ABSTRACT
Viral load monitoring for hepatitis C virus (HCV) is necessary to diagnose infection and monitor response to therapy, but the tests involved are currently confined to specialist institutions. There is a need for a fast, accurate assay with limited operator input to enhance the access to viral load monitoring. We evaluated the quantification of HCV RNA in serum and plasma by the Cepheid Xpert HCV Viral Load assay in comparison to the Abbott RealTime HCV assay. Serum and plasma samples were gathered from HCV-infected individuals at four international sites. These were tested with the Xpert HCV Viral Load assay, and results were compared to quantification by the Abbott RealTime HCV assay. An external quality assessment panel of eight samples was also tested. In total, 614 samples were analyzed in the study, and the qualitative results agreed on the two platforms for 588 (95.8%) samples. Further analysis of 396 samples quantified by both tests showed strong correlation (correlation coefficient r = 0.99) across the quantifiable range, with Bland-Altman plot data showing a mean difference (±1.96 standard deviation) of 0.03 ± 0.44 log10 IU/ml. In the external quality assessment panel, the Xpert HCV Viral Load assay results (quantified in log10 IU per milliliter) were within 1 standard deviation of the target value for all but one sample, which was also similarly misquantified by the Abbott RealTime HCV assay. The Xpert HCV Viral Load assay performs well compared to a market-leading HCV viral load test and should be considered for instances where rapid near-to-patient testing is required.
KEYWORDS: HCV RNA, hepatitis C virus, viral load
INTRODUCTION
Hepatitis C virus (HCV) is a leading burden on human health worldwide. Recent estimates suggest that there are over 185 million individuals living with chronic HCV infection and that HCV causes over 350,000 deaths every year (1, 2). HCV is a blood-borne virus that infects the liver; chronic infection causes liver disease and extrahepatic manifestations, cirrhosis, hepatocellular carcinoma, and death.
Detection of HCV RNA is necessary to diagnose infection. In addition, determination of HCV viral load is recommended prior to beginning antiviral therapy to establish a baseline, and subsequent monitoring is recommended to ascertain the response to treatment, particularly to assess the sustained virological response (3, 4). Currently, HCV viral load monitoring must be done in specialist centers, with a turnaround time of up to 1 week due to the need to batch samples (5, 6). The Xpert HCV Viral Load assay (Cepheid, Sunnyvale, CA) is a recently CE-marked in vitro diagnostic (IVD) test for HCV viral load determination in plasma and serum. Assay components are contained within a single-use cartridge which performs RNA extraction, reverse transcription, and real-time PCR targeting the 5′ untranslated region (UTR) of the HCV genome. The test cartridge contains internal controls to ensure accurate test performance and to quantify HCV viral load; results are interpreted by proprietary software. We describe a multicenter study assessing the performance of the Cepheid Xpert HCV Viral Load assay compared to that of the reference Abbott RealTime HCV assay.
(These results were presented as a poster at the European Society for Clinical Virology Meeting on 9 to 12 September 2015 in Edinburgh, United Kingdom [abstract 1530] [7]).
RESULTS
Testing of clinical samples.
In total, 636 samples from HCV-infected individuals were tested in this study. Thirty-three (5.2%) samples generated indeterminate results with the Xpert HCV Viral Load assay; no particular association was seen with genotype (6 genotype 1, 1 genotype 2, 1 genotype four, and 25 of unknown genotype). Assays of 11 samples were repeated and produced valid results; the remaining 22 samples had insufficient volume for repeat assays and so were excluded from analysis. Of the 614 samples analyzed, 588 (95.8%) had qualitative results that agreed on the two platforms (disagreement was seen in 8 genotype 1, 4 genotype 3, 3 genotype 4, and 11 unknown-genotype samples) (Table 1). The eight samples quantified by the Abbott RealTime HCV assay but not by the Xpert HCV Viral Load assay had mean ± standard deviation (SD) viral load values of 16.0 ± 4.6 IU/ml, and the four quantified by the Xpert HCV Viral Load assay but not by the Abbott RealTime HCV assay had mean ± SD viral load values of 16.0 ± 3.5 IU/ml. There were also 14 samples with detectable but unquantifiable HCV on one platform and with HCV not detected on the other; these discrepancies most likely represent Poisson distributions of low-level target nucleic acid.
TABLE 1.
