ABSTRACT
Breakpoints are used to predict whether an antifungal agent will be clinically effective against a particular fungal isolate. They are based on a combination of MIC values, pharmacokinetic/pharmacodynamic values, and clinical outcome data. For many fungus-antifungal combinations, these data might never be available. For these combinations, epidemiological cutoff values (ECVs) provide a methodology for categorizing isolates as either wild type (WT) or non-WT. In this review, we define ECVs, explain how they are generated using the CLSI methodology in standard M57, and describe how they can be used in clinical practice.
KEYWORDS: ECV, breakpoint, epidemiological cutoff value
INTRODUCTION
From the time that clinicians first had a choice in their selection of antimicrobials, there has been a need to predict which infections would and which would not respond to them. While a number of factors (described below) ultimately influence those predictions, the underlying susceptibility of a microorganism to an antimicrobial is one of the first factors to be considered. This need led to the standardization of antimicrobial susceptibility testing methodology and the use of the term “MIC.” An MIC is the lowest concentration of an antimicrobial agent that will prevent a predetermined amount of growth (generally a 50 to 100% decrease in growth relative to an untreated control) of a tested organism in vitro. The next step in the evolution of MIC testing was the development of MIC breakpoints. A breakpoint is used as a predictive MIC value for determining whether or not a microorganism is likely to respond in vivo to an achievable concentration of an antimicrobial at the site of infection when administered using a predefined dosing schedule. Breakpoints are determined by a consensus group using a preponderance of data which include the microbial MIC distribution, in vitro resistance markers, pharmacokinetic/pharmacodynamic values developed using animal models and human subjects, and patient treatment and outcome information based on controlled clinical trial data. It is difficult and expensive to generate the kind of data needed for breakpoint determination. For fungi, these problems are compounded. For most species of fungi, there are simply not enough clinical cases of infection available to make it economically feasible to perform a clinical trial within a reasonable time frame. Furthermore, persons who are susceptible to fungal infection generally have underlying conditions that make it difficult to determine whether their death is attributable specifically to the fungal infection. Practically, this means there will be many fungal pathogens that will never have defined antifungal breakpoints. However, there is an alternative value that, while not predictive of in vivo efficacy, can be used to determine whether an isolate is wild type (WT) in regard to its in vitro response to a given antifungal agent. This value is the epidemiological cutoff value (ECV). In this review, we define ECVs and explain how they are determined, how they differ from breakpoints, and how they can be used in clinical practice, specifically for fungi.
WHAT IS AN ECV?
The definition of an epidemiological cutoff value is the MIC or minimal effective concentration (MEC) (which is used only for echinocandins against molds as the concentrations that cause phenotypic changes in growth) that separates a population into isolates with and those without acquired or mutational resistance based on their phenotypic MIC value (1). An ECV is dependent on the testing methodology used to generate the MIC values upon which it is based; an ECV based on broth microdilution MIC values will not necessarily be the same as an ECV based on MIC values generated by another methodology, such as the Etest (bioMérieux, Inc., Hazelwood, MO) or VITEK 2 (bioMérieux, Inc., Hazelwood, MO) systems. To apply an ECV generated by the CLSI broth microdilution method to another testing system, either independent ECVs for the alternative testing system would need to be established or the alternative testing system would need to be validated in the testing laboratory using the CLSI broth microdilution method as the standard. An ECV is not, and should not be, considered the equivalent of a breakpoint.
HOW ARE ECVs DIFFERENT FROM BREAKPOINTS?
A breakpoint, by definition, is a predictor of the clinical success of a particular antifungal-fungus combination. In creating a breakpoint, the MIC distribution and the pharmacokinetic/pharmacodynamic (PK/PD) data of the antifungal in play are important, but perhaps most critical is the addition of outcome data, especially from a clinical trial. The outcome for a patient treated with a given antifungal agent is compared to the PK/PD values of the antifungal, the MIC of the isolate, and any known resistance mechanisms that are present. These data are pooled from all patients participating in the trial. The goal is to determine the highest MIC value for which an achievable nontoxic dose of the antifungal is possible, excluding isolates with known resistance mechanisms to that antifungal. This value is then balanced against the outcome data so that the breakpoint MIC value favors a positive/successful outcome for the patient being treated. When the only piece of data available is the MIC distribution, a breakpoint cannot be determined and an ECV is the only available tool that may provide some guidance for treatment (Table 1).
