin forming]; DJ41_566, putative acetoin reductase/2,3-butanediol dehydrogenase; and acoL, dihydrolipoyl dehydrogenase. (B) Genes involved in lipid metabolism. (C) Fimbrial biosynthetic cluster. Gene products are as follows: fimA, fimbrial subunit; fimC, P pilus assembly protein chaperone PapD; and fimD, outer membrane usher protein. (D) Efflux pump EmrA/B-encoding genes. (E) Genes for iron recycling from heme protein. (F) Flavohemoglobin-fumarate and nitrate reduction (FNR) genes and transcriptional regulator. Gene locus tags according to the A. baumannii ATCC 19606 database used as a reference are indicated below or within each gene (only the first and last are noted if they are correlative). Gene locus tag annotations according to the GenBank database are indicated in Table S1.
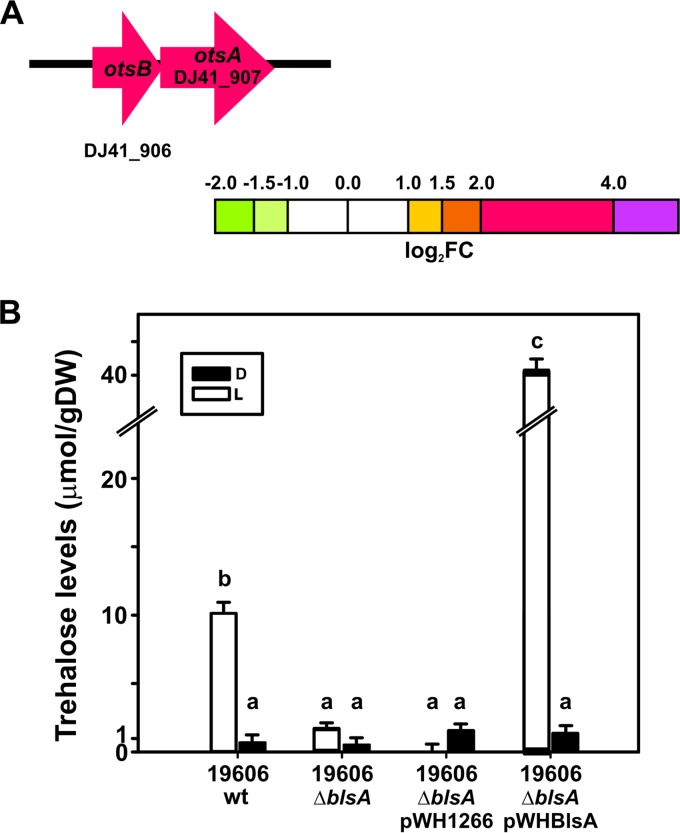
FIG 4.
Trehalose levels are increased under blue light. (A) Schematic representation of trehalose biosynthesis genes induced by light. Each arrow represents an individual gene to scale. The colors of the arrows represent relative degrees of differential gene expression under light versus darkness based on the color scale at the bottom. See the legend to Fig. 2 for further details. Gene products are as follows: otsA, trehalose-6-phosphate synthase, and otsB, trehalose-6-phosphate phosphatase. (B) Trehalose content was quantified in the indicated strains grown to the early exponential phase in LB medium under blue light (L) or in the dark (D). Bars indicate the micromoles of trehalose per gram (dry weight) (gDW) of bacterial pellet. Different letters indicate significant differences between treatments, as determined by analysis of variance (ANOVA) and Duncan multiple comparison test (P < 0.01). Data shown are the average ± SD for 4 biological replicates.
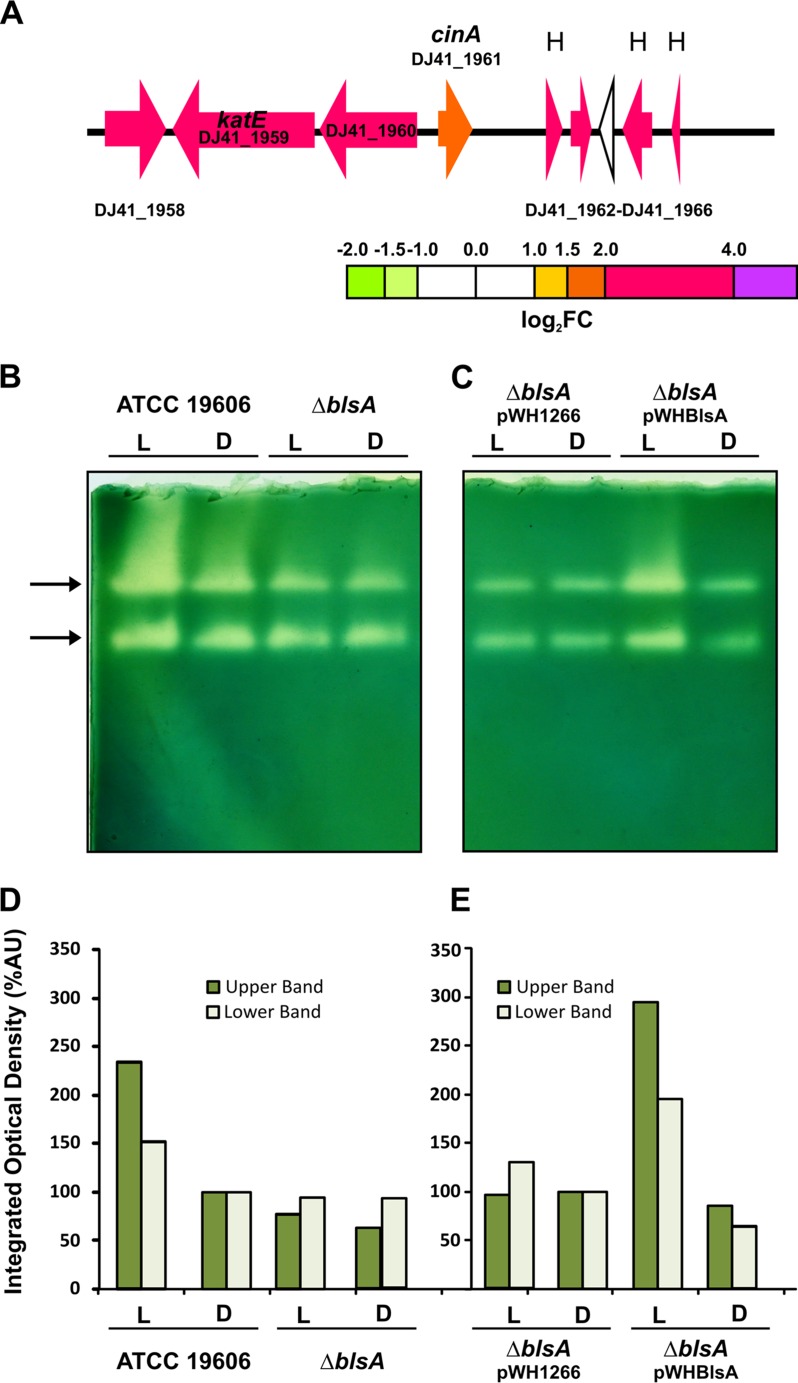
FIG 6.
