ABSTRACT
The emergence of new norovirus genotype GII.4 strains is associated with widespread norovirus epidemics. Extended periods of viral shedding can contribute to the epidemic potential of norovirus. To describe the duration of viral shedding in infections with novel emerging GII.4 strains versus infections with previously circulating strains, we performed a prospective cohort study of patients hospitalized with norovirus gastroenteritis during separate winter seasons. Rectal swab samples were obtained at the time of inclusion and weekly during follow-ups. The subgenotype strain was determined from capsid sequences. The outcome was defined by the detection of virus for >14 days (slow clearance) or by the detection of negative samples within 14 days (rapid clearance). Two major epidemic GII.4 strains emerged during the study period, GII.4 New Orleans 2009, in 2010, and GII.4 Sydney 2012, in 2012. From these two seasons, sequences were available from 24 cases where the duration of shedding could be determined. The median age of the patients was 83 years and 50% were women. The majority of patients were infected with virus that clustered with the respective season's epidemic strain (n = 19), whereas 5 patients had previously circulating strains (3 were Den Haag 2006b, in 2010, and 2 were New Orleans 2009, in 2012). Among the patients infected with an epidemic strain, the proportion who shed virus for >14 days was significantly higher (16/19 [84%] versus 1/5 [20%], P = 0.01). In summary, a slow clearance of norovirus from stool was more common in infections with novel epidemic GII.4 strains. This suggests that the average duration of shedding may be longer during seasons when new GII.4 strains have emerged.
KEYWORDS: elderly, gastroenteritis, norovirus, strain, viral shedding
INTRODUCTION
Norovirus is the most common cause of viral gastroenteritis (1). Every two to four years, a major new strain of norovirus genogroup II genotype 4 (GII.4) virus emerges through antigenic drift (2). The emergence of strains with changes in key antigenic sites is associated with more widespread norovirus epidemics (3).
After the resolution of gastroenteritis symptoms, virus can still be detected in stool for a period ranging from 1 week to several months (4, 5). An effective adaptive immune response appears critical for the clearance of norovirus from the gut (6, 7). Although extended periods of viral shedding seem important for the epidemic potential of norovirus (8), the impacts of antigenic drift and infection with different strains on viral clearance are not well described.
The objective of this study was to describe the duration of viral shedding following infection with novel, epidemic norovirus GII.4 strains compared with those of previously circulating strains in hospitalized patients with community-acquired norovirus gastroenteritis.
RESULTS
In this prospective cohort study of patients admitted with norovirus gastroenteritis, we were able to determine the duration of shedding in 34 patients who met the inclusion criteria, had a genogroup II infection, and had consented to participate in the longitudinal follow-up study. Two major new GII.4 strains emerged and spread in the area during the study period: GII.4 New Orleans 2009 (previously called GII.4-2010) in 2010, and GII.4 Sydney 2012 in 2012. Patients enrolled during the 2011 to 2012 winter season (n = 4), when no new GII.4 strain emerged, were excluded. Of the 30 participants who were included during the two epidemic winter seasons (2010 to 2011 and 2012 to 2013), three were infected with non-GII.4 genotypes, and sequencing was not possible in another three. Table 1 shows the characteristics of the 24 patients ultimately included in the analysis. The phylogenetic relations of the obtained GII.4 sequences to reference strains is presented in Fig. 1. The majority of patients were infected with a virus that clustered to the respective season's epidemic strain (14 patients with GII.4 New Orleans 2009 in 2010 to 2011, and 5 patients with GII.4 Sydney 2012 in 2012 to 2013). Five patients had virus that clustered with previously circulating strains (3 patients with GII.4 Den Haag 2006b in 2010 to 2011, and 2 patients with GII.4 New Orleans 2009 in 2012 to 2013).
TABLE 1.
