ABSTRACT
A novel selective agar (RGM medium) has been advocated for the isolation of rapidly growing mycobacteria from the sputa of cystic fibrosis (CF) patients. The aim of this study was to compare RGM medium to Burkholderia cepacia selective agar (BCSA) and a standard acid-fast bacillus (AFB) culture method for the isolation of nontuberculous mycobacteria (NTM) from patients with CF. The applicability of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) for the identification of NTM isolated on RGM medium was also assessed. Respiratory samples (n = 869) were collected from 487 CF patients and inoculated directly onto RGM medium and BCSA. Cultures were incubated at 30°C and examined for up to 28 days. A subset of 212 samples (from 172 patients) was also cultured by using a mycobacterial growth indicator tube (MGIT) and on Lowenstein-Jensen medium following dual decontamination. By using a combination of all methods, 98 mycobacteria were isolated from 869 samples (11.3%). The sensitivity of RGM medium (96.9%) was significantly higher than that of BCSA (35.7%) for the isolation of mycobacteria (P < 0.0001). The sensitivity of RGM medium was also superior to that of standard AFB culture for the isolation of mycobacteria (92.2% versus 47.1%; P < 0.0001). MALDI-TOF MS was effective for the identification of mycobacteria in RGM medium. RGM medium offers a simple and highly effective tool for the isolation of NTM from patients with CF. Extended incubation of RGM medium for 28 days facilitates the isolation of slow-growing species, including members of the Mycobacterium avium complex (MAVC).
KEYWORDS: culture, cystic fibrosis, mycobacterium, rapid grower, selective medium
INTRODUCTION
Nontuberculous mycobacteria (NTM) are recognized as significant respiratory pathogens in patients with cystic fibrosis (CF) (1). In the largest studies of CF patients, the prevalence of NTM in sputum has been estimated to be 6 to 13% (2) and is thought to be increasing (3–6). For the detection of NTM in sputum samples, international guidelines recommend the use of an automated liquid culture method (such as the mycobacterial growth indicator tube [MGIT]) and note that additional culture on solid media, such as Lowenstein-Jensen (LJ) medium, may increase recovery (1). These culture methods rely upon the initial decontamination of the sputum sample by using chemical treatments such as N-acetyl-l-cysteine (NALC) (0.5%) plus sodium hydroxide (NaOH) (2%), and decontamination is intended to eliminate nonmycobacterial species that might otherwise compromise the isolation of NTM. Such methods are problematic, as contaminated cultures may frequently occur, despite decontamination, or the number of viable mycobacteria may be reduced by the decontamination process (7–9).
Esther et al. demonstrated previously that extending the incubation time on Burkholderia cepacia selective agar (BCSA) from 5 to 14 days afforded an increased rate of recovery of NTM, and this has been used as an expedient method for the isolation of NTM (10). However, overgrowth of cultures with fungi and Gram-negative bacteria remains a problem, and this method has a lower sensitivity than that of standard acid-fact bacillus (AFB) culture (AFBC) methods (10). RGM medium is a novel agar-based medium that was specifically developed for the isolation of rapidly growing species of mycobacteria such as members of the Mycobacterium abscessus complex (MABSC) (11). In two European studies, RGM medium was shown to have a sensitivity superior to that of BCSA for the isolation of rapidly growing mycobacteria from the sputum samples of CF patients by using a 10-day incubation period for both media (11, 12). In a third European study, the use of RGM medium with a 10-day incubation period was shown to have a sensitivity equivalent to that of automated liquid culture using the MGIT (13). The aim of this study was to assess the utility of RGM medium in a clinical laboratory in the United States and to examine whether an extended incubation time from 10 to 28 days might enhance the isolation of slow-growing species of NTM such as members of the Mycobacterium avium complex (MAVC). The compatibility of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was also assessed with colonies of known species of mycobacteria cultured on RGM medium and BCSA.
(Part of this work was presented at ASM Microbe 2016, Boston, MA, USA.)
RESULTS
Comparison between BCSA and RGM medium incubated for 28 days.
A total of 869 respiratory samples from 487 CF patients were submitted for routine CF culture and/or culture for mycobacteria during the study period. The age range of patients was <1 year to 71 years, with a median age of 14.9 years, and 53.6% of the patients were female. The samples included 507 (58.4%) sputum/tracheal aspirates, 314 (36.1%) deep pharyngeal (DP) swabs, and 48 (5.5%) bronchial washing (BW)/bronchoalveolar lavage (BAL) fluid specimens.
