ABSTRACT
With the emerging Zika virus (ZIKV) epidemic, accessible real-time reverse transcription-PCR (rRT-PCR) assays are needed to streamline testing. The commercial Altona Diagnostics RealStar ZIKV rRT-PCR test kit (Altona PCR) has been approved for emergency use authorization by the U.S. FDA. Our aim was to verify the Altona PCR by comparing it to the CDC-designed dual-target ZIKV rRT-PCR reference assay (reference PCR) and describe the demographics of patients tested for ZIKV by rRT-PCR in Ontario, Canada. A large set of clinical specimens was tested for ZIKV by the Altona PCR and the reference PCR. Positive or equivocal specimens underwent PCR and Sanger sequencing targeting the ZIKV NS5 gene. A total of 671 serum specimens were tested by the reference PCR: 58 (8.6%) were positive, 193 (28.8%) were equivocal, and 420 (62.6%) were negative. Ninety percent of the reference PCR-positive patients were tested in the first 5 days after symptom onset. The Altona PCR was performed on 284/671 specimens tested by the reference PCR. The Altona PCR was positive for 53/58 (91%) reference PCR-positive specimens and 16/193 (8%) reference PCR-equivocal specimens; the ZIKV NS5 PCR was positive for all 68 Altona PCR-positive specimens and negative for all 181 Altona PCR-negative specimens that underwent the NS5 PCR. The Altona PCR has very good sensitivity (91%) and specificity (97%) compared to the reference PCR. The Altona PCR can be used for ZIKV diagnostic testing and has less extensive verification requirements than a laboratory-developed test.
KEYWORDS: diagnostic testing, PCR, Zika virus, flavivirus
INTRODUCTION
Zika virus (ZIKV) is a single-stranded, positive-sense RNA flavivirus (1) transmitted mostly by Aedes mosquitoes. In 2007, 60 years after its discovery in Uganda, ZIKV was for the first time recognized to be responsible for a febrile outbreak in Yap, Micronesia (2). ZIKV then spread eastward in the South Pacific Islands (3–5), before reaching Brazil at the end of 2014 (6). Since then, millions of persons have been infected, and active ZIKV transmission has been documented in all countries of North, Central, and South America except Chile, Uruguay, and Canada.
ZIKV diagnostic testing mainly relies on serology and reverse transcription-PCR (RT-PCR). IgM antibodies usually appear during the first week after symptom onset, and their appearance is rapidly followed by the appearance of IgG antibodies (7, 8). Because of cross-reactivity with other flaviviruses, such as dengue virus (DENV), a positive IgM serology requires confirmation with a plaque reduction neutralization test (PRNT) (9). RT-PCR can be performed on many specimens, such as blood, urine, saliva, and amniotic fluid (10), and ZIKV RNA detection by RT-PCR is considered the hallmark of acute infection, even if prolonged shedding has been reported, especially in semen (11, 12). The first RT-PCR assay widely used for ZIKV diagnosis during this outbreak was the dual-target real-time RT-PCR assay designed by the U.S. Centers for Disease Control and Prevention (CDC); this reference assay is based on the ZIKV strain responsible for the 2007 outbreak in Micronesia and targets the ZIKV envelope (E) and premembrane (prM) genes (7). As the clinical presentation of individuals with ZIKV infection may be similar to that of individuals with infections caused by other arboviruses, such as DENV and chikungunya virus (CHIKV), several multiplex assays have been developed, including the recent CDC-designed trioplex assay that has received emergency use authorization (EUA) from the U.S. FDA (13–15). Because of the need for easily accessible real-time RT-PCR (rRT-PCR) assays to streamline testing, commercial assays have been developed, including the Altona Diagnostics RealStar Zika virus rRT-PCR test kit (referred to here as the Altona PCR; Altona Diagnostics GmbH, Hamburg, Germany), which has been approved for EUA by the U.S. FDA for testing of blood and urine (16). The main aim of our study was to evaluate and verify the Altona rRT-PCR for routine ZIKV testing by comparing it to the CDC-designed reference assay with a large subset of clinical specimens with the aim of implementing the Altona ZIKV rRT-PCR assay at the Public Health Ontario Laboratory (PHOL), Ontario, Canada. The secondary aims were to describe the demographics of the patients tested for ZIKV by rRT-PCR in Ontario and evaluate the impact of the time following symptom onset to specimen collection on test positivity.