Characteristics of participating patients, gastroenteritis symptoms, and PCR resultsa
| Characteristic | All patients (n = 24) | Patients with infection caused by: |
P value | |
|---|---|---|---|---|
| Previously circulating GII.4 strain (n = 5) | Epidemic GII.4 strain (n = 19) | |||
| Age (years) | 83 (66–86) | 83 (57–86) | 81 (64–87) | >0.3b |
| Women | 12 (50) | 2 (40) | 10 (53) | >0.3c |
| Charlson score (comorbidity) | 1 (0–1) | 0 (0–3) | 1 (0–1) | >0.3b |
| Vesikari score (symptom severity) | 11 (9–13) | 11 (9–13) | 11 (8–13) | >0.3b |
| Disease duration (days) | 4 (2–5) | 3 (2–6) | 4 (2–5) | >0.3b |
| Fever | 6 (25) | 1 (20) | 5 (26) | >0.3c |
| CRPd (mg/L) | 20 (7–28) | 10 (6–111) | 24 (7–28) | >0.3b |
| qPCR cycle threshold, on admission | 21 (19–24) | 24 (19–27) | 20 (19–24) | 0.19b |
Values are median (interquartile range) or n (%).
Mann-Whitney U test.
Fisher's exact test.
CRP, C-reactive protein.
FIG 1.
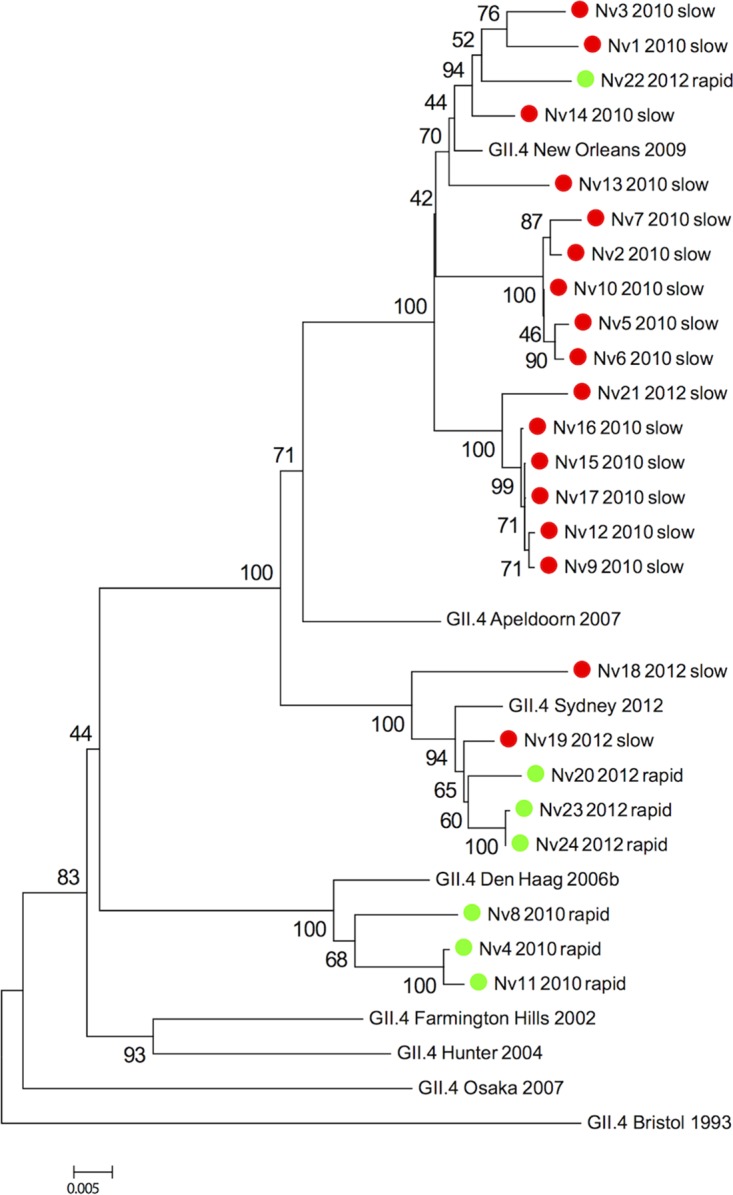
Phylogenetic tree of norovirus GII.4 polymerase-N/S capsid coding region sequences from samples obtained at the time of enrollment in the study. The analysis was performed with 1,000 bootstrap replicates. Scale bar represents the number of substitutions per site. Samples are denoted by number, season, and viral clearance time: green, norovirus cleared in <14 days (rapid); red, norovirus detected for >14 days (slow).