TABLE 1.
Differences between breakpoints and ECVs
| Breakpoints | ECVs |
|---|---|
| Require an MIC distribution, pharmacokinetic/pharmacodynamic data, and clinical treatment and outcome data | Require an MIC distribution |
| Can be used to predict clinical success | Do not predict clinical success |
| Can be used to identify isolates that may harbor mutations | Can be used to identify isolates that may harbor mutations |
| Extremely difficult to generate, as treatment and outcome data are rare | Can be generated against any species as long as enough isolates exist |
An ECV is not a predictor of clinical success. In order to make use of an ECV, a clinician must know whether a given antifungal agent is generally successful when used against a particular fungus. If a fungus has intrinsic resistance, such as Candida krusei has to fluconazole, having an MIC value lower than the ECV will not matter; the isolate will always be resistant to fluconazole. The usefulness of an ECV lies in its ability to predict for an isolate possible resistance to an antifungal agent that has known activity against the species but for which there are not enough data to establish breakpoints. For instance, there are no breakpoints for Candida guilliermondii and fluconazole. If the ECV is 8 μg/ml and the MIC is 2 μg/ml, then the isolate will be perceived to be wild type and to have no resistance to fluconazole, and a clinician may consider treatment with fluconazole. Alternatively, if the MIC is 16 μg/ml, and the ECV is 8 μg/ml, then the isolate will be perceived to be non-wild type and may have acquired resistance to fluconazole, in which case the clinician may want to consider other treatment options.
HOW ARE ECVs GENERATED?
Over the past few years, the methodology used to derive ECVs has evolved. The first method, the visual method, simply plots the distribution of MIC values, and a reviewer sets the ECV by sight, usually 1 to 2 dilutions beyond the modal MIC value but before the distribution endpoint, disregarding any tailing of the distribution (small numbers of MICs at the high end of the distribution) (2). In a more scientific approach, the MIC95 was used to determine ECVs (3). For this determination, the MIC that encompassed ≥95% of all MIC values in the distribution was designated the ECV. There are a few problems with using the MIC95 to set the ECV. First, if there is no tail at the high end of the distribution, then the ECV can be set too low. Second, if there are a significant number of resistant isolates in the population, then the MIC95 will be skewed toward the higher values and the ECV can encompass resistant (non-wild-type) isolates. An example of this would be with Candida glabrata and fluconazole, as ∼10% of isolates in any C. glabrata population are resistant to fluconazole. The 95% distribution would therefore include resistant MICs and be set artificially high. To overcome some of these problems, a statistical method for generating ECVs that gives more weight to isolates at the lower end of the distribution was developed (4), and this is the method that has been adopted by the CLSI as a standard method for ECV determination (1). The iterative method takes the log-normal distribution and fits it to different theoretical subsets of MIC values until the best fit that encompasses ≥97.5% of the theoretical distribution is found. The actual data set is fit to the Gaussian distribution that the data would be perceived to normally fit if all testing parameters were equal in all laboratories. ECVs that have been approved by the CLSI Subcommittee on Antifungal Susceptibility Testing can be found in CLSI publication M59 (5).
WHAT ARE THE CLSI RULES FOR GENERATING ECVs?
ECVs for a number of fungal species have been reported in the literature (3, 6–19); however, these ECVs were not generated in a systematic fashion following the same set of rules for ECV determination for each species. There are currently two organizations that have set international standards for susceptibility testing and the development of breakpoints and ECVs for fungi, the CLSI and the European Union Committee on Antimicrobial Susceptibility Testing (EUCAST). While the methodology used and the values derived by these two organizations are similar, they are not the same and cannot be used interchangeably. This review focuses on the newly published CLSI method for the development of ECVs (1).
Because not all isolate populations are the same, owing to different hosts and antifungal and other therapeutic exposures, the CLSI adopted a method meant to ensure that isolate diversity is represented in ECVs. To that end, ECVs established using the CLSI method must include MICs from a minimum of 100 unrelated isolates, must come from a minimum of three separate laboratories, and must not include MIC data from any one of the participating laboratories that account for more than 50% of the total data. Geographical separation of the labs is desirable but not essential, as it is not always possible. These rules are meant to ensure that data are not skewed by a particular lot of broth microdilution panels or by the way a particular technologist in one laboratory interprets the MIC data. In addition, all values used must come from testing runs in which quality control isolate MIC values are within the given CLSI range for the antifungal-fungus combination. Further, the modal MIC value for the data set from any given lab must not fall at either the highest or lowest value of the MIC range tested. This ensures that there is no truncation of the data in which values lower or higher than the mode are generated when additional higher or lower dilutions are tested.