Qualitative results of Xpert HCV Viral Load and Abbott RealTime HCV assays
| Assay and result | No. of samples with indicated Abbott RealTime HCV result |
Total no. of samples | ||
|---|---|---|---|---|
| Virus detected, ≥12 IU/ml | Virus detected, <12 IU/ml | Virus not detected | ||
| Xpert HCV Viral Load | ||||
| Virus detected, ≥10 IU/ml | 396 | 4 | 0 | 400 |
| Virus detected, <10 IU/ml | 8 | 24 | 13 | 45 |
| Virus not detected | 0 | 1 | 168 | 169 |
| Total | 404 | 29 | 181 | 614 |
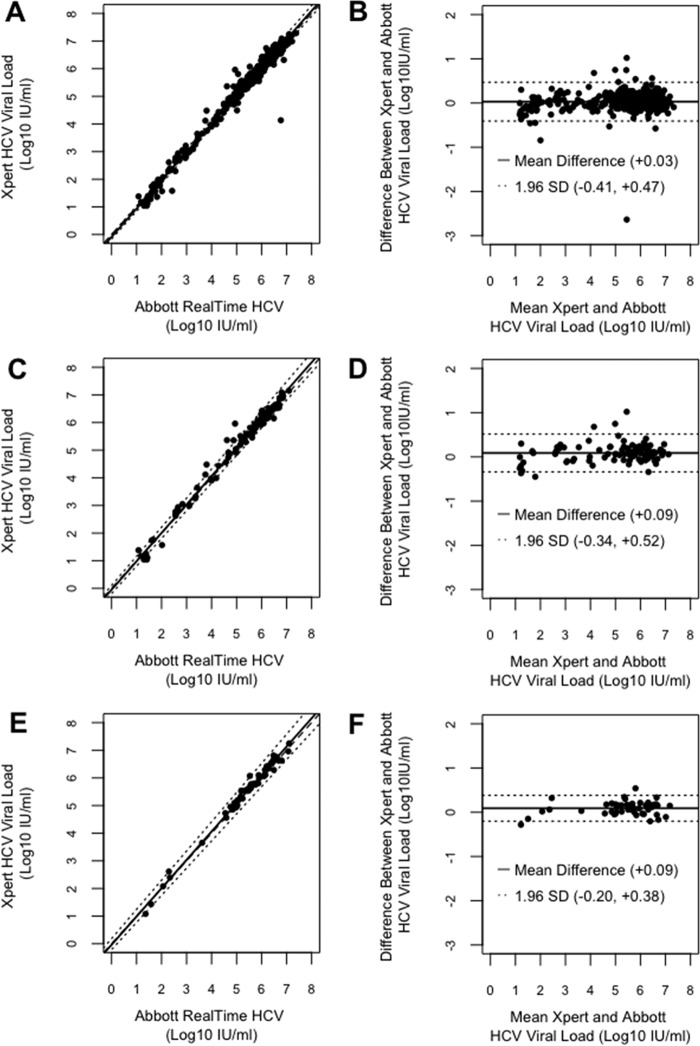
Further analysis of 396 quantified samples (including 169 with known genotype) by Deming regression (Fig. 1A) showed that the results from the two platforms correlated strongly across the quantifiable range (correlation coefficient [r], 0.99; Deming regression equation, Y = 1.02X + 0.09). The correlation between the two assays was strong in analyzing for genotype 1 and 3 samples separately (for genotype 1, r = 0.99, Deming regression equation Y = 1.03X + 0.05; for genotype 3, r = 0.99, Deming regression equation Y = 1.02X + 0.04) (Fig. 1C and E). The level of HCV quantification was higher with the Xpert HCV Viral Load assay in 232 (59.6%) samples, and the overall means ± 1.96 SD difference of results between the assays was 0.03 ± 0.44 log10 IU/ml, with a range of −2.64 to + 1.02 log10 IU/ml (Fig. 1B). A discrepancy in quantification between the two assays was detected for 13 (2.1%) samples that fell outside the estimated agreement interval, defined as ± 1.96 SD from the overall mean difference (Fig. 1B and Table 2). There was no remaining sample available for these to be investigated further. Further details of the discordant samples are shown in Table 2. The agreement between the results of the assays for genotypes 1 and 3 alone was also acceptable (Fig. 1D and F). Additionally, 100 HCV-negative blood donor samples all produced undetectable results when tested with the Xpert HCV Viral Load assay, indicating 100% specificity (95% confidence interval, 97.0% to 100.0%).
FIG 1.