Seven patients cleared the virus within 14 days, of whom five had negative samples on day 7. Norovirus was detected for 14 days in 17 patients, and in 15 of those it was detected for >21 days. The proportion of patients who shed virus for 14 days or more was significantly higher among those infected with a novel epidemic strain (Fig. 2). In a multivariate model that included age, comorbidity score, and GII.4 strain type, infection with a novel strain remained significantly associated with shedding of >14 days in duration (P = 0.02).
FIG 2.
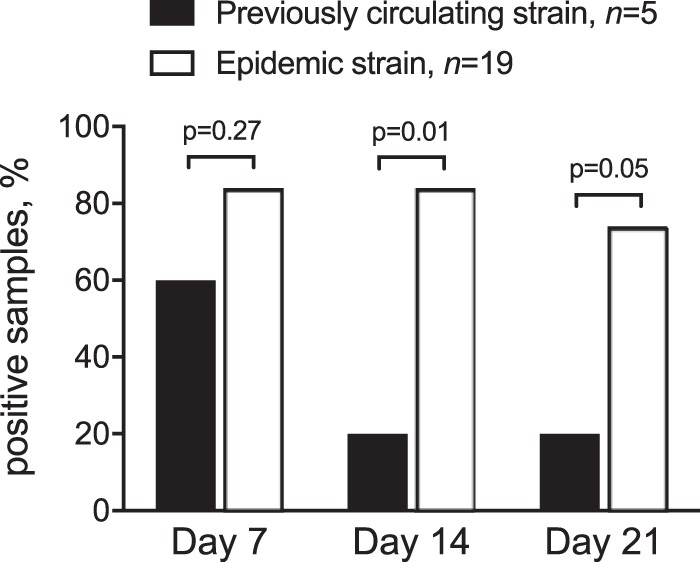
Proportion of patients with virus detected in rectal swab samples during follow-up after norovirus GII.4 infection. All patients were admitted with community-onset gastroenteritis, and norovirus was diagnosed with real-time PCR. Epidemic strain (empty bars) denotes patients with infections caused by a novel emerging GII.4 strain (GII.4 New Orleans 2009 in 2010 to 2011 and GII.4 Sydney 2012 in 2012 to 2013). Previously circulating strain (black bars) denotes patients with infections caused by an older GII.4 strain (GII.4 den Haag 2006b in 2010 to 2011 and GII.4 New Orleans 2009 in 2012 to 2013).
The cycle threshold (CT) values of reactive RT-qPCRs give a semiquantitative estimation of the amount of virus present in the sample. We were able to calculate the increases in CT, or slope, during the first week of follow-up for three patients with previously circulating strains who were still shedding virus on day 7 and for 15 patients with epidemic strains who submitted a positive sample on day 7 (Fig. 3). The CT slope during the first week was significantly steeper in patients with older strains compared with that in patients with epidemic strains, with average CT increases of 9.6 versus 6.2 cycles, respectively (95% confidence interval [CI] for the difference, 0.2 to 6.6, P = 0.04).
FIG 3.