Catalase activity is increased under blue light. (A) Schematic representation of the genomic region containing the catalase gene induced by light. Each arrow represents an individual gene to scale. The colors of the arrows represent relative degrees of differential gene expression under light versus darkness based on the color scale at the bottom. See the legend to Fig. 2 for further details. H, hypothetical protein. (B and C) Levels of catalase activities determined in gel for A. baumannii wild-type and derivative strains used in this study cultured in LB under blue light (L) or in the dark (D) at 24°C. Equal amounts of proteins corresponding to 30 μg of total soluble protein were resolved by nondenaturing PAGE and assayed for catalase activity according to established procedures (see Materials and Methods). Bands of catalase activities are indicated by arrows. One gel representative of three different experiments is shown. (D and E) Densitometric determinations for each sample are shown below the activity gels. Dark and light green bars represent the quantification of catalase activity for the slower and faster electrophoretic mobility bands, respectively. Values were normalized with respect to the intensity levels for the wild type in the dark or the ΔblsA mutant harboring the empty pWH1266 plasmid in the dark, which were assigned the arbitrary value of 100. %AU, percent arbitrary units.
FIG 9.
Light induces the type VI secretion system (T6SS). (A) Schematic representation of the T6SS cluster and the genes induced by light. Each arrow represents an individual gene to scale. The colors of the arrows represent relative degrees of differential gene expression under light versus darkness based on the color scale at the bottom. See the legend to Fig. 2 for further details. asa genes encode five Acinetobacter T6SS-associated proteins and have been identified only in A. baumannii T6SS gene clusters. (B) Quantification of the expression levels of tssM, tssD, and tssC genes under blue light (L) or in the dark (D) in A. baumannii 19606, as determined by real-time qRT-PCR assays. The means of the results obtained using three independent cultures grown under blue light or in the dark are shown. The y axis refers to the fold difference of a particular transcript level relative to the CT values corresponding to recA; the SD is shown. Asterisks indicate significant differences among light and dark treatments, as indicated by t test (P < 0.05).
FIG 3.
Light inhibits growth of A. baumannii ATCC 19606 in PAA and Phe. (A) Schematic representation of the PAA catabolic pathway and genes repressed by light. Each arrow represents an individual gene to scale. The colors of the arrows represent relative degrees of differential gene expression under light versus darkness based on the color scale at the bottom. See the legend to Fig. 2 for further details. (B) Quantification of the expression levels of paaK and paaE genes under blue light (L) or in the dark (D) in the wild-type (wt) A. baumannii 19606 and in ΔblsA mutant strains, as determined by real-time qRT-PCR assays. The means of the results obtained using three independent cultures grown under blue light or in the dark are shown. The y axis refers to the fold difference of a particular transcript level relative to the threshold cycle (CT) values corresponding to recA; the standard deviation (SD) is shown. Asterisks indicate significant differences among light and dark treatments, as indicated by t test (P < 0.05). (C) Growth curves of A. baumannii wild-type and derivative strains used in this study in M9 liquid minimal medium supplemented with 10 mM PAA as a sole carbon source and incubated stagnantly at 24°C under blue light or in the dark. Growth was measured by determining the optical density at 660 nm. (D) Growth curves of A. baumannii wild-type and derivative strains used in this study in M9 liquid minimal medium supplemented with 10 mM Phe as a sole carbon source and incubated stagnantly at 24°C under blue light or in the dark. Growth was measured by determining the optical density at 660 nm. Shown are representative results obtained from three independent experiments. Below are indicated the colors used to represent each curve for each strain used in this study.
FIG 8.
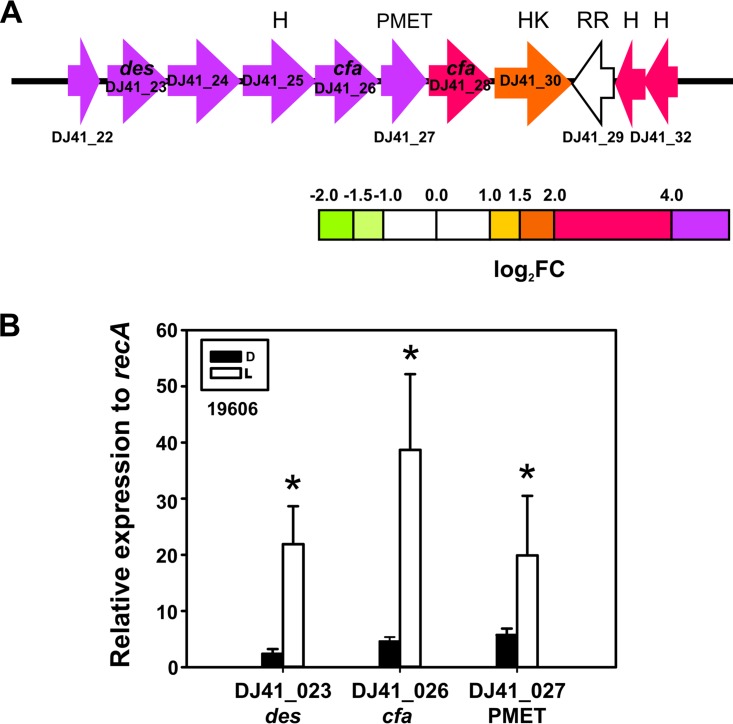
Light induces a gene cluster coding for lipid modification enzyme cluster. (A) Schematic representation of the lipid modification enzyme cluster and the genes induced by light. Each arrow represents an individual gene to scale. The colors of the arrows represent relative degrees of differential gene expression under light versus darkness based on the color scale at the bottom. See the legend to Fig. 2 for further details. Gene products are as follows: des, estearoyl CoA 9-desaturase; cfa, cyclopropane fatty-acyl synthase; PMET, protein with phospholipid methyltransferase domain; HK, histidine kinase putative two-component system sensor protein (ColS-like); RR, putative effector domain in response regulator (ColR-like); and H, hypothetical protein. (B) Quantification of the expression levels of the indicated genes under blue light (L) or in the dark (D) in A. baumannii 19606, as determined by real-time qRT-PCR assays. The means of the results obtained using three independent cultures grown under blue light or in the dark are shown. The y axis refers to the fold difference of a particular transcript level relative to the CT values corresponding to recA; the SD is shown. Asterisks indicate significant differences among light and dark treatments, as indicated by t test (P < 0.05).
FIG 7.
Light stimulates tolerance to ofloxacin. (A) Schematic representation of the genomic region containing a homolog to the dnpA gene induced by light. Each arrow represents an individual gene to scale. The colors of the arrows represent relative degrees of differential gene expression in light versus darkness based on the color scale at the bottom. See the legend to Fig. 2 for further details. (B) Relative persister fractions of cells exposed to blue light or incubated in the dark. Persister fraction is defined as the number of surviving cells after treatment with ofloxacin divided by the number of cells after control treatment, under blue light (L) or in the dark (D). Data shown are the average ± SEM for 3 biological replicates.
Finally, among highly differentially expressed genes, there were some that were solitary, i.e., that could not be incorporated into any pathway or cluster of genes (Table 1). Examples of these genes are DJ41_2234 and DJ41_2775, which code for RhtB proteins and may play a role in the density-dependent behavior of cell populations. Other examples include genes encoding a sulfate transporter family protein, a glutathione S-transferase, a porin, rubredoxin, and hypothetical proteins (Table 1).
TABLE 1.