Deming regression and Bland-Altman plot analysis of HCV quantification by the Abbott RealTime HCV and Xpert HCV Viral Load assays. (A) Deming regression of 396 samples quantified by both assays. (B) Bland-Altman plot of 396 samples quantified by both assays. (C) Deming regression of 92 HCV genotype 1 samples quantified by both assays. (D) Bland-Altman plot of 92 HCV genotype 1 samples quantified by both assays. (E) Deming regression of 51 HCV genotype 3 samples quantified by both assays. (F) Bland-Altman plot of 51 HCV genotype 3 samples quantified by both assays. The Deming regression plot shows the identity line (dashed line) and the Deming fit with a 95% confidence interval (black line and two dotted lines, respectively). The Bland-Altman plot shows the difference between the two assays in HCV quantification results plotted as a function of the mean of the two; the mean difference is highlighted by the black line, and dotted lines indicate ± 1.96 standard deviation.
TABLE 2.
Samples with discrepancy between assays
| Study no. | Storage | HCV Subtype(s) | Xpert HCV Viral Load result (log10 IU/ml) | Abbott RealTime HCV result (log10 IU/ml) | Difference (log10 IU/ml) |
|---|---|---|---|---|---|
| HCC229065 | Frozen | 1b | 5.96 | 4.94 | 1.02 |
| HCC207168 | Frozen | 1a | 5.36 | 4.61 | 0.75 |
| HCC111006 | Frozen | Unknown | 5.81 | 5.06 | 0.75 |
| HCC229207 | Fresh | 1b | 4.48 | 3.80 | 0.68 |
| HCV167007 | Frozen | 5a | 6.74 | 6.18 | 0.56 |
| W7897 | Frozen | 3a | 6.07 | 5.53 | 0.54 |
| HCC229071 | Frozen | 1b | 5.36 | 4.88 | 0.48 |
| W7930 | Frozen | 1b | 1.57 | 2.02 | −0.45 |
| HCC207027 | Frozen | Unknown | 1.36 | 1.82 | −0.46 |
| W7820 | Frozen | 2a/2c | 4.49 | 5.02 | −0.53 |
| HCC248052 | Frozen | Unknown | 6.31 | 6.89 | −0.58 |
| HCC111176 | Frozen | Unknown | 1.58 | 2.42 | −0.84 |
| HCC248043 | Frozen | Unknown | 4.13 | 6.77 | −2.64 |
EQA panel testing.
The external quality assessment (EQA) panel contained one HCV-negative sample, two HCV genotype 1b samples, and five HCV genotype 3a samples (Table 3). These were run twice using the Xpert HCV Viral Load assay and once using the Abbott RealTime HCV assay. A single sample tested using one Xpert HCV Viral Load assay run and the Abbott RealTime HCV assay run was quantified outside the target range, due to underquantification of genotype 1b HCV. All other samples were called within the target range by both platforms (Table 3).
TABLE 3.
Performance of Xpert HCV Viral Load and Abbott RealTime HCV assays with EQA samples
| Sample no. | Sample content or result | Target result |
Abbott RealTime HCV result |
Xpert HCV Viral Load result |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Run 1 |
Run 2 |
||||||||
| Valuea | SDa | Valuea | Within target range | Valuea | Within target range | Valuea | Within target range | ||
| HCVRNA14B-05 | HCV type 1b | 4.25 | 0.34 | 4.16 | Yes | 4.24 | Yes | 4.46 | Yes |
| HCVRNA14B-02 | HCV type 1b | 3.24 | 0.24 | 2.97 | No | 2.93 | No | 3.20 | Yes |
| HCVRNA14B-01 | HCV type 3a | 3.96 | 0.20 | 3.84 | Yes | 3.96 | Yes | 3.96 | Yes |
| HCVRNA14B-08 | HCV type 3a | 2.93 | 0.25 | 2.82 | Yes | 2.81 | Yes | 2.99 | Yes |
| HCVRNA14B-06 | HCV type 3a | 2.10 | 0.28 | 1.92 | Yes | 2.09 | Yes | 2.00 | Yes |
| HCVRNA14B-07 | HCV type 3a | 3.82 | 0.23 | 3.79 | Yes | 3.89 | Yes | 3.84 | Yes |
| HCVRNA14B-03 | HCV type 3a | 2.86 | 0.26 | 2.81 | Yes | 2.92 | Yes | 2.89 | Yes |
| HCVRNA14B-04 | Negative | NDb | ND | ND | ND | ||||
Log10 IU/ml.
ND, not detected.