A total of 98 isolates of NTM were recovered from the 869 patient samples (11.3%) by using a combination of all methods. These 98 isolates were recovered from 69 distinct patients, giving a prevalence of 14.2% among this population of patients. The overall sensitivities of RGM medium and BCSA were 96.9% and 35.7%, respectively (P < 0.0001) (Table 1). The M. abscessus complex accounted for the majority of mycobacterial isolates by using both methods (prevalence of 5.5%). RGM medium had a significantly higher sensitivity than that of BCSA for the recovery of MABSC organisms (100% versus 62.5%; P < 0.001), M. chelonae (100% versus 12.5%; P = 0.0005), and M. mucogenicum (100% versus 62.5%; P = 0.013). Of 35 mycobacteria isolated on BCSA, 32 isolates were detected on RGM medium, but 3 isolates of M. immunogenum were isolated only on BCSA. Slow-growing mycobacteria were recovered only from RGM medium, including MAVC organisms (n = 8), M. gordonae (n = 4), M. arupense-M. nonchromogenicum (n = 1), and M. nebraskense (n = 1). Almost all (34 of 35) isolates on BCSA were recovered within 14 days of incubation, a standard protocol for the screening of rapidly growing mycobacteria in our laboratory. An improved recovery of NTM was observed for all sample types. The proportions of positive DP swabs, sputum/tracheal aspirates, and BW/BAL fluid specimens with NTM increased from 0.6%, 5.1%, and 14.6% on BCSA to 5.4%, 12.2%, and 25% on RGM medium, respectively. As a result of extending the incubation time from 10 to 28 days, 35 additional isolates of NTM were recovered on RGM medium, including both rapidly growing species, e.g., MABSC organisms (n = 9), and slow-growing species, e.g., MAVC organisms (n = 6) (Fig. 1). Forty-eight NTM-positive patients had ≥2 sequential samples collected. Of these patients, 14 (29.2%) patients had ≥2 more positive cultures: 55% had MABSC organisms (11 of 20 patients), 10% had M. chelonae (1 of 10 patients), and 66.7% of MAVC organisms (2 of 3 patients). The remaining 34 patients had NTM isolated only once, including MABSC organisms (n = 9), M. chelonae (n = 9), M. immunogenum (n = 6), M. mucogenicum (n = 6), MAVC organisms (n = 1), M. arupense-M. nonchromogenicum (n = 1), M. nebraskense (n = 1), and M. gordonae (n = 1).
TABLE 1.
Mycobacteria from 869 respiratory samples of 487 patients with cystic fibrosis recovered on RGM medium and BCSA
| Organism | Total no. of mycobacteria | 28-day BCSA |
28-day RGM medium |
P value | ||
|---|---|---|---|---|---|---|
| No. of mycobacteria recovered | Sensitivity (%) | No. of mycobacteria recovered | Sensitivity (%) | |||
| M. abscessus complex | 48 | 30 | 62.5 | 48 | 100 | <0.0001 |
| M. chelonae | 16 | 2 | 12.5 | 16 | 100 | 0.0005 |
| M. immunogenum | 11 | 3 | 27.3 | 8 | 72.7 | 0.228 |
| M. mucogenicum | 8 | 0 | 0 | 8 | 100 | 0.013 |
| M. avium complex | 8 | 0 | 0 | 8 | 100 | 0.013 |
| M. gordonae | 4 | 0 | 0 | 4 | 100 | |
| M. arupense-M. nonchromogenicum | 1 | 0 | 0 | 1 | 100 | |
| M. nebraskense | 1 | 0 | 0 | 1 | 100 | |
| M. llatzerense | 1 | 0 | 0 | 1 | 100 | |
| Total | 98 | 35 | 35.7 | 95 | 96.9 | <0.0001 |
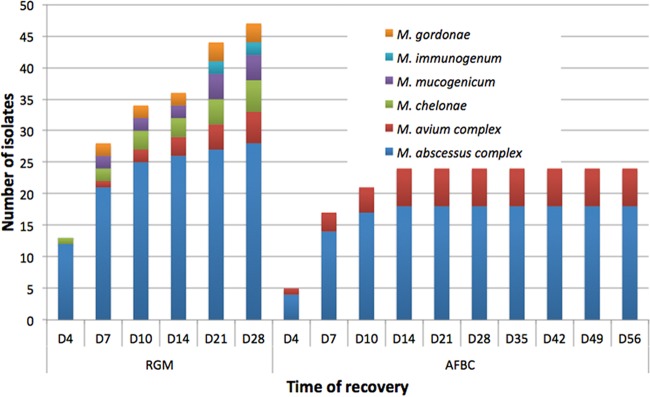
FIG 1.
Cumulative time to detection of NTM recovered on RGM medium and BCSA from 869 respiratory samples of 487 patients with cystic fibrosis.
Nonmycobacterial species were recovered from 243 BCSA cultures (28.1%) and 95 RGM medium cultures (10.9%) (Table 2). Gram-negative bacilli (n = 117) and fungi (n = 121) accounted for the majority of nonmycobacteria isolated on BCSA. On RGM medium, Gram-negative bacilli (n = 74) were the leading group of organisms, whereas fungi were mostly inhibited, except for 7 Candida species and 13 Trichosporon species.