Solitary genes differentially expressed in A. baumannii ATCC 19606 under blue light or in the dark
| Locus tag | Product or gene name | Description | Fold change (light vs dark) |
|---|---|---|---|
| DJ41_2234 | RhtB | lysE-type translocator family protein; threonine/homoserine/homoserine lactone efflux protein | 17.09 |
| DJ41_2775 | RhtB | lysE-type translocator family protein; threonine/homoserine/homoserine lactone efflux protein. | 5.85 |
| DJ41_817 | xapX | XapX domain protein | 10.59 |
| DJ41_3653 | Hypothetical protein | 6.44 | |
| DJ41_2560 | Hypothetical protein | 6.19 | |
| DJ41_2800 | Sulfate transporter family protein | 6.07 | |
| DJ41_1242 | Glutathione S-transferase | 5.00 | |
| DJ41_2155 | Hypothetical protein | 4.62 | |
| DJ41_157 | Hypothetical protein | 4.53 | |
| DJ41_2541 | rubA | Rubredoxin; participates in electron transfer in biological systems; nonheme iron binding domains containing an [Fe(SCys)4] center | 4.03 |
| DJ41_591 | Porin | 0.24 | |
| DJ41_502 | Hypothetical | 4.94 |
Growth on PAA degradation pathway substrates is inhibited by light.
Our results show that the genes coding for enzymes of the PAA catabolic pathway are repressed under blue light, from paaA (DJ41_2053) to paaK (DJ41_2044) genes, which are ordered in the same direction in an organization congruent with an operon (Fig. 3A and Table S1). paaZ (DJ41_2054), which is located in a divergent orientation, is also repressed (Fig. 3A), while the regulatory genes paaX and paaY are very slightly modulated (Fig. 3A). qRT-PCR experiments performed using wild-type A. baumannii confirmed that light represses the expression of genes of this pathway, such as paaK (DJ41_2044) and paaE (DJ41_2049) (Fig. 3B). Similar experiments using the isogenic ΔblsA mutant, in which the blsA coding sequence is interrupted by a kanamycin (Kn) cassette, show that the expression levels of these genes under blue light are restored to dark levels (Fig. 3B), suggesting the involvement of the BlsA photoreceptor in the response.
Sequence analyses show that synteny is conserved in other strains of A. baumannii, such as AB0057, 6013113, AB307-0294, AYE, 6013150, BJAB07104, and 6014059 (Fig. S2). The complete pathway seems to be absent in SDF and AB900 strains, while the organization of genes seems to be different for AB058, AB059, and AB056 (Fig. S2). The synteny is also conserved for Acinetobacter sp. strains SH024 and RUH2624 (not shown).
In agreement with transcriptome predictions, growth of A. baumannii wild-type cells in liquid minimal medium using 10 mM PAA as the sole carbon source at 24°C was severely compromised under blue light with respect to dark conditions (Fig. 3C and S3A). The ΔblsA mutant and the ΔblsA mutant carrying the empty pWH1266 plasmid did not show differences between light and dark (Fig. 3C and S3A, respectively) and behaved in the same manner as the wild type in the dark, indicating that light modulation of the PAA catabolic pathway depends on the BlsA photoreceptor. Finally, the genetic complementation of the ΔblsA mutant with pWHBLSA, which expresses a wild-type copy of BlsA, restored the differential growth in response to illumination with respect to dark conditions (Fig. 3C). Indeed, in this case, the abolition of growth was complete under blue light, most likely due to the higher copy number of the blsA allele in this plasmid-complemented derivative of the ΔblsA mutant. It should be noted that there was no effect of light on the viability of the cells under these conditions, as the growth curves were similar under blue light or in the dark for the ΔblsA mutant in minimal medium (Fig. 3C and S3A and B), as well as for the different strains in LB (Fig. S3D). Figures 3D and S3B show growth of the A. baumannii wild-type and derivative strains used in this work in liquid minimal medium with 10 mM phenylalanine (Phe) as the sole carbon source at 24°C. It should be noted that in bacteria, Phe is metabolized via PAA (13) and therefore represents another substrate of the PAA pathway. In this case, the results were similar to those obtained with PAA, despite the less-pronounced effect (Fig. 3D and S3B), further supporting the notion that the PAA catabolic pathway is modulated by light. Growth curves determined using 5 mM Phe or PAA (Fig. S3C) produced results consistent with those obtained at 10 mM. Finally, data obtained from growth curves in liquid medium were consistent with the patterns observed on solid medium both for PAA and Phe (Fig. S3E and F).
Blue light stimulates trehalose biosynthesis.
Our RNA-seq experiments revealed that light significantly induces the expression of otsA and otsB (Fig. 4A), which are involved in the biosynthesis of the disaccharide trehalose. The product of otsA, trehalose-6-phosphate synthase (TPS), is a glycosyltransferase that catalyzes the synthesis of alpha,alpha-1,1-trehalose-6-phosphate from glucose-6-phosphate using a UDP-glucose donor (14). The product of otsB, a trehalose-6-phosphate phosphatase, catalyzes the dephosphorylation of trehalose-6-phosphate to trehalose and orthophosphate (14).
In agreement with RNA-seq experiments, biosynthesis of trehalose is induced by light in A. baumannii wild-type cells, whose levels are very low in the dark (Fig. 4B). This response is completely dependent on BlsA, given that the mutant deficient in this photoreceptor loses photoregulation and behaves as in the dark. Again, the genetic complementation of the ΔblsA mutant with pWHBLSA restored the differential biosynthesis of trehalose in response to blue light, interestingly to much higher levels than those in the wild type (Fig. 4B), which most likely is a result of the extra BlsA doses.
Blue light stimulates extracellular surfactant production.
Recent reports indicate that members of the Acinetobacter genus can produce low-molecular-weight (LMW) surfactants from hydrophobic substrates (15, 16). We next analyzed the effect of blue light in surfactant production by acid treatment and solvent extraction of the extracellular media of A. baumannii wild-type cells grown in ethanol medium under blue light or in the dark at 24°C. Immediately after the addition of the organic phase, the supernatant of light-exposed cultures became crowded with bubbles, consistent with emulsifying activity, contrary to what happened in the supernatants of cultures incubated in the dark, which were much more translucent (see Fig. S4). The amount of synthesized extracellular surfactants (grams per liter) was determined gravimetrically after extraction from the supernatants, followed by evaporation, and normalized relative to the optical density values reached by the corresponding cells. Quantification of purified extracellular surfactant showed that light results in significant increases in its production by approximately 60% with respect to values obtained in the dark (Fig. 5).
FIG 5.

Extracellular surfactant production is increased under blue light. (A) Biosurfactants were extracted as described in Materials and Methods and quantified gravimetrically. The presented data are the milligrams of biosurfactant per liter of initial culture normalized relative to the OD reached by the cells under blue light or in the dark. Data shown are the average ± standard error of the mean (SEM) for 3 biological replicates. An asterisk indicates significant difference between light (L) and dark (D) treatments, as indicated by t test (P < 0.05).
Blue light induces catalase activity.
Another cluster of genes upregulated by light includes katE, which encodes catalase (Fig. 6A). Two catalase activity bands were detected in wild-type cells, and exposure to blue light appears to stimulate this activity (Fig. 6B and D). These two bands probably correspond to KatE and KatG, given that among the four catalase homologs described in A. baumannii, there is only evidence for the expression of these two (17). However, no clear assignment is available in the literature. In contrast, the ΔblsA mutant showed no differences in catalase activities between light and dark, displaying basal activity levels similar to those observed for the wild type in the dark (Fig. 6B and D) and therefore indicating that this response to light depends on BlsA. Moreover, genetic complementation of the ΔblsA mutant with pWHBLSA resulted in increased catalase activities under blue light with respect to the dark, while the ΔblsA mutant carrying the empty pWH1266 plasmid behaved as the ΔblsA mutant, without differences between light and dark (Fig. 6C and E).
