ABSTRACT
Tuberculous pleurisy is one of the most common types of extrapulmonary tuberculosis, but its diagnosis remains difficult. In this study, we report for the first time on the detection of cell-free Mycobacterium tuberculosis DNA in pleural effusion and an evaluation of a newly developed molecular assay for the detection of cell-free Mycobacterium tuberculosis DNA. A total of 78 patients with pleural effusion, 60 patients with tuberculous pleurisy, and 18 patients with alternative diseases were included in this study. Mycobacterial culture, the Xpert MTB/RIF assay, the adenosine deaminase assay, the T-SPOT.TB assay, and the cell-free Mycobacterium tuberculosis DNA assay were performed on all the pleural effusion samples. The cell-free Mycobacterium tuberculosis DNA assay and adenosine deaminase assay showed significantly higher sensitivities of 75.0% and 68.3%, respectively, than mycobacterial culture and the Xpert MTB/RIF assay, which had sensitivities of 26.7% and 20.0%, respectively (P < 0.01). All four of these tests showed good specificities: 88.9% for the adenosine deaminase assay and 100% for the remaining three assays. The T-SPOT.TB assay with pleural effusion showed the highest sensitivity of 95.0% but the lowest specificity of 38.9%. The cell-free Mycobacterium tuberculosis DNA assay detected as few as 1.25 copies of IS6110 per ml of pleural effusion and showed good accordance of the results between repeated tests (r = 0.978, P = 2.84 × 10−10). These data suggest that the cell-free Mycobacterium tuberculosis DNA assay is a rapid and accurate molecular test which provides direct evidence of Mycobacterium tuberculosis etiology.
KEYWORDS: Mycobacterium tuberculosis, cell-free DNA, pleural effusion
INTRODUCTION
Tuberculous pleurisy (TP) is one of the most common extrapulmonary manifestations of tuberculosis, but its diagnosis is quite challenging (1). The definite diagnosis of TP is made by detecting Mycobacterium tuberculosis from the pleural effusion (PE), sputum, or pleural tissue (1, 2). However, due to the paucity of M. tuberculosis organisms in PE, culture and molecular tests like the Xpert MTB/RIF assay show poor sensitivities (3–5).
Other assays for the immunochemical biomarkers mostly reported in PE are the adenosine deaminase assay (ADA), the interferon gamma (IFN-γ) assay, and IFN-γ release assays (IGRAs). Adenosine deaminase and IFN-γ are believed to be released during the immune response triggered by the presence of mycobacterial antigens in the pleural cavity. Recent meta-analyses show that adenosine deaminase and IFN-γ appear to be relatively accurate biomarkers for TP diagnosis. However, a wide range of diagnostic cutoff values may cause heterogeneity (6–9). IGRAs are T-cell-based assays that measure IFN-γ release by sensitized T cells in response to M. tuberculosis-specific antigens. The presence of IFN-γ is a strong indicator of M. tuberculosis infection, but the value of IGRAs for the diagnosis of TP is still controversial. Some studies show that IGRAs are useful for TP diagnosis even in areas where TB is highly endemic (10, 11). However, other studies show that assays for the detection of adenosine deaminase and IFN-γ are superior to IGRAs and do not recommend the use of IGRAs for TP diagnosis (12–15). In conclusion, new accurate and efficient methods are needed to improve TP diagnosis.
Cell-free DNA (cfDNA) refers to DNA fragments released from original cells into various body fluids. cfDNA was first found in human plasma in 1948 by Mandel and Metais (16). In 1997, Lo et al. discovered free fetal DNA from maternal blood, and this attracted great attention for cfDNA research in the diagnostic field (17). It is well-known that cell-free tumor cell DNA and fetal cfDNA can be detected in various body fluids, such as plasma, urine, and PE (18–20). Recently, the detection of cfDNA from pathogens, including fungi, bacteria, and parasites, has been reported (21–23). A nucleic acid amplification test (NAAT) detecting cfDNA from the pathogen has shown better sensitivity than just the detection of genomic DNA from the same pathogen. However, to our knowledge, cfDNA from M. tuberculosis has not been studied. Because of the paucity of pathogenic organisms in PE, detection of M. tuberculosis organisms or M. tuberculosis genomic DNA is not sufficient for TP diagnosis. We hypothesized that the cfDNA released from M. tuberculosis exists in the PE samples from TP patients and detection of M. tuberculosis cfDNA may improve the diagnostic sensitivity.