DISCUSSION
This study showed that the Xpert HCV Viral Load assay accurately quantifies HCV viral load compared to a leading commercial assay used worldwide. Also, the Xpert HCV Viral Load assay performed strongly in a well-respected EQA panel. These performance characteristics, aligned with limited hands-on time (5 min), short run time (105 min), random access testing, and the uncomplicated operator input of the Xpert HCV Viral Load assay, suggest that it can have an important role in diagnosis of HCV infection and HCV viral load monitoring. Our findings confirm those of Gupta et al. (8), who tested 118 plasma samples seropositive for HCV with the Xpert HCV Viral Load and Abbott RealTime HCV assays, Bland-Altman analysis showed a mean difference of 0.04 log10 IU/ml (1.96 SD, −0.42 to 0.49 log10 IU/ml), a result very similar to those reported in our multicenter study. We believe that the Xpert HCV Viral Load assay could be used for routine viral load monitoring as well as in situations such as organ donor testing or needle stick injuries, where a rapid evaluation of HCV status is required, and could even be used in a near-patient capacity in HCV clinics or prisons (9).
Although we found the Xpert HCV Viral Load assay result to be slightly higher overall than the Abbott RealTime HCV result (≤2 log10 IU/ml), the Xpert HCV Viral Load assay result appears to have been lower (Fig. 1B), with 3 of 29 samples falling outside a 1.96 SD from the mean difference. Also, the Abbott RealTime HCV assay quantified HCV in 8 samples that were detected by the Xpert HCV Viral Load assay at levels below the limit of quantification, compared to 4 samples quantified by the Xpert HCV Viral Load assay that were detected at levels below the limit of quantification by the Abbott assay (Table 1). However, no samples were quantified by the Abbott RealTime HCV assay but not detected by the Xpert HCV Viral Load assay. We believe that this finding will have limited impact in clinical use, as individuals generally have viral loads of >2 log10 IU/ml at diagnosis (8, 10), and the Xpert HCV Viral Load assay did not report any samples falsely negative in our sample collection. Overall, the Xpert HCV Viral Load assay detected HCV in 445 samples compared to 433 samples with HCV detected by the Abbott RealTime HCV assay (Table 1).
Results from one study site showed HCV viral loads significantly lower than those from the other sites by analysis of variance (ANOVA). That site was in France, where the level of access to antiviral treatment is high; the lower viral loads seen at that site were likely due to higher treatment rates as opposed to a methodological difference (11, 12). One drawback of this work was that most samples were tested retrospectively after being frozen, although samples were tested on each platform from the same freeze-thaw cycle. Another drawback was that the genotypes were not available for all samples, although we included 214 samples of 7 different genotypes covering those seen throughout the world (Table 4) (13). Eight of these samples, including one of the two genotype 5a samples, fell outside the agreement interval by Bland-Altman analysis (Fig. 1B and Table 2). Prior to implementation, laboratories should validate that the Xpert HCV Viral Load assay reliably quantifies the HCV types circulating locally. Further studies investigating the performance of the Xpert HCV Viral Load assay in samples with known genotypes, particularly genotype 5a, are warranted.
TABLE 4.
Known HCV genotypes tested
| Genotype and subgenotype | n | % |
|---|---|---|
| 1 | 110 | 51.4 |
| 1a | 47 | 22.0 |
| 1b | 33 | 15.4 |
| 1d | 1 | 0.5 |
| Subtype unknown | 29 | 13.6 |
| 2 | 15 | 7.0 |
| 2a/c | 6 | 2.8 |
| 2b | 3 | 1.4 |
| 2l | 1 | 0.5 |
| Subtype unknown | 5 | 2.3 |
| 3 | 61 | 28.5 |
| 3a | 47 | 22.0 |
| Subtype unknown | 14 | 6.5 |
| 4 | 21 | 9.8 |
| 4f | 1 | 0.5 |
| 4g | 1 | 0.5 |
| 4h | 1 | 0.5 |
| 4r | 1 | 0.5 |
| Subtype unknown | 17 | 7.9 |
| 5 | 2 | 0.9 |
| 5a | 2 | 0.9 |
| 6 | 4 | 1.9 |
| Subtype unknown | 4 | 1.9 |
| 1 or 6 | 1 | 0.5 |
| Total | 214 | 100.0 |
Disadvantages of the Xpert HCV Viral Load assay include the high cost per test cartridge (€35), the lack of external positive and negative controls in the reagent kit, and the relatively high sample input volume (1 ml). Also, one test cartridge in our study leaked after sample addition. Although a rare occurrence, this is a potentially serious issue. If the Xpert HCV Viral Load assay is to be used outside the laboratory setting, it will still be necessary to ensure that operators understand and adhere to appropriate risk assessments for handling infectious samples.