Blue light stimulates tolerance to fluoroquinolones.
Analyses of differentially expressed genes showed that some genes in close proximity to blsA are upregulated by light (Fig. 7A), in particular, a gene coding for a short putative membrane protein oriented tail to tail with respect to blsA (DJ41_1947), as well as a gene encoding an OB-fold protein (BOF) (Fig. 7A). The participation of BOF proteins in biofilm formation, motility, and virulence in response to quorum-sensing and hormonal signals (10) has been reported in some organisms. Moreover, a cluster of genes coding for a de-N-acetylase of the phosphatidylinositol-glycan biosynthesis class L protein (PIG-L) superfamily (DJ41_1943), followed by a methyltransferase gene (DJ41_1944) and a glycosyltransferase gene (DJ41_1955, which changes only 0.5-fold with respect to dark conditions), also located in the vicinity of blsA, are upregulated under blue light. It has been shown that a de-N-acetylase belonging to the PIG-L superfamily (encoded by the dnpA gene) is involved in noninherited fluoroquinolone tolerance in Pseudomonas aeruginosa (18). This phenomenon refers to the presence of antibiotic-insensitive persister cells, i.e., phenotypic variants of the wild type that are transiently capable of surviving prolonged periods of antibiotic treatment, without having developed or acquired resistance mechanisms to the drug (18). De-N-acetylases of the PIG-L superfamily typically display low overall sequence identity (∼25%) but share a catalytic site containing a number of conserved residues that are essential for enzymatic activity (18). The sequence of the putative enzyme present in A. baumannii ATCC 19606, encoded by DJ41_1943 and shown in this work to be induced by light (Fig. 7A), contains those conserved residues in the active site (18).
We next analyzed whether tolerance to fluoroquinolones was affected by light by comparing the persister fraction of A. baumannii wild-type cells used in this study under blue light or in the dark. Briefly, A. baumannii cells grown in LB at 24°C to exponential phase in the dark or under blue light were treated for 10 h with 3 times the MIC of ofloxacin or alternatively with water as a control. The number of CFU was then determined in each case. The persister fraction is defined as the number of surviving cells after treatment with ofloxacin divided by the number of cells after control treatment (18). Our results show that light effectively increases the persister fraction of this strain by about 80% compared to values obtained for cultures kept in the dark (Fig. 7B). It should be noted that A. baumannii ATCC 19606 displays the same MIC of ofloxacin under blue light as in the dark, at 0.315 μg/ml, excluding the involvement of fluoroquinolone resistance mechanisms. We further performed an ofloxacin stability assay in order to eliminate the possibility of degradation of the drug under the conditions used in this work (see Table S3 for details).
Genes coding for lipid modification enzymes are upregulated by light.
Our results show that a set of genes putatively coding for lipid modification enzymes are upregulated by light. In particular, genes DJ41_22 to DJ41_28, which are organized in the same direction in the genomic locus and likely constitute an operon, are all induced by light, according to RNA-seq experiments (Fig. 8A and Table S1) and as verified for DJ41_23, DJ41_26, and DJ41_27 by qRT-PCR (Fig. 8B). Sequence analyses showed that DJ41_22 codes for a lipocalin-like domain protein; DJ41_23 codes for a predicted stearoyl coenzyme A (stearoyl-CoA) 9 desaturase, which produces the monounsaturated fatty acid oleic acid (cis-9 octadecenoic acid) from the saturated fatty acid estearic acid; DJ41_24 codes for a dehydrogenase; DJ41_25 codes for a hypothetical protein containing an unidentified DUF1365 domain; DJ41_26 (cfa) is annotated as encoding a mycolic acid cyclopropane fatty acid synthetase family protein; DJ41_27 codes for a putative phospholipid methyltransferase (PMET), which could synthesize phosphatidylcholine from phosphatidylethanolamine; and DJ41_28 codes for another predicted cyclopropane fatty-acyl-phospholipid synthase and related methyltransferases (Fig. 8A). cfa genes have been shown to transfer a methylene group from S-adenosyl-l-methionine to the cis double bond of an unsaturated fatty acid chain, resulting in the replacement of the double bond with a methylene bridge (19). DJ41_30, which codes for a histidine kinase A (ColS-like) (Fig. 8A), follows in the genomic locus and is also upregulated, although to a lesser extent (Fig. 8A). In the opposite direction are located three genes, DJ41_29, DJ41_31, and DJ41_32, again oriented consecutively, suggesting a putative operon conformation (Fig. 8A). DJ41_31 and DJ41_32 code for hypothetical proteins and are highly upregulated (Fig. 8A). It should be noted that DJ41_29, which is not modulated by light, codes for a putative effector domain in a response regulator (ColR-like) arranged tail to tail with respect to the histidine kinase DJ41_30 (Fig. 8A). That DJ41_29 and DJ41_30 constitute a two-component system regulating these enzymes is a likely possibility that needs further elucidation. In summary, genes coding for putative enzymes that generate insaturations, cyclopropanation, and other phospholipid modifications, such as transfer of methyl groups, as well as a putative regulatory two-component system that could modulate their expression, are all contained in this gene cluster.
The T6SS is induced under blue light.
The T6SS is a class of macromolecular secretion machines that could be regarded as a potent weapon used by many Gram-negative bacteria to kill competitors (20). It has been shown that certain strains of A. baumannii produce a functional T6SS, which has been found to be implicated in virulence and host colonization in a strain-specific manner (21–24).
Our transcriptome analyses indicate that the complete T6SS gene cluster and also other genes located outside the cluster and encoding putative VgrG proteins, which are often secreted via the T6SS, are upregulated by light in A. baumannii (Fig. 9). Among the genes induced to the highest extent by light are DJ41_2093, DJ41_2094, DJ41_2095, DJ41_2096, and DJ41_2097 (Fig. 9A and Table S1), which include genes at the 5′ end of the gene cluster. qRT-PCR experiments confirmed RNA-seq results by showing that light significantly induces the expression of some representative genes, such as DJ41_2089, DJ41_2094, and DJ41_2095 (Fig. 9B).
DISCUSSION
In this work, we identified novel light-modulated physiological traits related to bacterial physiology and virulence in A. baumannii. Most interestingly, we show that light directly modulates metabolic pathways, such as the PAA catabolic pathway, which is repressed by blue light in strain ATCC 19606 in a BlsA-dependent manner. The paa genes encode proteins responsible for the metabolism of aromatic compounds and have been linked to virulence in A. baumannii (11, 25) and other human pathogens (26). It would therefore be expected that repression of the PAA pathway by light would contribute to reduce virulence. The results from our group show that light significantly stimulates virulence of A. baumannii ATCC 17978 against filaments of C. albicans (121-fold decrease in filament survival in light versus dark conditions), while this response is less pronounced in the case of ATCC 19606 (only 4-fold relative to the response displayed by ATCC 17978) (7). These strain-specific differences in the final virulence outcome in response to light probably reflect the balance between the extent of induction and repression of the different pathways related to virulence/competition (PAA catabolic pathway, T6SS, etc.) regulated by this stimulus in each strain. Indeed, the T6SS was found to be induced by light in ATCC 19606, which is in agreement with stimulation of virulence and or competition by light mentioned above (7). It is interesting to note that the PAA catabolic pathway has been recently shown to be controlled by the Gac system in A. baumannii ATCC 17978 (11). The GacA and BlsA regulatory systems could therefore be interconnected, or each could function in a strain-specific way. Heterogeneity between different strains of A. baumannii is common, and differential light perception has been already noted for different strains of this organism (7). Furthermore, the ability to use aromatic compounds is important in bioremediation, and in this work, we show that light should be regarded as a new variable that could be optimized for this purpose.