TABLE 2.
Nonmycobacterial organisms from 869 respiratory samples of 487 patients with cystic fibrosis recovered on RGM medium and BCSA
| Organism | No. of organisms recovered |
||
|---|---|---|---|
| 28-day culture on BCSA | 28-day culture on RGM medium | Total | |
| Burkholderia species | 54 | 42 | 54 |
| Achromobacter species | 16 | 17 | 22 |
| Pandoraea species | 10 | 10 | 10 |
| Pseudomonas aeruginosa | 9 | 0 | 9 |
| Stenotrophomonas maltophilia | 6 | 0 | 6 |
| Chryseobacterium species | 6 | 0 | 6 |
| Ralstonia species | 6 | 0 | 6 |
| Inquilinus species | 4 | 4 | 4 |
| Serratia marcescens | 3 | 1 | 3 |
| Providencia rettgeri | 1 | 0 | 1 |
| Morganella morganii | 1 | 0 | 1 |
| Staphylococcus aureus | 4 | 0 | 4 |
| Enterococcus faecalis | 2 | 0 | 2 |
| Streptomyces species | 0 | 1 | 1 |
| Mold | 55 | 0 | 55 |
| Candida species | 55 | 7 | 56 |
| Trichosporon species | 11 | 13 | 14 |
| Total | 243 | 95 | 254 |
Comparison between 28-day culture on RGM medium and 56-day culture for mycobacteria using MGIT and LJ medium (AFBC).
A subset of 212 samples, which comprised 164 (77.4%) sputum/tracheal aspirates, 4 (1.9%) DP swabs, and 44 (20.7%) BW/BAL fluid specimens, from 172 patients also had a simultaneous culture performed for mycobacteria in addition to culture on RGM medium and BCSA.
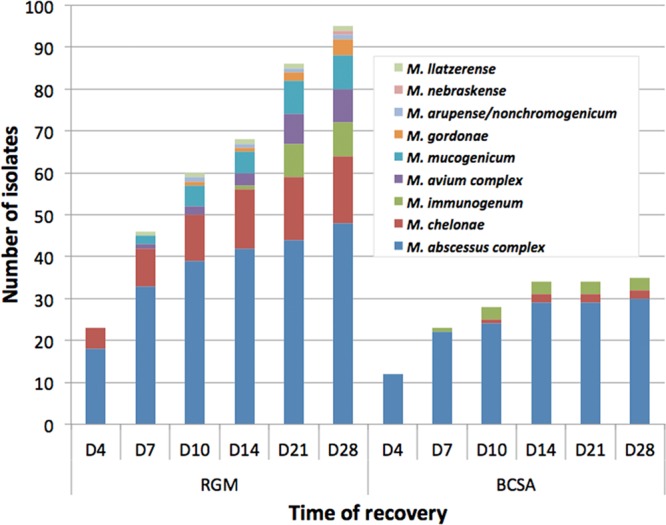
A total of 51 mycobacteria were recovered by using a combination of the three methods (24.1%). RGM medium showed a higher rate of recovery of NTM than did AFBC (92.2% versus 47.1%; P < 0.0001) (Table 3). The proportions of positive sputum/tracheal aspirates and BW/BAL fluid specimens with NTM increased from 12.8% and 4.5% upon AFBC to 20.7% and 18.1% on RGM medium, respectively. RGM medium missed one MABSC isolate and two MAVC isolates that were isolated by AFBC, while the use of AFBC was unable to recover 9 MABSC, 1 MAVC, and 14 other NTM isolates that grew on RGM medium. Fifteen samples had positive auramine-rhodamine stains. All smear-positive samples yielded mycobacteria by using RGM medium, whereas 13 (86.7%) of these samples yielded mycobacteria from AFBC. Only MABSC organisms and a mix of M. gordonae and M. mucogenicum could be isolated from the two cultures by using RGM medium. The rate of recovery of MABSC organisms was significantly higher by using RGM medium than by using AFBC (sensitivity of 96.6% versus 62.1%; P = 0.009). MAVC organisms were isolated from 7 samples by using a combination of methods (prevalence of 3.3%). The rates of recovery of MAVC organisms were comparable between RGM medium (5 of 7 isolates recovered) and AFBC (6 of 7 isolates recovered). The cumulative time to positivity is shown in Fig. 2, with time points being selected for AFBC that allow a direct comparison of the two methods. All mycobacteria that were isolated by AFBC were detected within 14 days of incubation, whereas only 36 (76.6%) of 47 isolates were recovered within 14 days by using RGM medium. The extended incubation time helped to detect 11 more NTM on RGM medium, including 2 MABSC and 2 MAVC organisms.
TABLE 3.