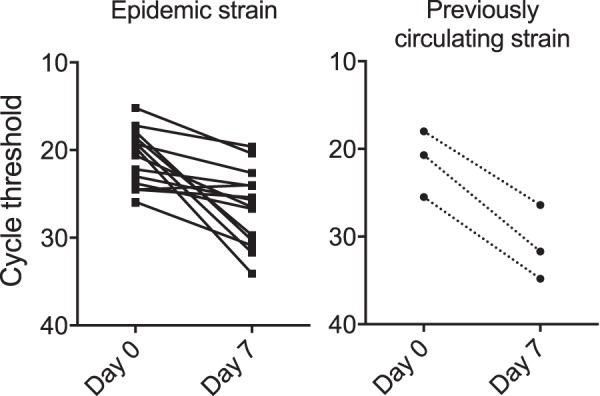
Cycle threshold (CT) values from rectal swab samples on enrollment and on day 7 of the study. The left panel shows the results of samples from patients infected with an epidemic norovirus GII.4 strain and the right panel shows the results from patients infected with a previously circulating GII.4 strain. Patients who did not provide positive samples on day 7 were not included in this analysis. The scales on the y axes are inverted to illustrate that lower CT values represent larger amounts of virus. The slope during the first week was steeper for previously circulating strains than for epidemic strains; the mean CT value increase was 3.4 (95% CI, 0.2 to 6.6) cycles larger, indicating that the virus decline was approximately 1 log10 greater for previously circulating strains.
DISCUSSION
In this longitudinal study of elderly patients, we found that infection with a novel epidemic strain of norovirus GII.4 was associated with a higher risk of viral shedding that lasted for more than 2 weeks.
Most published research on asymptomatic shedding in norovirus infections is based on outbreak investigations or experimental settings (5, 9, 10). Data from outbreak investigations, where all cases are caused by a single subtype strain, restrict the analysis to host factors. By contrast, we studied community-acquired infections in hospitalized patients over the course of separate norovirus seasons, during which new epidemic GII.4 strains emerged. This enabled a comparison, in a homogenous group of subjects, between infections caused by established and novel GII.4 strains. As expected, viruses related to the epidemic strains for each season caused the majority of infections. The emerging GII.4 strain effectively replaced the strain that previously dominated across Europe during both seasons (11, 12). Interestingly, rapid clearance was common among patients infected with previously circulating strains. An infection with a novel epidemic strain, on the other hand, was associated with a longer duration of shedding. A possible explanation is the lack of preexisting immunity to a novel strain that emerges through antigenic drift. An efficient specific immune response is required for the clearance of norovirus (6, 7), and there is limited cross-reactivity between epidemic GII.4 strains (13). A partial nonprotective immunity, derived from previous exposure, may affect the duration of shedding in infections with previously circulating nonepidemic subtypes. This hypothesis is supported by the observation that there were fewer participants with a longer duration of shedding among infected vaccine recipients than among placebo recipients in a comprehensive trial of a norovirus GII.4 vaccine (14). Moreover, in a recently published study of a cohort comparable to ours, infections with older GII.4 strains displayed lower viral loads than infections with strains that emerged during the study period (15). A similar trend, with lower cycle threshold values for the novel strain infections, was noted in our study. However, high initial viral load is not necessarily related to a longer duration of shedding (9). We were also able to relate the time of infection to the emergence of the infecting GII.4 strain in more detail. This enabled us to distinguish infections with emerging strains during an epidemic situation from infections with the same subtype strain in the setting of annually recurring norovirus GII.4 activity.
An alternative explanation for variations in the duration of shedding is that different norovirus strains elicit various immune responses, which has been shown for murine norovirus (16). It is possibly that innate immunity plays a prominent role for the viral clearance of specific strains (17). The virulence of novel subtype strains may also be gradually impaired over time, leading to lower peak viral titers and shorter durations of asymptomatic shedding. Attenuating single-amino-acid substitutions, located in the variable P2 domain of the capsid, have been described in murine norovirus (18). However, the variations in human GII.4 strains are located in a region of importance to the blocking ability of antibodies, which suggests that evasion of specific immunity is an important driving force for GII.4 evolution (19, 20).
Previous longitudinal studies of norovirus outbreaks have reported estimates of between 14 and 28 days for the median duration of shedding, with longer durations typically occurring among elderly patients (21, 22). In accordance with these findings, we found that over two-thirds of patients shed virus for more than 2 weeks, and over 60% shed virus for more than 3 weeks. We did not detect any significant associations between the duration of clearance and clinical variables, although additional weaker associations may be overlooked due to the limited sample size. Infection with a novel GII.4 strain remained independently associated with a slow clearance after adjusting for potential confounders (age and comorbidity score). The size of the study cohort was limited and the study period comprised few epidemic norovirus seasons. Regression models should be interpreted cautiously when few observations are included, and this finding needs to be further analyzed in a larger cohort over a longer period. The multivariate analysis in this study still supports the assumption that no major confounding factor explains the findings.