While not strictly a requirement, it is best if resistance mechanisms are known. Isolates with known molecularly characterized mechanisms of resistance may be included in the data set because the log-normal distribution takes into consideration values that fall outside the predicted range. Such isolates also provide a way to verify the ECV that is generated; if an isolate has a known resistance mechanism to an antifungal, then the MIC value for that isolate should be higher than the determined ECV.
Many species are now divided into species complexes. The cryptic species (sensu lato) will not necessarily have the same wild-type MIC range or resistance profile as the parental species (sensu stricto), as was recently shown for the Cryptococcus neoformans and Cryptococcus gattii species complexes (6). For that reason, all isolates, particularly those within a species complex, are identified to the species level using either molecular methods or matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) if the MICs for the isolates are to be used for generating an ECV.
Finally, as mentioned above, the iterative method of ECV determination is used to generate the final ECV (4). While this method is not perfect, as it forces the data into a curve in which 97.5% of the isolates are perceived to be wild type, it does provide precise and consistent ECVs.
HOW SHOULD ECVs BE USED?
While ECVs are not breakpoints and should not be used as breakpoints, having an ECV can provide useful data about an isolate. For example, an ECV can be used as a simple way to determine whether an isolate is or is not wild type with regard to its in vitro response to a given antifungal agent. Having a wild-type isolate does not guarantee a clinical response to or the efficacy of a given antifungal agent. However, having a non-wild-type isolate suggests that the isolate may not respond as expected. Thus, one of the best examples of a situation for which an ECV is useful is when a clinician is treating an infection with an antifungal for a species that is known to develop resistance but for which there are no established breakpoints, especially after long-term antifungal use. The ECV divides the wild-type and the non-wild-type population such that having an MIC value higher than the ECV is an indication that the isolate may have developed resistance to the given antifungal and that, therefore, using an alternative antifungal may be prudent.
Another use for ECVs is in the selection of an antifungal agent while awaiting MIC results for an isolate. As noted, the majority of wild-type isolates will fall below the ECV. Thus, knowing the ECV for the fungal species identified will enable the clinician to determine whether the expected MIC may be high or low in relationship to known achievable levels of a given drug. In that regard, the ECV is essentially an MIC95–99 which can be used to empirically rule in or out the use of various antifungal agents.
ECVs may also have utility for monitoring the epidemiology/emergence of drug resistance in a given fungal species. Testing populations of a fungal species recovered over time, or from a given cohort, can be used to monitor for an increase in the number of isolates with MICs higher than the ECV and, thus, can identify increasing and/or emerging resistance. Such isolates can be further molecularly interrogated to identify previously unidentified mechanisms of resistance. Such monitoring would be most useful for public health laboratories but may also be useful to infection control specialists in large hospitals that have significant antifungal drug use.
ARE ECVs USEFUL?
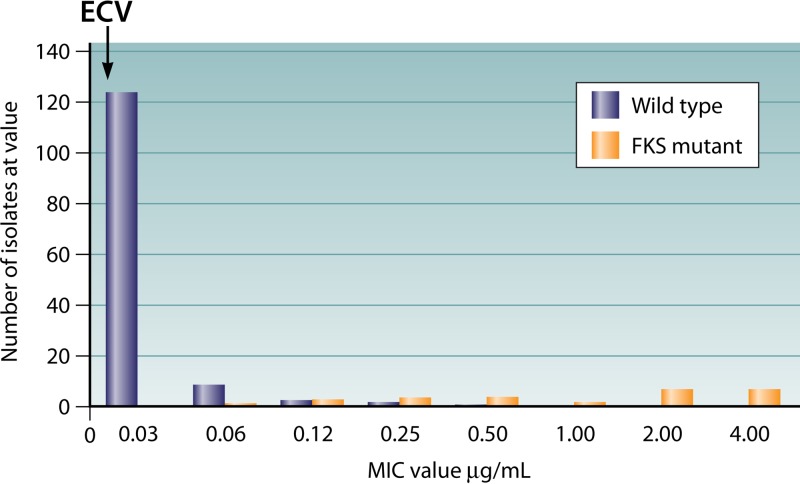
Like any tool, ECVs can be effective when used correctly. Figure 1 shows a data set for Candida glabrata isolates tested against micafungin (20). While a few of the wild-type isolates had MIC values higher than the ECV, no isolates with FKS mutations had an MIC lower than the ECV, indicating that all of the true non-wild-type isolates were identified using the ECV. For a fungus-antifungal combination for which there is a single target/resistance mechanism, such as C. glabrata and micafungin, ECVs can be a good predictor of which isolates are non-wild type.