Mycobacteria from 212 respiratory samples of 172 patients with cystic fibrosis recovered on RGM medium, BCSA, and conventional mycobacterial culture using MGIT and solid medium
| Organism | Total no. of mycobacteria | 28-day culture on BCSA |
56-day AFB culture |
28-day culture on RGM medium |
P valuea | |||
|---|---|---|---|---|---|---|---|---|
| No. of mycobacteria recovered | Sensitivity (%) | No. of mycobacteria recovered | Sensitivity (%) | No. of mycobacteria recovered | Sensitivity (%) | |||
| M. abscessus complex | 29 | 7 | 24.1 | 18 | 62.1 | 28 | 96.6 | 0.009 |
| M. chelonae | 5 | 3 | 60.0 | 0 | 0 | 5 | 100 | |
| M. immunogenum | 3 | 1 | 33.3 | 0 | 0 | 2 | 66.7 | |
| M. avium complex | 7 | 0 | 0 | 6 | 85.7 | 5 | 71.4 | |
| M. mucogenicum | 4 | 0 | 0 | 0 | 0 | 4 | 100 | |
| M. gordonae | 3 | 0 | 0 | 0 | 0 | 3 | 100 | |
| Total | 51 | 11 | 21.6 | 24 | 47.1 | 47 | 92.2 | <0.0001 |
Comparisons between 56-day AFBC and 28-day RGM medium culture.
FIG 2.
Cumulative time to detection of NTM recovered from 212 respiratory samples of 172 patients with cystic fibrosis using RGM medium and conventional mycobacterial culture (AFBC) with an MGIT and solid medium.
Nonmycobacterial organisms were recovered from 28 samples (13.2%) on RGM medium, including 24 Gram-negative bacilli and 4 fungi. There were 30 AFBCs (14.1%) from which mycobacteria could not be recovered due to the overgrowth of other organisms in both the MGIT and LJ medium. An additional 52 LJ and 2 MGIT samples were discarded due to overgrowth.
Performance of MALDI-TOF MS for identification of rapidly growing mycobacteria on BCSA and RGM medium.
All 41 archived isolates grew on RGM medium; however, 1 MABSC, 4 M. fortuitum complex, and 4 M. mucogenicum isolates did not grow on BCSA at 30°C in air, even if the incubation time was extended to 7 days. All NTM on both media could be correctly identified by MALDI-TOF MS with bioMérieux RUO SARAMIS Knowledge Base database scores of ≥75.0%, except for 1 M. immunogenum isolate on BCSA. However, all isolates could be identified by using the bioMérieux in vitro diagnostic (IVD) system with the updated v3.0 Knowledge Base database. Subspecies-level identification was not included in any of the databases used in this study.
DISCUSSION
Specific species of NTM such as MABSC organisms are considered significant CF pathogens and are associated with poorer clinical outcomes (14). The isolation of NTM from respiratory samples of CF patients is challenging because of bacterial and fungal overgrowth in AFB cultures. RGM medium, a novel selective medium designed for the isolation of MABSC organisms, has been shown to have promise for the isolation of rapidly growing mycobacteria from the sputa of CF patients (11–13). We are the first U.S. center to evaluate this medium with the largest number of samples to date. Extending the incubation time of RGM medium from 10 to 28 days and comparison between RGM medium and 8-week AFBC using an MGIT and LJ medium were intended to give a preliminary assessment of the ability to detect slowly growing mycobacteria on this medium.
RGM medium is superior to BCSA for the recovery of rapidly growing mycobacteria, with a sensitivity for the detection of NTM on RGM medium of 96.9%, compared with 35.7% on BCSA (P < 0.0001). A higher level of recovery was observed on RGM medium for MABSC, M. chelonae, M. mucogenicum, and MAVC isolates. The increased sensitivity is consistent with data from a previous study by Preece et al., who evaluated the recovery of rapidly growing mycobacteria from 502 sputum samples using a 10-day incubation of RGM medium and BCSA and reported sensitivities of 98% and 31%, respectively (P ≤ 0.0001) (11). Extending the incubation time from 10 to 28 days had a significant impact on the recovery of slow-growing mycobacteria but also on the recovery of rapidly growing mycobacteria (Fig. 1). Of 98 NTM recovered by using a combination of RGM medium and BCSA, only 60 isolates (61.2%) were recovered on RGM medium within 10 days of incubation, compared with 94 isolates (96.9%) being recovered on the same medium after 28 days of incubation (P < 0.001). The ability of RGM medium to recover slow-growing species such as members of the MAVC, an important pathogen in CF, raises the possibility of using RGM medium incubated for 28 days as a screening culture for all NTM in CF patients. RGM medium is also more selective than BCSA, as shown by fewer nonmycobacterial organisms detected (10.9% versus 28.1%); however, the inhibitory effect may be lost over the prolonged incubation time since 8 more isolates of Burkholderia species were recovered after 14 days of incubation.