RESULTS
Characteristics of patients.
In this study, 83 patients suspected of having tuberculous pleurisy (TP) were prospectively recruited, and 5 of them were excluded either because of incomplete clinical data (n = 3) or because the anti-TB treatment outcome was lacking (n = 2). The clinical features of the 78 patients finally included in the study are shown in Table 1. There were 32 patients with definite TP, 28 patients with possible TP, and 18 patients without TP (as control). All the patients with definite and possible TP were considered to have TP and were used as the “gold standard” for calculation of sensitivity and specificity. Among the 18 control patients, 10 had malignant diagnoses, 7 had other infectious diseases, and 1 had sarcoidosis. One of the TP patients was positive for HIV infection, and all the other TP patients and controls were HIV negative. Fifty-four patients (69.2%) were male, and the median age of the all patients was 44 years (range, 18 to 83 years).
TABLE 1.
Basic clinical features of the patients included in the study
| Patient group | No. of patients | Median (range) age (yr) | No. (%) of male patients | No. (%) of patients with exudate |
|---|---|---|---|---|
| Definite TPa | 32 | 46 (18–70) | 26 (81.3) | 31 (96.9) |
| Possible TP | 28 | 29 (18–83) | 19 (67.9) | 28 (100) |
| Subtotal for TP patients | 60 | 37 (18–83) | 45 (75.0) | 59 (98.3) |
| No TP | 18 | 57 (37–76) | 9 (50.0) | 18 (100) |
| Total | 78 | 44 (18–83) | 54 (69.2) | 77 (98.7) |
TP, tuberculous pleurisy.
Validation of cf-TB in pleural effusion.
In this study, we investigated whether cell-free M. tuberculosis DNA was detectable in PE. To avoid contamination with genomic DNA from intact bacilli, PE samples were filtered through a 0.22-μm-pore-size filter before cfDNA extraction. Forty-five PE samples from TP patients were positive by the assay for cell-free Mycobacterium tuberculosis DNA (cf-TB). The copy number of IS6110 in these positive samples ranged from 1.25 copies/ml PE to 2.09 × 104 copies/ml PE (Fig. 1A). We also randomly chose 15 PE samples from TP patients and repeated the test from the cfDNA extraction procedure. The results of two repeated tests were highly correlated (r = 0.978, P = 2.84 × 10−10). All 10 samples positive in the first test were also positive in the second test, while the other 5 samples, which were negative in the first test, were also negative in the second test (Fig. 1B).
FIG 1.
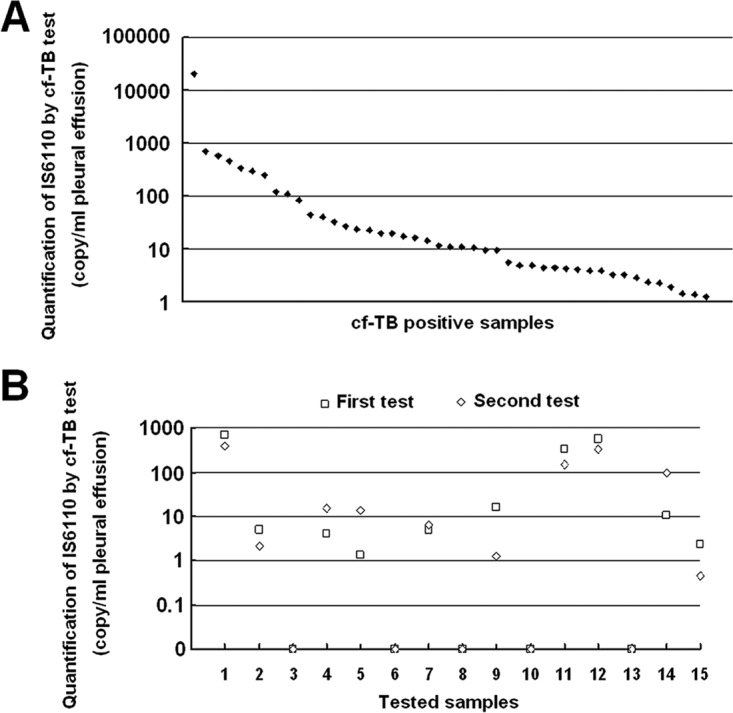
Validation of cell-free M. tuberculosis DNA test with tuberculous pleural effusion. (A) Copy number of IS6110 in 45 cell-free M. tuberculosis DNA test-positive pleural effusion samples; (B) copy number of IS6110 in 15 pleural effusion samples from tuberculous pleurisy patients by two repeated cell-free M. tuberculosis DNA tests.