Overall, the Xpert HCV Viral Load assay had performance comparable to that of a market-leading assay in testing samples from four countries and an international EQA panel. The Xpert HCV Viral Load assay has the potential to change how viral load monitoring is performed for HCV in the future.
MATERIALS AND METHODS
Clinical samples.
The study was a method comparison of the Xpert HCV Viral Load assay (Cepheid, Sunnyvale, CA) to the Abbott RealTime HCV assay (Abbott Molecular, Des Plaines, IL) in fresh and frozen human plasma (EDTA) and serum specimens. Remnant samples were collected at four health care sites in Europe and the United States after standard-of-care HCV viral load testing. Frozen (≤70°C) samples were tested within 6 months of freezing and were tested on both platforms from the same freeze-thaw cycle; after thawing, samples were held at 4°C for a maximum of 6 h prior to testing. Fresh samples were held at 4°C and tested within 72 h of collection. Samples were from known HCV-positive adults (18 to 89 years old) and were collected per the assay manufacturer's instructions. Different technicians performed sample randomization and testing, and the individuals running the Xpert HCV Viral Load assay were blind to the Abbott RealTime HCV assay result. The study methods were in accordance with local ethical guidelines, and written informed consent was not deemed necessary at each study site. Samples were processed with the Abbott RealTime HCV assay on an Abbott m2000 instrument according to the manufacturer's instructions; all sites are accredited to perform this test. For the Xpert HCV Viral Load assays, 1 ml of specimen was added to a test cartridge and the cartridge was loaded into a GeneXpert instrument. Three sites used the GeneXpert Dx XVI system, and one site used a GeneXpert Infinity 80 system. Acrometrix (Benicia, CA) external controls (HCV high-level, mid-level, and negative controls) were tested each day. The linear range of the Xpert HCV Viral Load assay is 10 IU/ml to 108 IU/ml; the linear range of the Abbott RealTime HCV assay is 12 IU/ml to 108 IU/ml. Results were reported as follows: HCV detected (with the associated quantitation reported in IU per milliliter); HCV not detected; HCV detected at <12 IU/ml (Abbott RealTime HCV) or <10 IU/ml (Xpert HCV Viral Load); and HCV quantified at ≥12 IU/ml (Abbott RealTime HCV) or ≥10 IU/ml (Xpert HCV Viral Load). Samples with error, invalid, or no-result outputs on the Xpert HCV Viral Load were classed as indeterminate.
A total of 589 samples were collected between August 2013 and October 2014. Of these, four were not collected in line with protocol, one leaked after loading of the Xpert HCV Viral Load assay cartridge, and one frozen sample was thawed for >24 h before Xpert testing; all these were excluded, leaving 583 samples (558 frozen, 25 fresh) for testing. The HCV genotype classification was available for samples from two participating centers; a further 53 frozen plasma samples of various genotypes were obtained from Diagnostic Laboratory Services (DLS; Los Osos, CA) and from one participating center (Table 4). Genotypes were determined in independent accredited centers by different methods (Versant HCV Genotype Assay [Innogenetics, Ghent, Belgium], Trugene HCV Genotyping kit [Siemens, Erlangen, Germany], RealTime HCV Genotype [Abbott], Hepatitis C Viral RNA Genotype LiPA [Quest Diagnostics, London, United Kingdom], AmpliSens HCV Genotype-FRT [InterLabService, Moscow, Russia], and in-house reverse transcription-PCR [RT-PCR] and sequencing). The 53 additional samples were collected between 2003 and 2016 and were stored as frozen samples as described above. Also, specimens were collected from 100 blood donors who were confirmed to be HCV seronegative by U.S. Food and Drug Administration-licensed antibody and nucleic acid tests.
External quality assessment panel testing.
Quality Control for Molecular Diagnostics (Glasgow, United Kingdom) is an external quality assessment (EQA) provider for routine clinical molecular diagnostic laboratories. The HCV panel contains eight simulated plasma samples containing different genotypes and target RNA concentrations (Table 3). The target range was defined as the mean ± one standard deviation (SD) of results from all respondents (n = 100) corresponding to the distribution (reference number HCVRNA14B).
Statistical analysis.