Eltringham et al. compared the sensitivity of RGM medium incubated for 10 days with that of 6-week MGIT cultures (13). A total of 187 sputum samples were directly inoculated onto RGM medium and were decontaminated by using 3% oxalic acid before inoculation into an MGIT. The level of detection of mycobacteria on RGM medium was equivalent to that with the MGIT (sensitivity of 82% versus 86%; P = 1.000). In the study presented here, there was also no statistical difference between AFBC and culture on RGM medium if results for RGM medium were analyzed after 10 days of incubation (P = 0.136). RGM medium had a higher sensitivity than that of standard AFB culture using a combination of the MGIT and LJ medium (92.2% versus 47.1%; P < 0.0001). The rate of recovery of MABSC organisms was significantly higher on RGM medium, and the rate of recovery of MAVC organisms was equivalent to that achieved by AFBC. In our laboratory, all specimens from CF patients undergo two-step decontamination with NALC-NaOH followed by 2.5% oxalic acid prior to culture. This harsh treatment may result in a reduced recovery of NTM on AFBC, especially with specimens containing low numbers of NTM. Bange et al. showed previously that 3 NTM from 406 specimens could not be isolated after two-step decontamination despite being recovered after decontamination with NALC-NaOH (15). It was notable that mycobacteria were recovered from two AFB smear-positive specimens on RGM medium but were not recovered from samples subjected to dual decontamination by using standard AFBC. RGM medium has potential benefits over AFBC because of its enhanced sensitivity and high specificity coupled with its relative simplicity. The use of RGM medium requires no decontamination and may reduce labor and material costs. Another potential advantage of RGM medium is the possibility of recovering MAVC organisms from a mixed culture. We isolated MAVC and MABSC organisms on RGM medium from a culture from which only MABSC organisms could be recovered by AFBC (Fig. 3). We also showed that NTM growing on RGM medium could be reliably identified by using MALDI-TOF MS. MALDI-TOF MS has been the method for the identification of rapidly growing mycobacteria in our laboratory since 2014 after thorough validation. However, further study to evaluate the performance of MALDI-TOF MS on RGM medium can be achieved by the selection of reference isolates that have been identified by a method other than MALDI-TOF MS. An additional application of this medium may be its use in the culture of specimens from patients with other chronic lung diseases, such as chronic obstructive pulmonary disease, or from patients with non-CF bronchiectasis. The single potential advantage of AFBC over culture on RGM medium would appear to be its capacity to accommodate the isolation of Mycobacterium tuberculosis, a pathogen that is rarely encountered in patients with CF. Although RGM medium is not yet commercially available, its formulation was previously reported (12). The time of stability of this medium was previously demonstrated to be at least 12 weeks when stored at +4°C (11).
FIG 3.
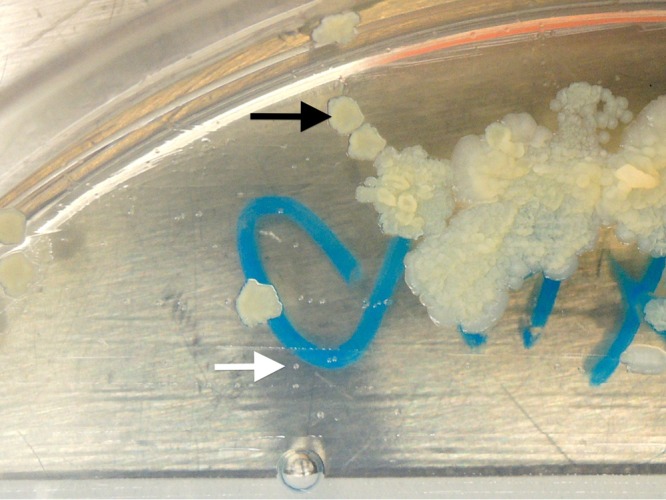
Colonies of M. abscessus complex (black arrow) and M. avium complex (white arrow) organisms on RGM medium after 14 days of incubation.
There are at least three shortcomings to this study. First, we do not have good longitudinal data, which is inherent in a study that was of only a 5-month duration. We found three NTM species, M. chelonae, M. immunogenum, and M. mucogenicum, that have only recently been recovered in our CF population largely because of the use of RGM medium. The clinical significance of these organisms is not understood and requires careful longitudinal study, as has been done with both MAVC organisms and M. abscessus, the other organisms found frequently in this study (16). Second, RGM medium is responsible for increased “noise” in these culture results. M. gordonae, an organism typically viewed as an environmental contaminant of mycobacterial cultures, was found in several RGM medium cultures but not in either BCSA or standard AFB cultures. Although RGM medium enhances recovery, it may also increase the likelihood of noise as well as “signal,” especially if M. chelonae, M. immunogenum, and M. mucogenicum isolates prove to be more likely to represent contamination or transient colonization. Third, although our data suggest that RGM medium may be used for the recovery of MAVC organisms from respiratory specimens, the number of isolates found in this study is too low to draw firm conclusions about its applicability for the recovery of slow-growing organisms, especially since they may grow better at 35 to 37°C than at 30°C, the temperature used in the present study (17). Additional studies under various incubation conditions are needed to answer this question. Additional technical issues regarding extended incubation should also be addressed thoroughly, including biosafety, stability of the culture medium, and the possibility of environmental contamination.