FIG 1.
MIC distribution of isolates of Candida glabrata tested against micafungin. The black bars represent wild-type isolates, and the gray bars represent isolates with a mutation in the FKS1 or FKS2 gene.
When a species has multiple mechanisms of generating resistance, the use of ECVs becomes more complicated. A good example is the relationship between Candida parapsilosis and fluconazole, for which both an ECV and a breakpoint have been established; the ECV is 1 μg/ml, while the breakpoint is 8 μg/ml. This means that a significant number of isolates (7% of C. parapsilosis isolates in the data set used to generate the ECV [12]) designated non-wild type using the ECV actually have an MIC value that, based on the breakpoint, predicts response to therapy. This is not unexpected for a fungus-antifungal combination where there can be an expression gradient for the resistance mechanism (meaning variability in the phenotypic expression), such as the one for the azole efflux pumps in Candida (21). This example shows that while an isolate may be non-wild type, it does not mean that it is nonsusceptible; careful consideration needs to be given to known mechanisms of resistance and how high the MIC is in relation to the ECV. For those fungus-antifungal combinations where both an ECV and a breakpoint exist, the breakpoint should always be used for clinical decision making.
HOW ARE ECVs REPORTED?
In the event that an ECV is to be reported, both the MIC and the ECV should be provided to the clinician rather than a categorical designation such as wild type or non-wild type. The CLSI M57 document provides specific language pertaining to how an ECV should be reported (1). Following the CLSI example, a laboratory can report the MIC for the organism and include a comment stating, “There are currently no breakpoints or interpretive criteria for species X and antifungal agent XYZ. The XYZ MIC for this isolate is above the Epidemiologic Cutoff Value (PQR μg/ml) for species X and antifungal agent XYZ which suggests that this isolate may have an acquired mechanism of resistance and could be considered non-wild type.” It is also recommended that the clinician consult with an infectious disease clinician and/or pharmacist in deciding whether a drug should be continued, the dosage changed, or an alternative agent considered.
SUMMARY
ECVs are the newest tools available to laboratories performing susceptibility testing and to clinicians treating infections. Like any new tool, their usefulness and best application are just beginning to be discovered. While ECVs are not meant to replace breakpoints, they may be a useful adjunct for determining the best course of antifungal treatment, particularly for fungal species and drug combinations for which breakpoints are not available. ECVs also provide another means for monitoring the emergence of drug resistance in any given fungal species.
ACKNOWLEDGMENTS
Barbara Alexander has received funding from Viamet and Astellas and is a consultant for Astellas, Cidara, Shire, and Scynexis. Mahmoud Ghannoum has received funding from Viamet, Cidara, and Scynexis. Shawn Lockhart has nothing to declare.