In this study, we used rectal swab samples instead of stool samples. Although previous studies have reported that rectal swabs contain approximately one log10, or 10 times, less virus than stool samples, swab samples are reliable for the detection of norovirus (23, 24). We did not quantify the fecal viral load in this study, but the cycle threshold (CT) values of reactive samples give a semiquantitative estimation of the amount of virus present. A large amount of virus gives a low CT value and, conversely, small amounts give high CT values. During the first week of follow-up, CT values increased more among patients infected with a previously circulating strain, which corresponds to a larger decrease in viral load. This observation supports the hypothesis that viral clearance was more effective in this group. The mean weekly CT increases of over 6 and 9 cycles indicate more rapid viral clearance than was previously reported (4, 9, 22). However, our models were based on limited sample sizes and should be interpreted cautiously. Moreover, in addition to viral load, crude cycle threshold values are dependent on the amount of fecal material in the sample and the efficiency of individual PCR runs. Rectal swab samples may thus lead to either over- or underestimation of the viral decay rate. Optimally, defined volumes from standardized whole stool samples obtained daily, analyzed together with internal standards of known concentrations, should be used for studies of norovirus shedding dynamics. Such an approach was beyond the scope of our study. Since the endpoint categories (rapid versus slow clearance) were broad and there were no differences in how samples were obtained between groups or over time, we regard the risk that the use of rectal swab samples affected the overall results as low.
We were not able to measure the infectivity of positive samples, and the risk of further spread associated with a long duration of virus shedding is unclear. The recently described culture systems for norovirus give possibilities for addressing this question in future studies (25, 26). We also lacked information on patients' histories of previous episodes of gastroenteritis illness and norovirus infections before their inclusion in the study. We did not have access to strain-specific serology or other methods used to describe or quantify the status of norovirus immunity among the participants. The most important limitation of this study is that few patients were infected with older previously circulating strains. The limited size of the study groups makes the outcome sensitive to random variation. Still, the results were statistically significant in a univariate analysis as well as in a tentative multivariate model. In our opinion, the original study design and findings are relevant if careful judgment is exercised when interpreting the data.
In conclusion, we found that a long duration of norovirus GII.4 shedding appears to be more common in infections with novel epidemic strains than in infections with previously circulating strains. These exploratory findings need to be confirmed in larger cohorts, preferably also including markers of general and strain-specific immunity, to clarify the relationships between emerging norovirus strains, preexisting immunity, and the duration of shedding.
MATERIALS AND METHODS
We performed a prospective cohort study at a 2,000-bed teaching hospital in Western Sweden during three consecutive winter seasons (2010 to 2011, 2011 to 2012, and 2012 to 2013). Adult patients who were admitted to the department of infectious diseases with gastroenteritis symptoms were screened for inclusion in the study. The inclusion criteria were an age of ≥18 years and with vomiting ≥2 times/24 h and/or ≥3 loose stools/24 h for ≤5 days. Patients with immunosuppressive treatment, with concurrent bacterial enteric infection, or who did not have gastroenteritis were excluded. Age, sex, the dates of the first symptom and symptom resolution, the dates of admission and discharge, gastroenteritis symptoms summarized as Vesikari scores (27), and Charlson comorbidity index scores (28) were recorded. Rectal swab samples were obtained at enrollment and on follow-up days 7, 14, and 21 to 28. Samples were collected in a standardized manner with a flocked swab (Copan Italia Spa, Brescia, Italy) and stored at −80°C. Rectal swab samples for bacterial culture were obtained at the time of admission in accordance with the clinical routine.