RGM medium offers a higher rate of recovery of NTM and is more selective than BCSA. The sensitivity for the recovery of NTM is also significantly higher than that of conventional mycobacterial culture. An extended incubation time on RGM medium of 28 days facilitates the isolation of both rapid-growing mycobacteria and slow-growing species, including MAVC organisms. There is the potential to replace mycobacterial culture of CF respiratory samples with the more simple and effective RGM medium.
MATERIALS AND METHODS
Patient samples.
Respiratory samples from adults and children with cystic fibrosis that were submitted to the Clinical Microbiology-Immunology Laboratories, UNC Health Care, Chapel Hill, NC, USA, for routine culture and culture for mycobacteria were prospectively collected from December 2015 to April 2016. Specimen types included DP swabs, sputum/tracheal aspirates, and BW and BAL fluids. This study was approved by the UNC Institutional Review Board (IRB) (approval number 15-2890).
Specimen processing and culture method for BCSA and RGM medium.
RGM medium was prepared at the Microbiology Department, Freeman Hospital, Newcastle upon Tyne, UK, as previously described and transported to UNC Health Care, Chapel Hill, NC (12). RGM medium is based on Middlebrook 7H9 medium supplemented with oleic acid-albumin-dextrose-catalase (OADC) and a mixture of four antimicrobials: fosfomycin, colistin, amphotericin B, and 9-chloro-9-[4-(diethylamino)phenyl]-9,10-dihydro-10-phenylacridine hydrochloride (C-390). DP swabs were inoculated directly onto BCSA (Remel, Lenexa, KS) and then RGM medium. Sputum samples and tracheal aspirates were plated onto BCSA and then RGM medium by using a sterile swab. A 100-μl aliquot of undiluted BW and BAL fluids was used for inoculation. Sputolysin (EMD Millipore, Billerica, MA) was added dropwise as needed to homogenize viscous BW/BAL fluid. A 100-μl aliquot of the Sputolysin-sample mixture was inoculated onto the two media after being vortexed and left at room temperature for 15 min. No treatment with Sputolysin was performed on sputum and tracheal aspirates. BCSA was initially incubated at 35°C in air for 4 days as part of the routine CF culture protocol and was then incubated at 30°C for 24 days. RGM medium was incubated at 30°C in air for 28 days. Both media were examined at 4, 7, 10, 14, 21, and 28 days.
Specimen processing and culture method for mycobacteria (AFBC).
A subset of samples had mycobacterial culture performed in addition to culture on BCSA and RGM medium. All CF respiratory samples underwent double decontamination with NALC–2% NaOH and oxalic acid before inoculation into an MGIT (Becton Dickinson, Sparks, MD) and LJ medium (Remel). Briefly, up to 5 ml of sputum/tracheal aspirate samples or concentrated BW/BAL fluid (centrifuged at 2,500 rpm for 15 min) was treated with an equal amount of the BBL MycoPrep reagent (NALC-NaOH; Becton Dickinson), vortexed, and incubated at room temperature for 15 min. BBL MycoPrep phosphate buffer (PB; Becton Dickinson) was added to give a final volume of 50 ml, and this mixture was centrifuged at 3,900 rpm for 15 min. The supernatant was discarded, and the sediment was resuspended in 2 ml of sterile PB. An equal amount (approximately 2 ml) of 5% oxalic acid (Remel) was added to make a final concentration of 2.5% oxalic acid after a smear for Truant auramine-rhodamine stain was prepared. The specimen was incubated at room temperature for 30 min, with vortexing at 10-min intervals. The steps of adding PB and centrifugation were repeated to obtain 2 ml of treated sample, and this sample was then adjusted to an optimal pH of between 6.9 and 7.2 by using 4% NaOH or 1 N HCl as required. A 0.5-ml aliquot of the processed specimen was inoculated into an MGIT containing OADC growth supplement (Becton Dickinson) and PANTA (polymyxin B-amphotericin B-nalidixic acid-trimethoprim-azlocillin) antimicrobial supplement (Becton Dickinson), prepared according to the manufacturer's protocol. The inoculated MGIT was loaded onto a Bactec MGIT 960 instrument (Becton Dickinson) and incubated at 37°C, with continuous monitoring of growth for 42 days. A solid medium (LJ medium) was also inoculated with 4 drops of the processed specimen. LJ medium was examined after 4, 7, 10, and 14 days and then weekly for up to 56 days.
Identification of mycobacteria and other microorganisms.