RESULTS
Seven hundred twenty-nine clinical specimens from 692 patients that met the criteria for ZIKV RT-PCR testing were submitted to PHOL. The median age of the patients was 33.9 years (interquartile range [IQR], 28.5 to 44.3 years). Sixty percent (414/681) of the patients were female, and 19% (55/297) of females for which the information was available were pregnant. Among the 729 specimens tested by the reference PCR, 671 (92%) were serum specimens and 50 (7%) were urine specimens; the remaining 8 specimens were cerebrospinal fluid (CSF; n = 3), placenta (n = 2), nasopharyngeal swab (n = 1), umbilical cord blood (n = 1), and autopsy fetal lung (n = 1) specimens.
Serum.
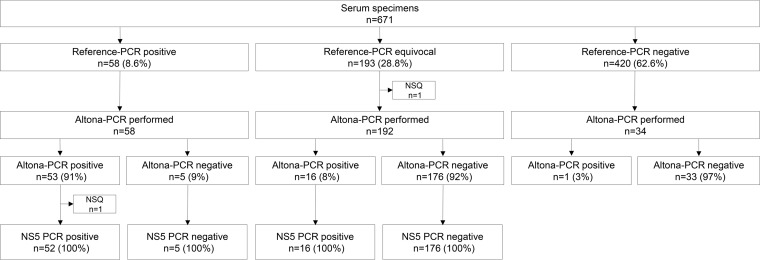
The reference PCR was positive for 58/671 (8.6%) blood specimens tested, equivocal for 193 (28.8%), and negative for 420 (62.6%) (Fig. 1). For reference PCR-positive serum specimens, median cycle threshold (CT) values were 36.5 (IQR, 33.5 to 37.0) for the prM gene target and 35.9 (IQR, 32.9 to 37.0) for the E gene target. Among the 193 reference PCR-equivocal serum specimens, the prM gene target was detected in 67 (34.7%) specimens (median CT value, 38.3 [IQR, 37.7 to 39.0]), whereas the E gene target was detected in 145 (75.1%) specimens (median CT value, 38.5 [IQR, 37.8 to 39.2]). DENV and CHIKV PCRs were performed on 669/671 serum specimens (specimen volumes were insufficient in two cases). The DENV PCR was positive for 7 specimens and negative for 662 specimens, whereas the CHIKV PCR was positive for 2 specimens, indeterminate for 1 specimen, and negative for 666 specimens.
FIG 1.
Distribution of serum specimens according to ZIKV reference PCR, Altona PCR, and NS5 gene PCR results. ZIKV, Zika virus; reference PCR, 2-step dual-target real-time reverse transcription-PCR, designed by the U.S. Centers for Disease Control and Prevention, targeting the Zika virus envelope (E) and premembrane (prM) genes; NS5, nonstructural protein 5; NSQ, insufficient quantity for testing.