We used an in-house real-time reverse transcriptase PCR (RT-qPCR) procedure for the detection of norovirus genogroup II, as described previously (29). For sequencing, RT-qPCR-positive samples were amplified using a seminested reverse transcription-PCR of the polymerase-N/S capsid coding region as described previously (30) with minor modifications using the primers listed in Table 2. The resulting chromatograms were visually inspected and contigs were prepared. Sequences were aligned with reference sequences from GenBank using the MacVector software (MacVector, Inc., Cambridge, UK). The accession numbers for reference strains were: X76716 (Bristol 1993), AY502023 (Farmington Hills 2002), DQ078814 (Hunter 2004), EF126965 (Den Haag 2006b), AB434770 (Osaka 2007), KM245072 (Apeldoorn 2007), GU445325 (New Orleans 2009), and KJ196296 (Sydney 2012). The phylogenetic analysis was performed with MEGA 5 software (31) to construct distance matrix trees after bootstrapping to 1,000 replicates.
TABLE 2.
List of primers used for RT-qPCR and sequencing
| Primer | Polarity | Oligonucleotide sequence (5′ to 3′) | Reference |
|---|---|---|---|
| RT-qPCR | |||
| GIIFP | + | TGGAYTTTTAYGTGCCCAG | Nenonen et al. (32) |
| GIIRP | − | CGACGCCATCTTCATTCAC | Nenonen et al. (32) |
| GIIprobe | + | AGCCAGATTGCGATCGCCC | Nenonen et al. (32) |
| Sequencing | |||
| JV12Y | + | ATACCACTATGATGCAGAYTA | Vennema et al. (33) |
| G2SKR | − | CCRCCNGCATRHCCRTTRTACATa | Kojima et al. (34) |
| COG2F | + | CARGARBCNATGTTYAGRTGGATGAG | Yan et al. (35) |
| GII_Fou | + | AGCCAATGTTCAGATGGATGAG | |
| GII_Fin2 | + | TTTGTGAATGAAGATGGCGTC | |
| GII.4_Rin | − | CCCGTTCCATTTCCCATGG | |
| GII.4_R | − | CAGCAAAGAAAGCTCCAGCCAT | |
| Caps2ndpart_F | + | CACGACTGATGGCGTGCT | |
| Caps1stpart_R | − | CTGAAGGTGCAGATGTTGACA |
R = G or A.
For univariate comparisons, we used the Mann-Whitney U test for continuous variables and Fisher's exact test for proportions, with P values (two-tailed) of < 0.05 considered significant. Cycle threshold value slopes were compared with Welch's t test. Multivariate analysis was performed with one-step logistic regression, with likely confounders included a priori as covariables. All calculations were made with SPSS Statistics 22 software (IBM Corp., Armonk, NY).
The study was approved by the regional ethical review board in Gothenburg, Sweden (Dnr 136-10). Written informed consent was obtained from all enrolled patients.
ACKNOWLEDGMENTS
We thank Vilma Molnegren, Anna Rosnäs, and Helén Christiansen for expert technical assistance.
This work was supported by grants from the Regional Research Funds for Western Sweden and the Swedish Society of Medicine.