Another novel trait related to metabolism modulated by light identified in this work is the biosynthesis of trehalose, which is also dependent on BlsA. Regarding the physiological importance of this sugar, most bacteria use trehalose as a general osmo- and thermoprotectant (27). Also, it has been reported that trehalose is involved in colonization and pathogenesis of phytopathogens (12, 28, 29). In addition, mycobacteria produce trehalose-containing glycolipids which are virulence factors (27). Trehalolipids have gained increased interest for their potential applications in a number of fields due to their ability to lower interfacial tension and increase the pseudosolubility of hydrophobic compounds (14). In this sense, it was proposed (15, 30) that enhancement of surfactant production could be associated with an increase in trehalolipid synthesis, which correlates with increases in trehalose phosphate synthase activity as well as other enzymes in Rhodococcus erythropolis EK-1 and possibly in Acinetobacter calcoaceticus K-4 (15, 30). In this work, we show that light also induces the production of surfactants, and we raise the possibility that increases in trehalose synthesis and surfactant production are related. Further research will contribute to unveil the role(s) that trehalose plays in A. baumannii. Since trehalose finds industrial and pharmaceutical applications, enhanced accumulation of trehalose in bacteria seems advantageous for commercial production (14). In this work, we have identified not only that light is a variable that could be optimized for enhanced production of trehalose but also that strains engineered to overproduce BlsA result in elevated levels of this compound. The enhanced production of surfactants observed under blue light is interesting considering our previous findings indicating the inhibition of biofilm formation in ATCC 17978 under this condition. Indeed, biosurfactants of microbial origin have been reported to have antiadhesive and biofilm-disrupting abilities (31–34); therefore, these two responses observed under blue light in A. baumannii might be related.
We further show that another novel trait modulated by light is catalase production, which increases in the presence of blue light in a BlsA-dependent manner. Given that light absorption by photosensitizer molecules present in bacterial cells, such as flavins and tetrapyrroles, can generate reactive species, catalase expression could be induced to counteract this effect (35). Interestingly, catalases are present in the dark, in accordance with the microorganism being typified as catalase positive, but their expression is adjusted under blue light by BlsA to achieve extra levels that could contribute to effectively managing possible reactive oxygen species (ROS) produced under light.
It is also interesting that an efflux pump of the type EmrAB is upregulated by light in the context of the reduced susceptibility to minocycline (MIN) and tigecycline (TIG) we have observed under blue light (8). EmrAB can function as components of tripartite systems together with TolC, for example (e.g., EmrAB-TolC), and in this context, it has been recently shown that the TolC homolog AbuO is involved in tigecycline resistance in Acinetobacter baumannii (36). EmrAB is therefore an interesting candidate to explain the reduction in susceptibility to these antibiotics in response to blue light, in addition to induction of adeABC genes previously described under this condition (8). Moreover, in this context, we present evidence here indicating that light stimulates tolerance to the fluoroquinolone ofloxacin, as is suggested by the induction of the expression of a de-N-acetylase of the PIG family by light. These aspects could have a profound effect on the success of the microorganism in the hospital environment.
Finally, in this work, we identified a novel arrangement of genes coding for lipid-modifying enzymes not found in other organisms and not reported before for Acinetobacter, which was found to be upregulated by light. Interestingly, the genes coding for cyclopropane fatty acid (CFA) and PMET enzymes, DJ41_26, DJ41_27, and DJ41_28, showed approximately 80, 70, and 80% identity, respectively, with genes present in Bacillus mycoides, therefore suggesting acquisition of these genes by horizontal transfer events from Gram-positive bacteria. Cyclopropanation plays a role in the pathogenesis of Mycobacterium tuberculosis, and pathogenic Escherichia coli strains have higher CFA contents and are more resistant to acid shock than nonpathogenic strains (19). Interestingly, this set of genes is well conserved in synteny in many strains of A. baumannii and in other members of the Acinetobacter genus. Moreover, many members of the pathway are present in Achromobacter xylosoxidans, Alteromonas macleodii, Agrobacterium tumefaciens, Rhizobium leguminosarum, Mycobacterium tuberculosis, Mycobacterium bovis, and Mycobacterium canettii, among others. In A. baumannii, the presence of CFA synthase and desaturase genes in genomic islands in epidemic strains has been reported (37).
Overall, in this work, we disclosed a plethora of novel physiological traits whose expression levels are modulated by light in A. baumannii, many of which could be demonstrated in this work to the level of phenotype. Many of these traits show that light modulates metabolic pathways, such as the PAA catabolic pathway, trehalose biosynthesis, and acetoin degradation, and also clusters related to lipid metabolism, competition, tolerance, and resistance to antibiotics. In this way, it is clear that in this bacterium, light influences diverse processes determining its lifestyle. Overall, these findings open a new perspective regarding the lifestyle of A. baumannii, highlighting the importance of light perception in this microorganism and challenging future research for further confirmation of the other predicted traits. The lipid metabolism cluster falls in this category, in which other conditions possibly faced by the microorganism, such as contact with the host or life under adverse environmental conditions, could constitute situations in which modulation by light of this putative pathway becomes evident.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains used in this work include A. baumannii ATCC 19606 and its derivatives: the isogenic ΔblsA mutant, which harbors a Kn cassette inserted within its coding sequence (8); the ΔblsA mutant carrying the empty pWH1266 plasmid (this work); and the ΔblsA mutant containing pWHBlsA, which expresses a wild-type copy of blsA (this work). Luria-Bertani (LB) broth and agar (Difco) were used to grow and maintain bacterial strains. Broth cultures were incubated either statically or with shaking at 200 rpm at 24°C or 37°C.
Blue light treatments.
Blue light treatments were performed as described in our previous studies (7–9). Briefly, cells were incubated for 26 h (or else as specified) at 24°C in the dark or under blue light emitted by 9-light-emitting diode (LED) arrays, with an intensity of 6 to 10 μmol photons · m−2 · s−1. Each array was built using 3-LED module strips emitting blue light, with emission peaks centered at 462 nm, determined using a LI-COR LI-1800 spectroradiometer (7).
Isolation of RNA.
A. baumannii cells were grown stagnantly in multiwell microplates in LB Difco broth until they reached an optical density at 600 nm (OD600) of 0.4 at 24°C in the presence or absence of blue light. Cells were pelleted and immediately mixed with 1 ml of lysis buffer (0.1 M sodium acetate, 10 mM EDTA, 1% SDS) in a boiling-water bath. Cell lysates were extracted twice at 60°C with one volume of phenol, which was adjusted to pH 4.0 with 50 mM sodium acetate, and then once with chloroform at room temperature. The RNA precipitated overnight at −20°C with 2.5 volumes of ethanol was collected by centrifugation, washed with 70% ethanol, and dissolved in diethyl pyrocarbonate (DEPC)-treated deionized water. RNA was cleaned, and residual genomic DNA was removed using Quick-RNA miniprep, according the manufacturer's recommendation (Zymo Research). RNA quality and quantity were evaluated by gel electrophoresis and by determination of the relationship of absorbance at 260 nm to absorbance at 280 nm.