Comparison of five different testing methods.
The performance of mycobacterial culture (culture), the Xpert MTB/RIF assay (Xpert), the T-SPOT.TB assay (T-SPOT), the adenosine deaminase assay (ADA), and cf-TB are listed in the Table 2. According to the clinical classification (all patients with definite and possible TP were considered to have TP), the sensitivity of the assays from the highest to the lowest was T-SPOT, cf-TB, ADA, culture, and Xpert, with the sensitivity values being 95.0%, 75.0%, 68.3%, 26.7%, and 20.0%, respectively. The sensitivity of cf-TB was 2.8-fold and 3.8-fold higher than the sensitivities of culture and Xpert, respectively; these are the methods most commonly used to confirm the existence of M. tuberculosis. The sensitivity of cf-TB was higher than that of ADA, but the difference was not significant (P > 0.05). Culture, Xpert, and cf-TB each showed a perfect specificity of 100%, and ADA also showed a decent specificity of 88.9%. However, T-SPOT gave positive results for 61.1% (11/18) of the patients in the no TP group (Table 2; see also Fig. S1A in the supplemental material), indicating a very poor specificity of 38.9%. T-SPOT is believed to a good indicator for M. tuberculosis infection but not for active TB. The high false-positive rate for T-SPOT in this study may be result of the high prevalence of M. tuberculosis infection in the population.
TABLE 2.
Performance of five different tests for diagnosis of TP
| Diagnostic method | % sensitivity | % specificity | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|
| Mycobacterial culture | 26.7 (15.2–38.2)a | 100 (100–100) | 100 (100–100) | 29.0 (17.4–40.7) |
| Xpert MTB/RIF assay | 20.0 (9.6–30.4) | 100 (100–100) | 100 (100–100) | 27.3 (16.2–38.3) |
| Adenosine deaminase assay | 68.3 (56.2–80.5) | 88.9 (72.8–100) | 95.4 (88.8–100) | 45.7 (28.4–63.1) |
| T-SPOT.TB assay | 95.0 (89.3–100) | 38.9 (13.9–63.8) | 83.8 (74.8–92.8) | 70.0 (35.4–100) |
| Cell-free M. tuberculosis DNA assay | 75.0 (63.7–86.3) | 100 (100–100) | 100 (100–100) | 54.6 (36.6–72.5) |
Values in parentheses are 95% confidence intervals.
The rates of positivity of the different patient groups by the five testing methods are shown in Fig. 2. The sensitivities of ADA, T-SPOT, and cf-TB were stable for the different TP groups. The sensitivity of cf-TB was significantly higher than those of culture and Xpert even for the definite TP group (P < 0.05).
FIG 2.

Comparison of five diagnostic tests with samples from different patient groups. Pearson's chi-square test was used to determine the significance among categorical variables, except that Fisher's exact test was used to determine the significance between cf-TB and T-SPOT in the group with definite TP. NS, not significant (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001; Δ, analysis was by Fisher's exact test.
All these results indicate that cf-TB showed the best accuracy among the tests evaluated. Culture and Xpert showed poor sensitivities, and T-SPOT showed a poor specificity. The accuracy of ADA was the closest to that of cf-TB, but both the sensitivity and the specificity of ADA were lower than those of cf-TB.
DISCUSSION
Our study reports for the first time on the detection of M. tuberculosis cfDNA in PE and its value for TP diagnosis. Compared to the sensitivities of culture and Xpert, cf-TB showed a greatly improved sensitivity (75.0%) over the sensitivities of culture and Xpert of 26.7% and 20.0%, respectively. It has been reported that cfDNA has many origins, such as apoptosis, necrosis, active release, phagocytosis, and exocytosis (24). Detection of cfDNA thus requires tests different from conventional tests that just detect genomic DNA from intact bacteria. cf-TB also targets the IS6110 insertion sequence, which is specific for the M. tuberculosis complex and which is present at 10 to 12 copies in various strains of M. tuberculosis (25). The ability to detect cfDNA of various origins and with various circulatory properties, along with the detection of the multicopy insertion sequence IS6110, might be the reason why cf-TB was more sensitive than culture and Xpert.