The findings and conclusions in this report are those of the authors and do not necessarily represent official positions of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Clinical and Laboratory Standards Institute. 2016. Principles and procedures for the development of epidemiological cutoff values for antifungal susceptibility testing. CLSI supplement M57. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 2.Kahlmeter G, Brown DF, Goldstein FW, MacGowan AP, Mouton JW, Osterlund A, Rodloff A, Steinbakk M, Urbaskova P, Vatopoulos A. 2003. European harmonization of MIC breakpoints for antimicrobial susceptibility testing of bacteria. J Antimicrob Chemother 52:145–148. doi: 10.1093/jac/dkg312. [DOI] [PubMed] [Google Scholar]
- 3.Pfaller MA, Diekema DJ, Ghannoum MA, Rex JH, Alexander BD, Andes D, Brown SD, Chaturvedi V, Espinel-Ingroff A, Fowler CL, Johnson EM, Knapp CC, Motyl MR, Ostrosky-Zeichner L, Sheehan DJ, Walsh TJ; Clinical and Laboratory Standards Institute Antifungal Testing Subcommittee. 2009. Wild-type MIC distribution and epidemiological cutoff values for Aspergillus fumigatus and three triazoles as determined by the Clinical and Laboratory Standards Institute broth microdilution methods. J Clin Microbiol 47:3142–3146. doi: 10.1128/JCM.00940-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnidge J, Kahlmeter G, Kronvall G. 2006. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin Microbiol Infect 12:418–425. doi: 10.1111/j.1469-0691.2006.01377.x. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2016. Epidemiological cutoff values for antifungal susceptibility testing. CLSI supplement M59. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 6.Espinel-Ingroff A, Aller AI, Canton E, Castanon-Olivares LR, Chowdhary A, Cordoba S, Cuenca-Estrella M, Fothergill A, Fuller J, Govender N, Hagen F, Illnait-Zaragozi MT, Johnson E, Kidd S, Lass-Florl C, Lockhart SR, Martins MA, Meis JF, Melhem MS, Ostrosky-Zeichner L, Pelaez T, Pfaller MA, Schell WA, St-Germain G, Trilles L, Turnidge J. 2012. Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole. Antimicrob Agents Chemother 56:5898–5906. doi: 10.1128/AAC.01115-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espinel-Ingroff A, Alvarez-Fernandez M, Canton E, Carver PL, Chen SC, Eschenauer G, Getsinger DL, Gonzalez GM, Govender NP, Grancini A, Hanson KE, Kidd SE, Klinker K, Kubin CJ, Kus JV, Lockhart SR, Meletiadis J, Morris AJ, Pelaez T, Quindos G, Rodriguez-Iglesias M, Sanchez-Reus F, Shoham S, Wengenack NL, Borrell Sole N, Echeverria J, Esperalba J, Gomez-G de la Pedrosa E, Garcia I, Linares MJ, Marco F, Merino P, Peman J, Perez Del Molino L, Rosello Mayans E, Rubio Calvo C, Ruiz Perez de Pipaon M, Yague G, Garcia-Effron G, Guinea J, Perlin DS, Sanguinetti M, Shields R, Turnidge J. 2015. Multicenter study of epidemiological cutoff values and detection of resistance in Candida spp. to anidulafungin, caspofungin, and micafungin using the Sensititre YeastOne colorimetric method. Antimicrob Agents Chemother 59:6725–6732. doi: 10.1128/AAC.01250-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espinel-Ingroff A, Chowdhary A, Gonzalez GM, Guinea J, Hagen F, Meis JF, Thompson GR III, Turnidge J. 2015. Multicenter study of isavuconazole MIC distributions and epidemiological cutoff values for the Cryptococcus neoformans-Cryptococcus gattii species complex using the CLSI M27-A3 broth microdilution method. Antimicrob Agents Chemother 59:666–668. doi: 10.1128/AAC.04055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espinel-Ingroff A, Cuenca-Estrella M, Fothergill A, Fuller J, Ghannoum M, Johnson E, Pelaez T, Pfaller MA, Turnidge J. 2011. Wild-type MIC distributions and epidemiological cutoff values for amphotericin B and Aspergillus spp. for the CLSI broth microdilution method (M38-A2 document). Antimicrob Agents Chemother 55:5150–5154. doi: 10.1128/AAC.00686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espinel-Ingroff A, Diekema DJ, Fothergill A, Johnson E, Pelaez T, Pfaller MA, Rinaldi MG, Canton E, Turnidge J. 2010. Wild-type MIC distributions and epidemiological cutoff values for the triazoles and six Aspergillus spp. for the CLSI broth microdilution method (M38-A2 document). J Clin Microbiol 48:3251–3257. doi: 10.1128/JCM.00536-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espinel-Ingroff A, Fothergill A, Fuller J, Johnson E, Pelaez T, Turnidge J. 2011. Wild-type MIC distributions and epidemiological cutoff values for caspofungin and Aspergillus spp. for the CLSI broth microdilution method (M38-A2 document). Antimicrob Agents Chemother 55:2855–2859. doi: 10.1128/AAC.01730-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espinel-Ingroff A, Pfaller MA, Bustamante B, Canton E, Fothergill A, Fuller J, Gonzalez GM, Lass-Florl C, Lockhart SR, Martin-Mazuelos E, Meis JF, Melhem MS, Ostrosky-Zeichner L, Pelaez T, Szeszs MW, St-Germain G, Bonfietti LX, Guarro J, Turnidge J. 2014. Multilaboratory study of epidemiological cutoff values for detection of resistance in eight Candida species to fluconazole, posaconazole, and voriconazole. Antimicrob Agents Chemother 58:2006–2012. doi: 10.1128/AAC.02615-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfaller MA, Boyken L, Hollis RJ, Kroeger J, Messer SA, Tendolkar S, Diekema DJ. 2011. Wild-type MIC distributions and epidemiological cutoff values for posaconazole and voriconazole and Candida spp. as determined by 24-hour CLSI broth microdilution. J Clin Microbiol 49:630–637. doi: 10.1128/JCM.02161-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaller MA, Castanheira M, Diekema DJ, Messer SA, Jones RN. 2011. Wild-type MIC distributions and epidemiologic cutoff values for fluconazole, posaconazole, and voriconazole when testing Cryptococcus neoformans as determined by the CLSI broth microdilution method. Diagn Microbiol Infect Dis 71:252–259. doi: 10.1016/j.diagmicrobio.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Pfaller MA, Boyken L, Hollis RJ, Kroeger J, Messer SA, Tendolkar S, Jones RN, Turnidge J, Diekema DJ. 2010. Wild-type MIC distributions and epidemiological cutoff values for the echinocandins and Candida spp. J Clin Microbiol 48:52–56. doi: 10.1128/JCM.01590-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfaller MA, Espinel-Ingroff A, Bustamante B, Canton E, Diekema DJ, Fothergill A, Fuller J, Gonzalez GM, Guarro J, Lass-Florl C, Lockhart SR, Martin-Mazuelos E, Meis JF, Ostrosky-Zeichner L, Pelaez T, St-Germain G, Turnidge J. 2014. Multicenter study of anidulafungin and micafungin MIC distributions and epidemiological cutoff values for eight Candida species and the CLSI M27-A3 broth microdilution method. Antimicrob Agents Chemother 58:916–922. doi: 10.1128/AAC.02020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfaller MA, Espinel-Ingroff A, Canton E, Castanheira M, Cuenca-Estrella M, Diekema DJ, Fothergill A, Fuller J, Ghannoum M, Jones RN, Lockhart SR, Martin-Mazuelos E, Melhem MS, Ostrosky-Zeichner L, Pappas P, Pelaez T, Peman J, Rex J, Szeszs MW. 2012. Wild-type MIC distributions and epidemiological cutoff values for amphotericin B, flucytosine, and itraconazole and Candida spp. as determined by CLSI broth microdilution. J Clin Microbiol 50:2040–2046. doi: 10.1128/JCM.00248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espinel-Ingroff A, Chakrabarti A, Chowdhary A, Cordoba S, Dannaoui E, Dufresne P, Fothergill A, Ghannoum M, Gonzalez GM, Guarro J, Kidd S, Lass-Florl C, Meis JF, Pelaez T, Tortorano AM, Turnidge J. 2015. Multicenter evaluation of MIC distributions for epidemiologic cutoff value definition to detect amphotericin B, posaconazole, and itraconazole resistance among the most clinically relevant species of Mucorales. Antimicrob Agents Chemother 59:1745–1750. doi: 10.1128/AAC.04435-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espinel-Ingroff A, Colombo AL, Cordoba S, Dufresne PJ, Fuller J, Ghannoum M, Gonzalez GM, Guarro J, Kidd SE, Meis JF, Melhem TM, Pelaez T, Pfaller MA, Szeszs MW, Takahaschi JP, Tortorano AM, Wiederhold NP, Turnidge J. 2016. International evaluation of MIC distributions and epidemiological cutoff value (ECV) definitions for Fusarium species identified by molecular methods for the CLSI broth microdilution method. Antimicrob Agents Chemother 60:1079–1084. doi: 10.1128/AAC.02456-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaller MA, Diekema DJ, Andes D, Arendrup MC, Brown SD, Lockhart SR, Motyl M, Perlin DS; CLSI Subcommittee for Antifungal Testing. 2011. Clinical breakpoints for the echinocandins and Candida revisited: integration of molecular, clinical, and microbiological data to arrive at species-specific interpretive criteria. Drug Resist Updat 14:164–176. doi: 10.1016/j.drup.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Berkow EL, Manigaba K, Parker JE, Barker KS, Kelly SL, Rogers PD. 2015. Multidrug transporters and alterations in sterol biosynthesis contribute to azole antifungal resistance in Candida parapsilosis. Antimicrob Agents Chemother 59:5942–5950. doi: 10.1128/AAC.01358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]