Analyses of gene expression by qRT-PCR.
Retrotranscription and qRT-PCR analysis were done as described in reference 8, using primers listed in Table S2 in the supplemental material. recA was used as normalizer gene in qRT-PCR experiments. We confirmed that recA is an adequate reference gene with stable expression under our experimental conditions using three other reference genes (16S rRNA, rpoB, and gyrB).
Transcriptome studies by RNA-seq.
For RNA-seq experiments, RNA samples were quantified using Quant-iT RiboGreen RNA reagent (Thermo Fisher Scientific, USA). For RNA quality determination, a 2100 Bioanalyzer (Agilent Technologies, Inc., USA) was used in combination with an Agilent RNA 6000 Pico kit (Agilent Genomics, USA). All the samples presented very good quality parameters, with an RNA integrity number (RIN) higher than 8 (38). Five micrograms of each total RNA was further processed by removing 23S and 16S rRNAs using the Ribo-Zero magnetic kit compatible with A. baumannii (Illumina, USA). RNA was then chemically fragmented and cDNA was synthesized, followed by end repair, adenylation, and finally adapter ligation and enrichment according to protocols described in the TruSeq RNA sample preparation guide (Illumina). Final concentrations and purity grades of the libraries were determined using a NanoDrop ND-1000 (Thermo Scientific) and a Bioanalyzer 2100 (Agilent Technologies, Inc.) in combination with a DNA 1000 kit (Agilent Genomics). The libraries were composed of double-stranded DNA fragments of 260 bp on average and were quantified by quantitative PCR (LightCycler 480; Roche) using the Kapa library quantification kit (Kapa Biosystems, USA).
Paired-end 2 × 100-bp reads were generated with HiSeq 1500 (Illumina). The generated data presented very good quality parameters, analyzed with the FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), with 95.9% of the reads with a Q score of >30. The bioinformatic analysis of the RNA samples was performed using the Trinity software (https://github.com/trinityrnaseq/trinityrnaseq/wiki). Short reads were aligned against the complete genome of A. baumannii ATCC 19606 (http://www.ncbi.nlm.nih.gov/Traces/wgs/?val=JMRY01) using Bowtie 2 (39), allowing 0 mismatches within the first 22 bases. The abundance of the transcripts was estimated using RSEM (http://deweylab.github.io/RSEM/) and eXpress (http://bio.math.berkeley.edu/eXpress/). Both processes were carried out using the “align_and_estimate_abundance.pl” script from the software Trinity (https://github.com/trinityrnaseq/trinityrnaseq/wiki), with the default parameters. Analyses were performed to verify that biological replicates under each condition correlate appropriately. Finally, analyses of differentially expressed genes were performed using the statistic packages edgeR (40) and DESeq (41). DESeq performs a count normalization to control the variation in the number of reads sequenced across samples. After normalization, fold changes and their significance (P values), indicating differential expression, were determined after a negative binomial distribution.
Blast2GO (43) was used for the functional reannotation of genes, the mapping of gene ontology terms, and the description of biological processes, molecular functions, cellular components, and metabolic pathways associated with the biofilm expression profiles.
For construction of figures showing pathways/clusters of genes differentially expressed by light according to RNA-seq experiments (Fig. 2 to 4 and 6 to 9), a color coding bar was designed in which colors vary according to the parameter log2 fold change (log2FC).
Growth of A. baumannii on different carbon sources.
To test the ability of the A. baumannii wild-type and derivative strains used in this work to grow on a given carbon source, 1/100 dilutions of overnight cultures grown in LB Difco were washed and inoculated in M9 liquid medium supplemented with 5 or 10 mM phenylalanine (Phe) or phenylacetic acid (PAA) or in LB Difco medium and grown stagnantly at 24°C under blue light or in the dark. At the times indicated in the figures, an aliquot was taken to record the A600 of the culture.
Quantification of trehalose.
The strains were grown in LB Difco medium until they reached an OD660 of 0.4 under blue light or in the dark, and sugars were extracted from bacterial cells as described previously (44). After sugar extraction, a bacterial pellet was dried at 80°C, and the number of grams (dry weight) (gDW) was measured. The trehalose content was determined by quantifying glucose increments after trehalase (Sigma) treatment (44). Briefly, trehalase treatments were performed in 50-μl reaction mixtures containing 30 μl of 135 mM citric acid buffer (pH 5.7), 10 μl of cell extract, and 10 μl of trehalase enzyme solution, which was prepared by dissolving 0.2 units of enzyme per ml in cold 135 mM citric acid buffer (pH 5.7). The reaction mixtures were incubated for 1 h at 37°C, and reactions were terminated by the addition of 50 μl of 500 mM Tris-HCl (pH 7.5). Calibration curves were generated using 5, 10, 25, 50, 75, 100, 150, 200, and 250 nmol glucose (Sigma) as standards and were determined in parallel for each set of samples analyzed. Glucose was measured using a glucose oxidase kit (Glicemia enzimática AA líquida) under the recommendations of the manufacturer (Wiener, Argentina) before and after trehalase treatment. All along the calibration curve range, absorbance versus glucose concentration was linear, and the sample values fell within this range. The content of trehalose is half of the difference between these two values, given that trehalose is composed of two glucose monomers. Fifty nanomoles of trehalose (Sigma) was also included as a control of the technique. The results were expressed as micromoles of trehalose per gram of bacterial pellet (dry weight).
Evaluation of surfactant production.
Cell supernatants were obtained from 100 ml of liquid culture of cells grown in ethanol medium (15) stagnantly at 24°C under blue light or in the dark to an OD660 of 1 by centrifuging liquid at 5,000 × g for 20 min. Extracellular surfactants were isolated as follows. Each supernatant was placed into a 500-ml separatory funnel, and 20 ml of 1 M hydrochloric acid was added. The funnel was closed and shaken for 3 min. Then, an additional 15 ml of 1 M HCl and 65 ml of a 2:1 chloroform-methanol mixture were added, and the funnel was shaken for 5 min. The mixture in the funnel was set aside until phase separation. The bottom phase (organic extract 1) was released, and the water phase was extracted once more for 5 min after the addition of 35 ml of 1 M HCl and 65 ml of a 2:1 chloroform-methanol mixture to obtain organic extract 2. The third extraction was conducted with 100 ml of a 2:1 chloroform-methanol mixture to obtain organic extract 3. Extracts 1 to 3 were combined and evaporated in an R II Büchi Rotavapor (Switzerland) at room temperature at 8,000 Pa until complete desiccation (15) and finally dried under high vacuum. The amount of synthesized extracellular surfactants (in grams per liter) was then determined gravimetrically with a precision analytical balance (Mettler, Switzerland) (15) and normalized relative to the optical density values reached by the cells.
Catalase activity assay.