Another important point worth mentioning is that cfDNA is less stable than genomic DNA, which may contribute to the high specificity of cf-TB. It has been reported that circulating fetal DNA is rapidly cleared from maternal plasma and urine. The half-life of circulating fetal DNA is within hours postpartum, and fetal DNA is not detectable by about 1 to 2 days postpartum (26, 27). cfDNA from pathogens has also been reported to be cleared rapidly from plasma after treatment (28, 29). These studies suggest that the M. tuberculosis cfDNA detected in PE has very likely been released into the body very recently and reflects the present disease condition. In this study, cf-TB did not give any positive results for the 18 control samples from patients with other diseases, although 11 (61.1%) of them were positive for latent M. tuberculosis infection according to T-SPOT (Table 2 and Fig. S1A in the supplemental material). As a kind of NAAT like Xpert, cf-TB also targets an M. tuberculosis-specific gene, which provides direct evidence for the existence of M. tuberculosis in the lesion.
Adenosine deaminase is the most widely used biomarker for TP diagnosis, and an ADA was also evaluated in this study. The sensitivity and specificity of cf-TB (75.0% and 100%, respectively) were better than those of ADA (68.3% and 88.9%, respectively). The main drawback of ADA is that some diseases other than TP, such as empyema, parapneumonic effusions, lymphomas, and rheumatoid arthritis, also cause elevations in adenosine deaminase levels in PE (1, 30). In this study, 1 case with lung cancer and 1 case with pneumonia showed adenosine deaminase levels of >40 IU/liter in PE. The limitation of this study is the absence of some control diseases, such as lymphomas and rheumatoid arthritis, which are rare diseases in the Beijing Chest Hospital. Although samples from patients with such diseases that cause elevations in adenosine deaminase levels in PE were lacking as controls, cf-TB still showed better sensitivity and specificity than ADA. These results suggest that cf-TB might have an accuracy superior to that of ADA for TP diagnosis.
Another immunologic method, T-SPOT, was also evaluated in this study. First, T-SPOT showed a very poor specificity. Although the specificity of T-SPOT was significantly different between the TP and no TP groups, 11 out of 18 (61.1%) control samples were positive by T-SPOT using the cutoff value suggested by the manufacturer (Fig. 2 and S1). The high number of false-positive results strongly suggested that the IGRA result should be carefully interpreted when it is used for diagnosis in areas where tuberculosis is epidemic due to the high rates of latent tuberculosis infection. Second, analysis of the specificity of T-SPOT in TP patients showed interesting results (Fig. S1C). The specificity of T-SPOT was significantly higher with the cf-TB-positive PE samples than with the cf-TB-negative ones. However, there were no significant differences in the specificity of T-SPOT between the culture-positive and -negative groups. The specificity of T-SPOT was even significantly higher for Xpert-negative PE samples than for Xpert-positive ones. The pathogenesis of tuberculous PE is thought to result from the hypersensitivity reaction induced by the direct access of M. tuberculosis or the proteins secreted or lysed from M. tuberculosis (30). cfDNA is a kind of small DNA fragment which, like proteins, can be released and circulated into the pleural space. Considering the paucity of M. tuberculosis organisms in PE, M. tuberculosis cfDNA may be more effective in triggering immune responses than the bacterium itself.
In conclusion, cell-free M. tuberculosis DNA is detectable in pleural effusion by a real-time PCR technique, and this is a rapid and accurate method for TP diagnosis.
MATERIALS AND METHODS
Study population.
This was a prospective study. Inpatients aged ≥18 years who had evidence of pleural effusion (PE) demonstrated by X-ray, who were suspected of having tuberculous pleurisy (TP), and who agreed to participate in the study were enrolled from June 2015 to July 2016 at the Beijing Chest Hospital, Beijing, China. The study was approved by the Ethical and Institutional Review Boards for Human Investigation of the Beijing Chest Hospital.