REFERENCES
- 1.Tam CC, Rodrigues LC, Viviani L, Dodds JP, Evans MR, Hunter PR, Gray JJ, Letley LH, Rait G, Tompkins DS, O'Brien SJ, IID2 Study Executive Committee. 2012. Longitudinal study of infectious intestinal disease in the UK (IID2 study): incidence in the community and presenting to general practice. Gut 61:69–77. doi: 10.1136/gut.2011.238386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siebenga JJ, Vennema H, Renckens B, de Bruin E, van der Veer B, Siezen RJ, Koopmans M. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J Virol 81:9932–9941. doi: 10.1128/JVI.00674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasing ME, Lee BE, Preiksaitis JK, Tellier R, Honish L, Senthilselvan A, Pang XL. 2013. Emergence of a new norovirus GII.4 variant and changes in the historical biennial pattern of norovirus outbreak activity in Alberta, Canada, from 2008 to 2013. J Clin Microbiol 51:2204–2211. doi: 10.1128/JCM.00663-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirby AE, Shi J, Montes J, Lichtenstein M, Moe CL. 2014. Disease course and viral shedding in experimental Norwalk virus and Snow Mountain virus infection. J Med Virol 86:2055–2064. doi: 10.1002/jmv.23905. [DOI] [PubMed] [Google Scholar]
- 5.Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Graham DY. 2008. Norwalk virus shedding after experimental human infection. Emerg Infect Dis 14:1553–1557. doi: 10.3201/eid1410.080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chachu KA, LoBue AD, Strong DW, Baric RS, Virgin HW. 2008. Immune mechanisms responsible for vaccination against and clearance of mucosal and lymphatic norovirus infection. PLoS Pathog 4:e1000236. doi: 10.1371/journal.ppat.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomov VT, Osborne LC, Dolfi DV, Sonnenberg GF, Monticelli LA, Mansfield K, Virgin HW, Artis D, Wherry EJ. 2013. Persistent enteric murine norovirus infection is associated with functionally suboptimal virus-specific CD8 T cell responses. J Virol 87:7015–7031. doi: 10.1128/JVI.03389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milbrath MO, Spicknall IH, Zelner JL, Moe CL, Eisenberg JN. 2013. Heterogeneity in norovirus shedding duration affects community risk. Epidemiol Infect 141:1572–1584. doi: 10.1017/S0950268813000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai CC, Wang YH, Wu CY, Hung CH, Jiang DD, Wu FT. 2013. A norovirus outbreak in a nursing home: norovirus shedding time associated with age. J Clin Virol 56:96–101. doi: 10.1016/j.jcv.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Teunis PF, Sukhrie FH, Vennema H, Bogerman J, Beersma MF, Koopmans MP. 2015. Shedding of norovirus in symptomatic and asymptomatic infections. Epidemiol Infect 143:1710–1717. doi: 10.1017/S095026881400274X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arana A, Cilla G, Montes M, Gomariz M, Perez-Trallero E. 2014. Genotypes, recombinant forms, and variants of norovirus GII.4 in Gipuzkoa (Basque Country, Spain), 2009–2012. PLoS One 9:e98875. doi: 10.1371/journal.pone.0098875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen DJ, Adams NL, Aladin F, Harris JP, Brown DW. 2014. Emergence of the GII-4 norovirus Sydney2012 strain in England, winter 2012–2013. PLoS One 9:e88978. doi: 10.1371/journal.pone.0088978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindesmith LC, Donaldson EF, Baric RS. 2011. Norovirus GII.4 strain antigenic variation. J Virol 85:231–242. doi: 10.1128/JVI.01364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein DI, Atmar RL, Lyon GM, Treanor JJ, Chen WH, Jiang X, Vinje J, Gregoricus N, Frenck RW Jr, Moe CL, Al-Ibrahim MS, Barrett J, Ferreira J, Estes MK, Graham DY, Goodwin R, Borkowski A, Clemens R, Mendelman PM. 2015. Norovirus vaccine against experimental human GII.4 virus illness: a challenge study in healthy adults. J Infect Dis 211:870–878. doi: 10.1093/infdis/jiu497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costantini VP, Cooper EM, Hardaker HL, Lee LE, Bierhoff M, Biggs C, Cieslak PR, Hall AJ, Vinje J. 2016. Epidemiologic, virologic, and host genetic factors of norovirus outbreaks in long-term care facilities. Clin Infect Dis 62:1–10. doi: 10.1093/cid/civ747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu S, Regev D, Watanabe M, Hickman D, Moussatche N, Jesus DM, Kahan SM, Napthine S, Brierley I, Hunter RN III, Devabhaktuni D, Jones MK, Karst SM. 2013. Identification of immune and viral correlates of norovirus protective immunity through comparative study of intra-cluster norovirus strains. PLoS Pathog 9:e1003592. doi: 10.1371/journal.ppat.1003592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, Diamond MS, Virgin HW. 2015. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey D, Thackray LB, Goodfellow IG. 2008. A single amino acid substitution in the murine norovirus capsid protein is sufficient for attenuation in vivo. J Virol 82:7725–7728. doi: 10.1128/JVI.00237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindesmith LC, Costantini V, Swanstrom J, Debbink K, Donaldson EF, Vinje J, Baric RS. 2013. Emergence of a norovirus GII.4 strain correlates with changes in evolving blockade epitopes. J Virol 87:2803–2813. doi: 10.1128/JVI.03106-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen DJ, Gray JJ, Gallimore CI, Xerry J, Iturriza-Gomara M. 2008. Analysis of amino acid variation in the P2 domain of the GII-4 norovirus VP1 protein reveals putative variant-specific epitopes. PLoS One 3:e1485. doi: 10.1371/journal.pone.0001485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aoki Y, Suto A, Mizuta K, Ahiko T, Osaka K, Matsuzaki Y. 2010. Duration of norovirus excretion and the longitudinal course of viral load in norovirus-infected elderly patients. J Hosp Infect 75:42–46. doi: 10.1016/j.jhin.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Tu ET, Bull RA, Kim MJ, McIver CJ, Heron L, Rawlinson WD, White PA. 2008. Norovirus excretion in an aged-care setting. J Clin Microbiol 46:2119–2121. doi: 10.1128/JCM.02198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arvelo W, Hall AJ, Estevez A, Lopez B, Gregoricus N, Vinje J, Gentsch JR, Parashar U, Lindblade KA. 2013. Diagnostic performance of rectal swab versus bulk stool specimens for the detection of rotavirus and norovirus: implications for outbreak investigations. J Clin Virol 58:678–682. doi: 10.1016/j.jcv.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabayiza JC, Andersson ME, Welinder-Olsson C, Bergstrom T, Muhirwa G, Lindh M. 2013. Comparison of rectal swabs and faeces for real-time PCR detection of enteric agents in Rwandan children with gastroenteritis. BMC Infect Dis 13:447. doi: 10.1186/1471-2334-13-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. 2016. Replication of human noroviruses in stem cell-derived human enteroids. Science 353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones MK, Grau KR, Costantini V, Kolawole AO, de Graaf M, Freiden P, Graves CL, Koopmans M, Wallet SM, Tibbetts SA, Schultz-Cherry S, Wobus CE, Vinje J, Karst SM. 2015. Human norovirus culture in B cells. Nat Protoc 10:1939–1947. doi: 10.1038/nprot.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruuska T, Vesikari T. 1990. Rotavirus disease in Finnish children: use of numerical scores for clinical severity of diarrhoeal episodes. Scand J Infect Dis 22:259–267. doi: 10.3109/00365549009027046. [DOI] [PubMed] [Google Scholar]
- 28.Charlson ME, Pompei P, Ales KL, MacKenzie CR. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 40:373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 29.Gustavsson L, Westin J, Andersson LM, Lindh M. 2011. Rectal swabs can be used for diagnosis of viral gastroenteritis with a multiple real-time PCR assay. J Clin Virol 51:279–282. doi: 10.1016/j.jcv.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 30.Nenonen NP, Hannoun C, Svensson L, Toren K, Andersson LM, Westin J, Bergstrom T. 2014. Norovirus GII.4 detection in environmental samples from patient rooms during nosocomial outbreaks. J Clin Microbiol 52:2352–2358. doi: 10.1128/JCM.00266-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nenonen NP, Hannoun C, Olsson MB, Bergstrom T. 2009. Molecular analysis of an oyster-related norovirus outbreak. J Clin Virol 45:105–108. doi: 10.1016/j.jcv.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Vennema H, de Bruin E, Koopmans M. 2002. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J Clin Virol 25:233–235. doi: 10.1016/S1386-6532(02)00126-9. [DOI] [PubMed] [Google Scholar]
- 34.Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods 100:107–114. doi: 10.1016/S0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- 35.Yan H, Yagyu F, Okitsu S, Nishio O, Ushijima H. 2003. Detection of norovirus (GI, GII), sapovirus and astrovirus in fecal samples using reverse transcription single-round multiplex PCR. J Virol Methods 114:37–44. doi: 10.1016/j.jviromet.2003.08.009. [DOI] [PubMed] [Google Scholar]