Cell extracts were prepared from 20-ml cultures grown in LB stagnantly at 24°C and harvested by centrifugation at 10,000 × g for 10 min at 4°C. Bacteria were washed and resuspended in 500 μl of ice-cold 50 mM potassium phosphate buffer (pH 7.0) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and then disrupted by intermittent sonication. Suspensions were clarified by centrifugation at 12,000 × g for 20 min at 4°C. Protein concentrations in soluble cell extracts were determined by the Bradford method (45), with bovine serum albumin as a standard. For catalase activity determination in gels, aliquots of cell extracts containing 30 μg of soluble protein were electrophoresed on 8% (wt/vol) nondenaturing polyacrylamide gels and stained for catalase activity, as described by Scandalios (46).
MIC determination in liquid medium.
MIC determinations were performed in multiwell microplates using LB broth at 24°C incubated under blue light or in the dark, according to the procedures described by Ramírez et al. (8). For these assays, we used ofloxacin (Alfa Aesar, USA), which was subjected to serial half dilutions starting from 10 μg/ml. A. baumannii cells were resuspended in physiological solution and adjusted to an OD660 of 0.1, diluted 1:10 in LB medium, and applied to the wells. Identical microplates were incubated overnight in the dark or under blue light using the devices described above.
Ofloxacin stability assay under blue light.
LB Difco broth was supplemented with ofloxacin at a final concentration of 0.945 μg/ml (which corresponds to three times the MIC obtained for ATCC 19606) and incubated for 10 h at 24°C under blue light or in the dark under conditions similar to those used for the persistence assay (see below). After the stipulated time, 10 or 50 μl of liquid from each condition was used to load blank commercial disks (Oxoid, USA), which were then placed on the surface of Mueller-Hinton plates (Difco) previously inoculated with a standardized inoculum (0.5 McFarland standard) of a sensitive bacterium (Escherichia coli ATCC 25922). These plates were then incubated at 37°C in the dark, and the diffusion halo was read after 18 h. The experiments were performed in triplicate.
Persistence assay.
The persistence assay was performed as described previously (18), with some modifications. Briefly, exponential-phase cultures of A. baumannii grown in LB under blue light or in the dark were treated with ofloxacin at a final concentration of 0.945 μg/ml for 10 h stagnantly at 24°C in 96-well microplates. A control treatment with sterile water was performed in parallel. The number of CFU was determined by plate counts in each case. The persister fraction is defined as the number of surviving cells after treatment with ofloxacin divided by the number of cells after control treatment (water). The experiments were performed in triplicate.
Accession number(s).
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (42) and are accessible through GEO series accession number GSE90630.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Wiener lab for their gift of a kit for determining glucose and Natalia Gottig for providing trehalase. We acknowledge the LABGeM and the National Infrastructure France Genomique for the platform MAGE (Magnifying Genomes), which has been very helpful for the genomic context analyses described in this work. We thank Ramiro Rodriguez and Carla Schommer for critical reading of the manuscript. We thank Vincent Nguyen for excellent technical assistance.
This work was supported by grants from Agencia Nacional de Promoción Científica y Tecnológica (PICT 2013-0018 and PICT 2014-1161) to M.A.M. G.L.M., P.C., L.E.C., S.A., and M.A.M. are career investigators of CONICET. M.T. and M.M.A. are fellows from the same institution.
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00011-17.
REFERENCES
- 1.Spellberg B, Bonomo RA. 2014. The deadly impact of extreme drug resistance in Acinetobacter baumannii. Crit Care Med 42:1289–1291. doi: 10.1097/CCM.0000000000000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roca I, Espinal P, Vila-Farres X, Vila J. 2012. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front Microbiol 3:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McConnell MJ, Actis L, Pachon J. 2013. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev 37:130–155. doi: 10.1111/j.1574-6976.2012.00344.x. [DOI] [PubMed] [Google Scholar]
- 4.Bassetti M, Righi E. 2015. New antibiotics and antimicrobial combination therapy for the treatment of Gram-negative bacterial infections. Curr Opin Crit Care 21:402–411. doi: 10.1097/MCC.0000000000000235. [DOI] [PubMed] [Google Scholar]
- 5.Solomkin J, Hershberger E, Miller B, Popejoy M, Friedland I, Steenbergen J, Yoon M, Collins S, Yuan G, Barie PS, Eckmann C. 2015. Ceftolozane/tazobactam plus metronidazole for complicated intra-abdominal infections in an era of multidrug resistance: results from a randomized, double-blind, phase 3 trial (ASPECT-cIAI). Clin Infect Dis 60:1462–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mussi MA, Limansky AS, Relling V, Ravasi P, Arakaki A, Actis LA, Viale AM. 2011. Horizontal gene transfer and assortative recombination within the Acinetobacter baumannii clinical population provide genetic diversity at the single carO gene, encoding a major outer membrane protein channel. J Bacteriol 193:4736–4748. doi: 10.1128/JB.01533-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mussi MA, Gaddy JA, Cabruja M, Arivett BA, Viale AM, Rasia R, Actis LA. 2010. The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J Bacteriol 192:6336–6345. doi: 10.1128/JB.00917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramírez MS, Traglia GM, Perez JF, Muller GL, Martinez MF, Golic AE, Mussi MA. 2015. White and blue light induce reduction in susceptibility to minocycline and tigecycline in Acinetobacter spp. and other bacteria of clinical importance. J Med Microbiol 64:525–537. doi: 10.1099/jmm.0.000048. [DOI] [PubMed] [Google Scholar]
- 9.Golic A, Vaneechoutte M, Nemec A, Viale AM, Actis LA, Mussi MA. 2013. Staring at the cold sun: blue light regulation is distributed within the genus Acinetobacter. PLoS One 8:e55059. doi: 10.1371/journal.pone.0055059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramírez MS, Muller GL, Perez JF, Golic AE, Mussi MA. 2015. More than just light: clinical relevance of light perception in the nosocomial pathogen Acinetobacter baumannii and other members of the genus Acinetobacter. Photochem Photobiol 91:1291–1301. doi: 10.1111/php.12523. [DOI] [PubMed] [Google Scholar]
- 11.Cerqueira GM, Kostoulias X, Khoo C, Aibinu I, Qu Y, Traven A, Peleg AY. 2014. A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J Infect Dis 210:46–55. doi: 10.1093/infdis/jiu024. [DOI] [PubMed] [Google Scholar]
- 12.Piazza A, Zimaro T, Garavaglia BS, Ficarra FA, Thomas L, Marondedze C, Feil R, Lunn JE, Gehring C, Ottado J, Gottig N. 2015. The dual nature of trehalose in citrus canker disease: a virulence factor for Xanthomonas citri subsp. citri and a trigger for plant defence responses. J Exp Bot 66:2795–2811. doi: 10.1093/jxb/erv095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, Eisenreich W, Haehnel W, Fuchs G. 2010. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci U S A 107:14390–14395. doi: 10.1073/pnas.1005399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iturriaga G, Suarez R, Nova-Franco B. 2009. Trehalose metabolism: from osmoprotection to signaling. Int J Mol Sci 10:3793–3810. doi: 10.3390/ijms10093793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pirog TP, Shevchuk TA, Konon AD, Dolotenko E. 2012. Production of surfactants by Acinetobacter calcoaceticus K-4 grown on ethanol with organic acids. Prikl Biokhim Mikrobiol 48:631–639. (In Russian.) [PubMed] [Google Scholar]