Clinical categories of TB pleurisy.
Patients were divided into three groups according to the previously published definition of a TP diagnosis (1). Patients were classified as having definite TP if they were positive for the detection of M. tuberculosis in the sputum, PE, or pleural tissue. Patients were classified as having possible TP if a pathological examination demonstrated granulomas in the pleural biopsy specimens but M. tuberculosis was not identified, the ADA level in PE was over 40 U/liter, a good response to anti-TB chemotherapy was observed, and a diagnosis of active tuberculosis could not be excluded. All cases of definite and possible TP were considered to be TP and were used as the gold standard for the calculation of sensitivity and specificity. Patients were classified as not having TP if an alternative diagnosis was made or there was clinical improvement in the absence of anti-TB chemotherapy.
Collection of clinical samples.
All PE samples collected from the patients were simultaneously sent for mycobacterial culture (culture), Xpert MTB/RIF assay (Xpert), adenosine deaminase assay (ADA), T-SPOT.TB assay (T-SPOT), and cell-free M. tuberculosis DNA assay (cf-TB). Culture, Xpert, ADA, and T-SPOT were done within 1 to 4 h after PE collection. PE samples for cf-TB were centrifuged at 3,000 × g for 10 min within 1 h after collection. The supernatants were stored at −80°C until cfDNA extraction and detection.
Mycobacterial culture and Xpert MTB/RIF assay.
Sediments from 10 ml PE were cultured in a mycobacterial growth indicator tube for 6 weeks (MGIT 960 system; Becton Dickinson, Sparks, MD). All positive cultures were subjected to Ziehl-Neelsen staining to confirm the presence of acid-fast bacilli. Then, the presence of M. tuberculosis was confirmed by a real-time PCR for detection of the IS6110 insertion sequence.
Sediments from 40 ml PE were tested by the Xpert MTB/RIF system (Cepheid, Sunnyvale, CA) according to the manufacturer's instructions.
T-SPOT.TB and adenosine deaminase assay.
Sediments from 40 ml PE were resuspended in 5 ml phosphate buffer. The following procedure was performed according to the instructions of the manufacturer of the T-SPOT.TB assay (Oxford Immunotec Ltd., Abingdon, UK).
Adenosine deaminase activity was determined using supernatant from 1 ml of PE and an adenosine deaminase assay kit (Beijing Strong Biotechnologies, Beijing, China) according to the manufacturer's instructions. The most widely accepted cutoff value of 40 U/liter was used for TP diagnosis.
Cell-free M. tuberculosis DNA test.
cfDNA was extracted from PE utilizing a QIAamp circulating nucleic acid kit (Qiagen, Hilden, Germany). Six milliliters of PE supernatant was thawed and centrifuged at 16,000 × g for 10 min. The supernatant was then filtered through a 0.22-μm-pore-size filter. The extraction procedure was carried out exactly according to the manufacturer's instructions.
An M. tuberculosis DNA-specific PCR was performed immediately after cfDNA extraction, utilizing a Mycobacterium tuberculosis fluorescent PCR diagnostic kit (DaAn Gene, Guangzhou, China) according to the manufacturer's instructions. This PCR kit detects the IS6110 insertion sequence, which is specific for the M. tuberculosis complex and which is absent from nontuberculous mycobacteria and other bacteria.
Statistical analysis.
Data were analyzed with the SPSS software package (v17.0; SPSS Inc., Chicago, IL). Pearson's chi-square test and Fisher's exact test were used to determine the significance among categorical variables. Differences were considered statistically significant when P was <0.05. The accordance of the results of repeated tests was analyzed by Pearson's correlation test. The correlation was considered statistically significant when P was <0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank the patients who participated in the study.
We all meet the criteria for authorship according to our contributions to the work. N.C., X.Y., and X.C. came up with the concept for the study and designed the study; N.C., X.Y., Z.L., and K.L. acquired the data; N.C., X.Y., and X.C. analyzed and interpreted the data; N.C. drafted the manuscript; N.C. and X.C. critically revised the manuscript for important intellectual content; and N.C. performed the statistical analysis.