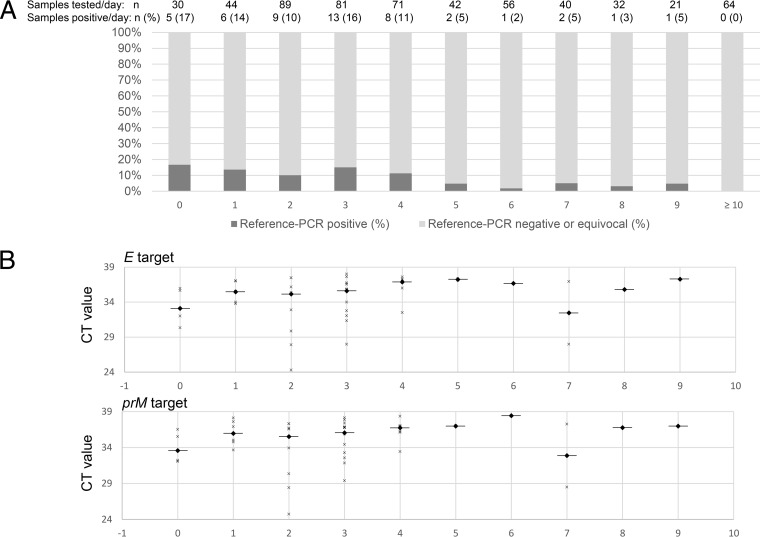
Among patients for whom the date of symptom onset was available, the median time between symptom onset and specimen collection was 3 days (IQR, 2 to 4 days); 89.6% (43/48) of reference PCR-positive specimens were collected during the first 5 days since symptom onset; the proportion of positive specimens remained stable from day 0 to day 4 and then rapidly decreased (Fig. 2A). The distribution of the CT values for the E and prM gene targets by the number of days since symptom onset is shown in Fig. 2B.
FIG 2.
(A) Serum ZIKV reference PCR positivity by day following symptom onset. (B) CT values of E and prM gene targets by day following symptom onset. ZIKV, Zika virus; reference PCR, 2-step dual-target real-time reverse transcription-PCR, designed by the U.S. Center for Disease Control and Prevention, targeting the Zika virus envelope (E) and premembrane (prM) genes; CT, cycle threshold. The horizontal bars represent median CT values.
The Altona PCR was performed on 284 (42%) serum specimens selected from among the 671 serum specimens previously tested by the reference PCR. Among the 58 reference PCR-positive specimens, the Altona PCR was positive for 53/58 (91%) and negative for 5 (9%) (Fig. 1); the results of the NS5 PCR showed 100% positive percent agreement with those of the Altona PCR among the 52 Altona PCR-positive (and reference PCR-positive) specimens with a sufficient quantity to undergo the NS5 gene PCR. Among the 193 reference PCR-equivocal specimens, the Altona PCR was positive for 16 (8%). Among the reference PCR-negative specimens, the Altona PCR was performed on 34 specimens, of which only 1 was positive.
Among the 284 serum specimens selected for testing by the Altona PCR, 70 were positive; the median CT value was 34.4 (IQR, 31.9 to 35.8). Of these 70 Altona PCR-positive specimens, the reference PCR was positive for 53 (76%), equivocal for 16 (23%), and negative for 1 (1%) (Fig. 1). The NS5 PCR was positive for 68/68 (100%) of the Altona PCR-positive specimens with a sufficient quantity to undergo the NS5 PCR. Among the 214 Altona PCR-negative specimens, the reference PCR was positive for 5 (2.3%), equivocal for 176 (82.2%), and negative for 33 (15.5%); the NS5 PCR was positive for 0/181 (0%) of the Altona PCR-negative specimens tested by this additional method. As was observed with the reference PCR, most (85%; 53/62) of the Altona PCR-positive specimens for which the date of symptom onset was available were collected during the first 5 days of illness. However, 4 specimens collected between day 8 and day 21 after symptom onset were Altona PCR positive and reference PCR equivocal (n = 3) or negative (n = 1).
All 284 specimens tested by the Altona PCR were CHIKV PCR negative, except for 1 specimen that was CHIKV PCR indeterminate; it was Altona PCR negative, reference PCR indeterminate, and NS5 PCR negative. Four specimens were DENV PCR positive, and among these specimens, the Altona PCR was negative for all 4, whereas the reference PCR was positive for 1, equivocal for 2, and negative for 1. The NS5 PCR was negative for all 3 specimens positive (n = 2) or equivocal (n = 1) by the reference PCR. Of the 4 specimens positive by the DENV PCR, 1 was positive for DENV serotype 1 (CT, 19.68) and another was positive for DENV serotype 4 (CT, 21.25); because of the high CT values in the DENV PCR (CT > 36), we were not able to serotype the virus in the 2 remaining specimens.