- 16.Zhao Z, Wong JW. 2009. Biosurfactants from Acinetobacter calcoaceticus BU03 enhance the solubility and biodegradation of phenanthrene. Environ Technol 30:291–299. doi: 10.1080/09593330802630801. [DOI] [PubMed] [Google Scholar]
- 17.Sun D, Crowell SA, Harding CM, De Silva PM, Harrison A, Fernando DM, Mason KM, Santana E, Loewen PC, Kumar A, Liu Y. 2016. KatG and KatE confer Acinetobacter resistance to hydrogen peroxide but sensitize bacteria to killing by phagocytic respiratory burst. Life Sci 148:31–40. doi: 10.1016/j.lfs.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liebens V, Defraine V, Van der Leyden A, De Groote VN, Fierro C, Beullens S, Verstraeten N, Kint C, Jans A, Frangipani E, Visca P, Marchal K, Versees W, Fauvart M, Michiels J. 2014. A putative de-N-acetylase of the PIG-L superfamily affects fluoroquinolone tolerance in Pseudomonas aeruginosa. Pathog Dis 71:39–54. doi: 10.1111/2049-632X.12174. [DOI] [PubMed] [Google Scholar]
- 19.Cronan JE., Jr 2002. Phospholipid modifications in bacteria. Curr Opin Microbiol 5:202–205. doi: 10.1016/S1369-5274(02)00297-7. [DOI] [PubMed] [Google Scholar]
- 20.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carruthers MD, Nicholson PA, Tracy EN, Munson RS Jr. 2013. Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One 8:e59388. doi: 10.1371/journal.pone.0059388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber BS, Ly PM, Irwin JN, Pukatzki S, Feldman MF. 2015. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci U S A 112:9442–9447. doi: 10.1073/pnas.1502966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber BS, Miyata ST, Iwashkiw JA, Mortensen BL, Skaar EP, Pukatzki S, Feldman MF. 2013. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS One 8:e55142. doi: 10.1371/journal.pone.0055142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Repizo GD, Gagne S, Foucault-Grunenwald ML, Borges V, Charpentier X, Limansky AS, Gomes JP, Viale AM, Salcedo SP. 2015. Differential role of the T6SS in Acinetobacter baumannii virulence. PLoS One 10:e0138265. doi: 10.1371/journal.pone.0138265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhuiyan MS, Ellett F, Murray GL, Kostoulias X, Cerqueira GM, Schulze KE, Mahamad Maifiah MH, Li J, Creek DJ, Lieschke GJ, Peleg AY. 2016. Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc Natl Acad Sci U S A 113:9599–9604. doi: 10.1073/pnas.1523116113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Law RJ, Hamlin JN, Sivro A, McCorrister SJ, Cardama GA, Cardona ST. 2008. A functional phenylacetic acid catabolic pathway is required for full pathogenicity of Burkholderia cenocepacia in the Caenorhabditis elegans host model. J Bacteriol 190:7209–7218. doi: 10.1128/JB.00481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodruff PJ, Carlson BL, Siridechadilok B, Pratt MR, Senaratne RH, Mougous JD, Riley LW, Williams SJ, Bertozzi CR. 2004. Trehalose is required for growth of Mycobacterium smegmatis. J Biol Chem 279:28835–28843. doi: 10.1074/jbc.M313103200. [DOI] [PubMed] [Google Scholar]
- 28.Freeman BC, Chen C, Beattie GA. 2010. Identification of the trehalose biosynthetic loci of Pseudomonas syringae and their contribution to fitness in the phyllosphere. Environ Microbiol 12:1486–1497. [DOI] [PubMed] [Google Scholar]
- 29.Djonović S, Urbach JM, Drenkard E, Bush J, Feinbaum R, Ausubel JL, Traficante D, Risech M, Kocks C, Fischbach MA, Priebe GP, Ausubel FM. 2013. Trehalose biosynthesis promotes Pseudomonas aeruginosa pathogenicity in plants. PLoS Pathog 9:e1003217. doi: 10.1371/journal.ppat.1003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pirog TP, Shevchuk TA, Klimenko IuA. 2010. Intensification of surfactant synthesis in Rhodococcus erythropolis EK-1 cultivated on hexadecane. Prikl Biokhim Mikrobiol 46:651–658. (In Russian.) [PubMed] [Google Scholar]
- 31.Boles BR, Thoendel M, Singh PK. 2005. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol 57:1210–1223. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- 32.Irie Y, O'Toole GA, Yuk MH. 2005. Pseudomonas aeruginosa rhamnolipids disperse Bordetella bronchiseptica biofilms. FEMS Microbiol Lett 250:237–243. doi: 10.1016/j.femsle.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 33.Díaz De Rienzo MA, Stevenson P, Marchant R, Banat IM. 2016. Antibacterial properties of biosurfactants against selected Gram-positive and -negative bacteria. FEMS Microbiol Lett 363:fnv224. doi: 10.1093/femsle/fnv224. [DOI] [PubMed] [Google Scholar]
- 34.Díaz De Rienzo MA, Stevenson PS, Marchant R, Banat IM. 2016. Pseudomonas aeruginosa biofilm disruption using microbial surfactants. J Appl Microbiol 120:868–876. doi: 10.1111/jam.13049. [DOI] [PubMed] [Google Scholar]
- 35.Glaeser J, Nuss AM, Berghoff BA, Klug G. 2011. Singlet oxygen stress in microorganisms. Adv Microb Physiol 58:141–173. doi: 10.1016/B978-0-12-381043-4.00004-0. [DOI] [PubMed] [Google Scholar]
- 36.Srinivasan VB, Vaidyanathan V, Rajamohan G. 2015. AbuO, a TolC-like outer membrane protein of Acinetobacter baumannii, is involved in antimicrobial and oxidative stress resistance. Antimicrob Agents Chemother 59:1236–1245. doi: 10.1128/AAC.03626-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Nocera PP, Rocco F, Giannouli M, Triassi M, Zarrilli R. 2011. Genome organization of epidemic Acinetobacter baumannii strains. BMC Microbiol 11:224. doi: 10.1186/1471-2180-11-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Götz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugawara M, Cytryn EJ, Sadowsky MJ. 2010. Functional role of Bradyrhizobium japonicum trehalose biosynthesis and metabolism genes during physiological stress and nodulation. Appl Environ Microbiol 76:1071–1081. doi: 10.1128/AEM.02483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 46.Scandalios JG. 1968. Genetic control of multiple molecular forms of catalase in maize. Ann N Y Acad Sci 151:274–293. doi: 10.1111/j.1749-6632.1968.tb11896.x. [DOI] [PubMed] [Google Scholar]
- 47.Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Falin LJ, Grabmueller C, Humphrey J, Kerhornou A, Khobova J, Aranganathan NK, Langridge N, Lowy E, McDowall MD, Maheswari U, Nuhn M, Ong CK, Overduin B, Paulini M, Pedro H, Perry E, Spudich G, Tapanari E, Walts B, Williams G, Tello-Ruiz M, Stein J, Wei S, Ware D, Bolser DM, Howe KL, Kulesha E, Lawson D, Maslen G, Staines DM. 2016. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res 44:D574–D580. doi: 10.1093/nar/gkv1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallenet D, Belda E, Calteau A, Cruveiller S, Engelen S, Lajus A, Le Fevre F, Longin C, Mornico D, Roche D, Rouy Z, Salvignol G, Scarpelli C, Thil Smith AA, Weiman M, Medigue C. 2013. MicroScope—an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res 41:D636–D647. doi: 10.1093/nar/gks1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.