This work was supported by grants from the National Natural Science Foundation of China (grant number 81572077), the Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special (grant numbers XMLS201506 and ZYLX201304), the Beijing Health System Training Program for High Level Technique Talents (grant numbers 2014-3-082 and 2014-3-083), the Capital Health Research and Development of Special (grant number 2014-4-2161), the Beijing Municipal Administration of Hospitals' Ascent Plan (grant number DFL20151501), and the Key Project of the Department of Science and Technology, Beijing, China (grant number D141107005214003).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.02473-16.
REFERENCES
- 1.Porcel JM. 2009. Tuberculous pleural effusion. Lung 187:263–270. doi: 10.1007/s00408-009-9165-3. [DOI] [PubMed] [Google Scholar]
- 2.Zhai K, Lu Y, Shi HZ. 2016. Tuberculous pleural effusion. J Thorac Dis 8:E486–E494. doi: 10.21037/jtd.2016.05.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rufai SB, Singh A, Kumar P, Singh J, Singh S. 2015. Performance of Xpert MTB/RIF assay in diagnosis of pleural tuberculosis by use of pleural fluid samples. J Clin Microbiol 53:3636–3638. doi: 10.1128/JCM.02182-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christopher DJ, Schumacher SG, Michael JS, Luo R, Balamugesh T, Duraikannan P, Pollock NR, Pai M, Denkinger CM. 2013. Performance of Xpert MTB/RIF on pleural tissue for the diagnosis of pleural tuberculosis. Eur Respir J 42:1427–1429. doi: 10.1183/09031936.00103213. [DOI] [PubMed] [Google Scholar]
- 5.Porcel JM, Palma R, Valdés L, Bielsa S, San-José E, Esquerda A. 2013. Xpert® MTB/RIF in pleural fluid for the diagnosis of tuberculosis. Int J Tuberc Lung Dis 17:1217–1219. doi: 10.5588/ijtld.13.0178. [DOI] [PubMed] [Google Scholar]
- 6.Gui X, Xiao H. 2014. Diagnosis of tuberculosis pleurisy with adenosine deaminase (ADA): a systematic review and meta-analysis. Int J Clin Exp Med 7:3126–3135. [PMC free article] [PubMed] [Google Scholar]
- 7.Liang QL, Shi HZ, Wang K, Qin SM, Qin XJ. 2008. Diagnostic accuracy of adenosine deaminase in tuberculous pleurisy: a meta-analysis. Respir Med 102:744–754. doi: 10.1016/j.rmed.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Jiang J, Shi HZ, Liang QL, Qin SM, Qin XJ. 2007. Diagnostic value of interferon-gamma in tuberculous pleurisy: a metaanalysis. Chest 131:1133–1141. doi: 10.1378/chest.06-2273. [DOI] [PubMed] [Google Scholar]
- 9.Greco S, Girardi E, Masciangelo R, Capoccetta GB, Saltini C. 2003. Adenosine deaminase and interferon gamma measurements for the diagnosis of tuberculous pleurisy: a meta-analysis. Int J Tuberc Lung Dis 7:777–786. [PubMed] [Google Scholar]
- 10.Li ZZ, Qin WZ, Li L, Wu Q, Wang YJ. 2015. Accuracy of enzyme-linked immunospot assay for diagnosis of pleural tuberculosis: a meta-analysis. Genet Mol Res 14:11672–11680. doi: 10.4238/2015.September.28.19. [DOI] [PubMed] [Google Scholar]
- 11.Liu F, Gao M, Zhang X, Du F, Jia H, Yang X, Wang Z, Zhang L, Ma L, Wu X, Xie L, Zhang Z. 2013. Interferon-gamma release assay performance of pleural fluid and peripheral blood in pleural tuberculosis. PLoS One 8:e83857. doi: 10.1371/journal.pone.0083857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keng LT, Shu CC, Chen JY, Liang SK, Lin CK, Chang LY, Chang CH, Wang JY, Yu CJ, Lee LN. 2013. Evaluating pleural ADA, ADA2, IFN-γ and IGRA for diagnosing tuberculous pleurisy. J Infect 67:294–302. doi: 10.1016/j.jinf.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Dheda K, van Zyl-Smit RN, Sechi LA, Badri M, Meldau R, Meldau S, Symons G, Semple PL, Maredza A, Dawson R, Wainwright H, Whitelaw A, Vallie Y, Raubenheimer P, Bateman ED, Zumla A. 2009. Utility of quantitative T-cell responses versus unstimulated interferon-γ for the diagnosis of pleural tuberculosis. Eur Respir J 34:1118–1126. doi: 10.1183/09031936.00005309. [DOI] [PubMed] [Google Scholar]