Descriptive statistics are presented as means ± SD. We used Deming regression to compare the results of quantification by the two assays, using the Method Comparison Regression (MCR) R package (Roche, Basel, Switzerland). Deming regression takes account of measurement errors for both methods and so is suited to comparing laboratory assays. We also use Bland-Altman plot analysis to highlight the differences between the assays, where the differences between the two assays are plotted against the mean of the results from the two assays. Bland-Altman analysis was performed with the BlandAltmanLeh R package (University of Greifswald, Griefswald, Germany).
ACKNOWLEDGMENT
We thank Cepheid for financially supporting this study.
REFERENCES
- 1.Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. 2006. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol 45:529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 2.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. 2013. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 3.EASL. 2017. European Association for the Study of the Liver recommendations on treatment of hepatitis C. 2016. J Hepatol 66:153–194. doi: 10.1016/j.jhep.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 4.AASLD-IDSA. 27 September 2016. Recommendations for testing, managing, and treating hepatitis C. www.hcvguidelines.org Accessed 25 November 2016.
- 5.Michelin BDA, Muller Z, Stelzi E, Marth E, Kessler HH. 2007. Evaluation of the Abbott RealTime HCV assay for quantitative detection of hepatitis C virus RNA. J Clin Virol 38:96–100. doi: 10.1016/j.jcv.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Vermehren J, Yu ML, Monto A, Yao JD, Anderson C, Bertuzis R, Scheider G, Sarrazin C. 2011. Multi-center evaluation of the Abbott RealTime HCV assay for monitoring patients undergoing antiviral therapy for chronic hepatitis C. J Clin Virol 52:133–137. doi: 10.1016/j.jcv.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 7.McHugh M, Wu A, Pawlotsky JM, Chevaliez S, Hallin M, Templeton K. 2015. Evaluation of the Cepheid Xpert® HCV Viral Load assay, abstr 1530, poster 2. Abstr Eur Soc Clin Virol Meet, Edinburgh, United Kingdom, 9 to 12 September 2015. [Google Scholar]
- 8.Gupta E, Agarwala P, Kumar G, Maiwall R, Sarin SK. 2017. Point-of-care testing (POCT) in molecular diagnostics: performance evaluation of GeneXpert HCV RNA test in diagnosing and monitoring of HCV infection. J Clin Virol 88:46–51. doi: 10.1016/j.jcv.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Rich JD, Beckwith CG, Macmadu A, Marshall BDL, Brinkley-Rubinstein L, Amon JJ, Milloy MJ, King MRF, Sanchez J, Atwoli L, Altice FL. 2016. Clinical care of incarcerated people with HIV, viral hepatitis, or tuberculosis. Lancet 388:1103–1114. doi: 10.1016/S0140-6736(16)30379-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alonso R, Pérez-García F, Ampuero D, Reigada E, Bouza E. 2017. New direct-acting antivirals for patients with chronic HCV infection: can we monitor treatment using an HCV core antigen assay? Diagn Microbiol Infect Dis 87:243–246. doi: 10.1016/j.diagmicrobio.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Geri G, Maynard M, Rosenthal E, Fontaine H, Lacombe K, Slama L, Goujard C, Loustaud-Ratti V, Bergmann JF, Morlat P, Vittecoq D, Alric L, Cacoub P, for the GERMIVIC Group. 2014. Care of hepatitis C virus infection in France: modifications in three consecutive surveys between 1995 and 2010. Liver Int 34:1349–1357. doi: 10.1111/liv.12388. [DOI] [PubMed] [Google Scholar]
- 12.Razavi H, Waked I, Sarrazin C, Myers RP, Idilman R, Calinas F, Vogel W, Mendes Correa MC, Hézode C, Lázaro P, Akarca U, Aleman S, Balık I, Berg T, Bihl F, Bilodeau M, Blasco AJ, Brandão Mello CE, Bruggmann P, Buti M, Calleja JL, Cheinquer H, Christensen PB, Clausen M, Coelho HS, Cramp ME, Dore GJ, Doss W, Duberg AS, El-Sayed MH, Ergör G, Esmat G, Falconer K, Félix J, Ferraz ML, Ferreira PR, Frankova S, García-Samaniego J, Gerstoft J, Giria JA, Gonçales FL Jr, Gower E, Gschwantler M, Guimarães Pessôa M, Hindman SJ, Hofer H, Husa P, Kåberg M, et al. 2014. The present and future disease burden of hepatitis C virus (HCV) infection with today's treatment paradigm. J Viral Hepat 21:34–59. doi: 10.1111/jvh.12248. [DOI] [PubMed] [Google Scholar]
- 13.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. 2015. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 61:77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]