A Kinyoun stain was prepared from any colonies consistent with mycobacteria growing on BCSA, RGM medium, and LJ medium and from broth from any positive MGIT. AFB-positive colonies and AFB-positive MGIT samples were subcultured on Middlebrook 7H11 agar (Remel) and incubated at 30°C in air for subsequent identification. Any AFB-negative bacterial colonies recovered on BCSA and RGM medium were subcultured on Trypticase soy agar with 5% sheep blood (Becton Dickinson) and were further identified by using the MALDI-TOF MS (Vitek MS, bioMérieux, Durham, NC) IVD system with the Knowledge Base version 2.0 database after 24 h of incubation at 35°C in 5% CO2. Candida spp. and Trichosporon spp. were presumptively identified by colony and Gram stain morphologies. There was no further identification of molds or Kinyoun-negative organisms from positive MGIT and LJ medium cultures. The Vitek MS research-use-only (RUO) system with the SARAMIS v4.12 database (bioMérieux) was used to identify mycobacteria that were subcultured on Middlebrook 7H11 agar after 72 to 96 h of incubation according to the manufacturer's recommendations (18). Confidence scores of ≥75% were acceptable for species identification according to our previous validation study and our laboratory protocol. Slowly growing mycobacteria and bacteria that could not be identified by MALDI-TOF MS were identified by using partial sequencing of the 16S rRNA gene as previously described (19).
Evaluation of MALDI-TOF MS for identification of rapidly growing mycobacteria on BCSA and RGM medium.
Forty-one archived isolates were used to evaluate the performance of MALDI-TOF MS for the identification of rapidly growing mycobacteria on BCSA and RGM medium. The isolates included Mycobacterium abscessus subsp. abscessus (n = 11), M. abscessus subsp. massiliense (n = 11), M. chelonae (n = 5), the M. fortuitum complex (n = 6), M. mucogenicum (n = 5), and M. immunogenum (n = 3). The species of all strains were previously identified by MALDI-TOF MS from colonies grown on Middlebrook 7H11 agar using the method described above (18). The subspecies of M. abscessus was identified by whole-genome sequencing at the Wellcome Trust Sanger Institute (Hinxton, UK) as described previously (20). The isolates were stored at −80°C in Middlebrook 7H9 broth with glycerol (Becton Dickinson). A 10-μl aliquot of frozen isolates was subcultured on Trypticase soy agar with 5% sheep blood (Becton Dickinson) and incubated at 30°C in air. The isolates were then subcultured on BCSA and RGM medium and incubated at 30°C in air. MALDI-TOF MS was performed on the isolates after 72 to 96 h of incubation depending on the growth rate in accordance with the methods described above by using the Vitek MS RUO system with the SARAMIS database. Additional spectrum analysis was performed by using the new v3.0 Knowledge Base database for the Vitek MS system (bioMérieux).
Statistical analysis.
The differences in the performances of the culture methods were analyzed by using McNemar's test with the continuity correction applied. Statistical significance was taken as a P value of <0.05.
ACKNOWLEDGMENTS
All authors participated in the preparation of the manuscript.
We thank staff members of the Clinical Microbiology-Immunology Laboratories, UNC Health Care, for their diligent work on CF respiratory and mycobacterial cultures. We also thank David H. Pincus from bioMérieux, Inc., for analysis of MALDI-TOF MS spectra.
No outside funding supported this study. The Freeman Hospital Microbiology Department (represented by C.L.P. and J.D.P.) receives funding from bioMérieux for the development and evaluation of culture media, and J.D.P. has performed consultancy work for the same company. The other authors have no conflicts to declare.