- 14.Aggarwal AN, Agarwal R, Gupta D, Dhooria S, Behera D. 2015. Interferon gamma release assays for diagnosis of pleural tuberculosis: a systematic review and meta-analysis. J Clin Microbiol 53:2451–2459. doi: 10.1128/JCM.00823-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Q, Chen YQ, Qin SM, Tao XN, Xin JB, Shi HZ. 2011. Diagnostic accuracy of T-cell interferon-γ release assays in tuberculous pleurisy: a meta-analysis. Respirology 16:473–480. doi: 10.1111/j.1440-1843.2011.01941.x. [DOI] [PubMed] [Google Scholar]
- 16.Mandel P, Metais P. 1948. Les acides nucléiques du plasma sanguin chez l'homme. C R Seances Soc Biol Fil 142:241–243. [PubMed] [Google Scholar]
- 17.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. 1997. Presence of fetal DNA in maternal plasma and serum. Lancet 350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 18.Patel KM, Tsui DW. 2015. The translational potential of circulating tumour DNA in oncology. Clin Biochem 48:957–961. doi: 10.1016/j.clinbiochem.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Devaney SA, Palomaki GE, Scott JA, Bianchi DW. 2011. Noninvasive fetal sex determination using cell-free fetal DNA: a systematic review and meta-analysis. JAMA 306:627–636. doi: 10.1001/jama.2011.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jian G, Songwen Z, Ling Z, Qinfang D, Jie Z, Liang T, Caicun Z. 2010. Prediction of epidermal growth factor receptor mutations in the plasma/pleural effusion to efficacy of gefitinib treatment in advanced non-small cell lung cancer. J Cancer Res Clin Oncol 136:1341–1347. doi: 10.1007/s00432-010-0785-z. [DOI] [PubMed] [Google Scholar]
- 21.Weerakoon KG, McManus DP. 2016. Cell-free DNA as a diagnostic tool for human parasitic infections. Trends Parasitol 32:378–391. doi: 10.1016/j.pt.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 22.De Vlaminck I, Martin L, Kertesz M, Patel K, Kowarsky M, Strehl C, Cohen G, Luikart H, Neff NF, Okamoto J, Nicolls MR, Cornfield D, Weill D, Valantine H, Khush KK, Quake SR. 2015. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A 112:13336–13341. doi: 10.1073/pnas.1517494112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinakaran V, Rathinavel A, Pushpanathan M, Sivakumar R, Gunasekaran P, Rajendhran J. 2014. Elevated levels of circulating DNA in cardiovascular disease patients: metagenomic profiling of microbiome in the circulation. PLoS One 9:e105221. doi: 10.1371/journal.pone.0105221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. 2016. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev 35:347–376. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cave MD, Eisenach KD, McDermott PF, Bates JH, Crawford JT. 1991. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol Cell Probes 5:73–80. doi: 10.1016/0890-8508(91)90040-Q. [DOI] [PubMed] [Google Scholar]
- 26.Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. 1999. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet 64:218–224. doi: 10.1086/302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu SC, Lee SW, Jiang P, Leung TY, Chan KC, Chiu RW, Lo YM. 2013. High-resolution profiling of fetal DNA clearance from maternal plasma by massively parallel sequencing. Clin Chem 59:1228–1237. doi: 10.1373/clinchem.2013.203679. [DOI] [PubMed] [Google Scholar]
- 28.To EW, Chan KC, Leung SF, Chan LY, To KF, Chan AT, Johnson PJ, Lo YM. 2003. Rapid clearance of plasma Epstein-Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin Cancer Res 9:3254–3259. [PubMed] [Google Scholar]
- 29.Disch J, Oliveira MC, Orsini M, Rabello A. 2004. Rapid clearance of circulating Leishmania kinetoplast DNA after treatment of visceral leishmaniasis. Acta Trop 92:279–283. doi: 10.1016/j.actatropica.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Light RW. 2010. Update on tuberculous pleural effusion. Respirology 15:451–458. doi: 10.1111/j.1440-1843.2010.01723.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