REFERENCES
- 1.Floto RA, Olivier KN, Saiman L, Daley CL, Herrmann J-L, Nick JA, Noone PG, Bilton D, Corris P, Gibson RL, Hempstead SE, Koetz K, Sabadosa KA, Sermet-Gaudelus I, Smyth AR, van Ingen J, Wallace RJ, Winthrop KL, Marshall BC, Haworth CS. 2016. US Cystic Fibrosis Foundation and European Cystic Fibrosis Society consensus recommendations for the management of non-tuberculous mycobacteria in individuals with cystic fibrosis: executive summary. Thorax 71:88–90. doi: 10.1136/thoraxjnl-2015-207983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martiniano SL, Nick JA, Daley CL. 2016. Nontuberculous mycobacterial infections in cystic fibrosis. Clin Chest Med 37:83–96. doi: 10.1016/j.ccm.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Qvist T, Gilljam M, Jönsson B, Taylor-Robinson D, Jensen-Fangel S, Wang M, Svahn A, Kötz K, Hansson L, Hollsing A, Hansen CR, Finstad PL, Pressler T, Høiby N, Katzenstein TL, Scandinavian Cystic Fibrosis Study Consortium. 2015. Epidemiology of nontuberculous mycobacteria among patients with cystic fibrosis in Scandinavia. J Cyst Fibros 14:46–52. doi: 10.1016/j.jcf.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seddon P, Fidler K, Raman S, Wyatt H, Ruiz G, Elston C, Perrin F, Gyi K, Bilton D, Drobniewski F, Newport M. 2013. Prevalence of nontuberculous mycobacteria in cystic fibrosis clinics, United Kingdom, 2009. Emerg Infect Dis 19:1128–1130. doi: 10.3201/eid/1907.120615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar-On O, Mussaffi H, Mei-Zahav M, Prais D, Steuer G, Stafler P, Hananya S, Blau H. 2015. Increasing nontuberculous mycobacteria infection in cystic fibrosis. J Cyst Fibros 14:53–62. doi: 10.1016/j.jcf.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Raidt L, Idelevich EA, Dübbers A, Küster P, Drevinek P, Peters G, Kahl BC. 2015. Increased prevalence and resistance of important pathogens recovered from respiratory specimens of cystic fibrosis patients during a decade. Pediatr Infect Dis J 34:700–705. doi: 10.1097/INF.0000000000000714. [DOI] [PubMed] [Google Scholar]
- 7.Bange F-C, Böttger EC. 2002. Improved decontamination method for recovering mycobacteria from patients with cystic fibrosis. Eur J Clin Microbiol Infect Dis 21:546–548. doi: 10.1007/s10096-002-0760-y. [DOI] [PubMed] [Google Scholar]
- 8.Ferroni A, Vu-Thien H, Lanotte P, Le Bourgeois M, Sermet-Gaudelus I, Fauroux B, Marchand S, Varaigne F, Berche P, Gaillard J-L, Offredo C. 2006. Value of the chlorhexidine decontamination method for recovery of nontuberculous mycobacteria from sputum samples of patients with cystic fibrosis. J Clin Microbiol 44:2237–2239. doi: 10.1128/JCM.00285-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buijtels PCAM, Petit PLC. 2005. Comparison of NaOH-N-acetyl cysteine and sulfuric acid decontamination methods for recovery of mycobacteria from clinical specimens. J Microbiol Methods 62:83–88. doi: 10.1016/j.mimet.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Esther CR, Hoberman S, Fine J, Allen S, Culbreath K, Rodino K, Kerr A, Gilligan P. 2011. Detection of rapidly growing mycobacteria in routine cultures of samples from patients with cystic fibrosis. J Clin Microbiol 49:1421–1425. doi: 10.1128/JCM.02379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preece CL, Perry A, Gray B, Kenna DT, Jones AL, Cummings SP, Robb A, Thomas MF, Brodlie M, O'Brien CJ, Bourke SJ, Perry JD. 2016. A novel culture medium for isolation of rapidly-growing mycobacteria from the sputum of patients with cystic fibrosis. J Cyst Fibros 15:186–191. doi: 10.1016/j.jcf.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Preece CL, Wichelhaus TA, Perry A, Jones AL, Cummings SP, Perry JD, Hogardt M. 2016. Evaluation of various culture media for detection of rapidly growing mycobacteria from patients with cystic fibrosis. J Clin Microbiol 54:1797–1803. doi: 10.1128/JCM.00471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eltringham I, Pickering J, Gough H, Preece CL, Perry JD. 2016. Comparison of the mycobacterial growth indicator tube (MGIT) with culture on RGM selective agar for detection of mycobacteria in sputum samples from patients with cystic fibrosis. J Clin Microbiol 54:2047–2050. doi: 10.1128/JCM.00630-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilligan PH. 2014. Infections in patients with cystic fibrosis: diagnostic microbiology update. Clin Lab Med 34:197–217. doi: 10.1016/j.cll.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bange F-C, Kirschner P, Böttger EC. 1999. Recovery of mycobacteria from patients with cystic fibrosis. J Clin Microbiol 37:3761–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esther CR, Esserman DA, Gilligan P, Kerr A, Noone PG. 2010. Chronic Mycobacterium abscessus infection and lung function decline in cystic fibrosis. J Cyst Fibros 9:117–123. doi: 10.1016/j.jcf.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simner P, Stenger S, Richter E, Brown-Elliott B, Wallace R, Wengenack N. 2015. Mycobacterium: laboratory characteristics of slowly growing mycobacteria, p 570–594. In Jorgensen J, Pfaller M, Carroll K, Funke G, Landry M, Richter S, Warnock D (ed), Manual of clinical microbiology, 11th ed ASM Press, Washington, DC. [Google Scholar]
- 18.Mather CA, Rivera SF, Butler-Wu SM. 2014. Comparison of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization–time of flight mass spectrometry systems for identification of mycobacteria using simplified protein extraction protocols. J Clin Microbiol 52:130–138. doi: 10.1128/JCM.01996-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo PCY, Ng KHL, Lau SKP, Yip K, Fung AMY, Leung K, Tam DMW, Que T, Yuen K. 2003. Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles. J Clin Microbiol 41:1996–2001. doi: 10.1128/JCM.41.5.1996-2001.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]