Among the 249 serum specimens tested by both the NS5 PCR and the Altona PCR, the Altona PCR was positive for 68 (27.3%) and negative for 181 (72.7%) (Fig. 1). The Altona PCR showed 100% positive and negative percent agreement with the NS5 PCR for all 249 specimens tested by both methods.
Urine.
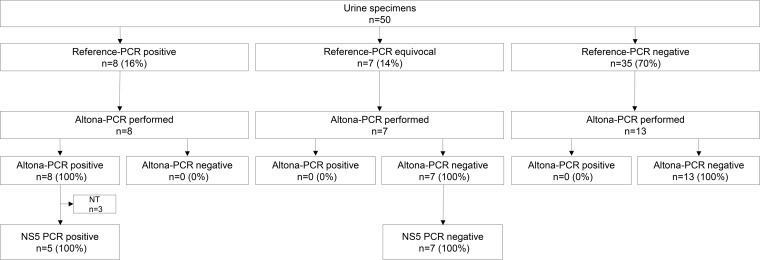
Among the 50 urine specimens tested by the reference PCR, 8 (16%) were positive, 7 (14%) were equivocal, and 35 (70%) were negative. For the positive urine specimens, the time interval from symptom onset to specimen collection was longer than that for the blood specimens, with the median value being 4 days after symptom onset (IQR, 1 to 11 days); 2/7 of the positive urine samples were collected >10 days after symptom onset (11 and 12 days, respectively). Among the eight reference PCR-positive urine specimens, all were also positive by the Altona PCR, as were the 5/8 with sufficient remaining specimen retested by NS5 PCR (Fig. 3). Among the 14 ZIKV-negative urine specimens that were spiked with ZIKV RNA, the detection rate was 100% using the Altona PCR (median CT value, 32.8 [IQR, 32.6 to 32.9]). All reference PCR-equivocal and -negative specimens that were tested by the Altona PCR assay were negative.
FIG 3.
Distribution of urine specimens according to ZIKV reference PCR, Altona PCR, and NS5 gene PCR results. ZIKV, Zika virus; reference PCR, 2-step dual-target real-time reverse transcription-PCR, designed by the U.S. Centers for Disease Control and Prevention, targeting the Zika virus envelope (E) and premembrane (prM) genes; NS5, nonstructural protein 5; NT, not tested.
Paired urine and serum specimens.
Twenty-six paired specimens of urine and serum from 25 patients were submitted for PCR testing. Among the 17 paired specimens tested during the first 5 days of illness using the reference PCR, both the blood and urine of 3 paired specimens were positive and only the urine of 2 paired specimens was positive (the 2 paired specimens were collected on days 2 and 5 after symptom onset). For the 9 paired specimens collected >5 days after symptom onset, only 1 urine specimen collected 11 days after symptom onset was ZIKV positive; ZIKV RNA was not detected by either the reference PCR or the Altona PCR in blood specimens from this subset of patients if the specimens were collected >5 days after symptom onset.
Analytical sensitivity, specificity, and LOD.
Compared to the reference PCR, the sensitivity and specificity of the Altona PCR for serum specimens were 91.4% (53/58; 95% confidence interval [CI], 84.2% to 98.6%) and 97.1% (33/34; 95% CI, 91.4% to 100%), respectively. We did not determine the analytical sensitivity and specificity for urine, given the small number of specimens available for inclusion in this study. The Altona PCR was negative for all 4 serum specimens known to be positive for DENV RNA. The R2 value generated from the dilution series was 0.9975. These data informed the inter- and intra-assay reproducibility studies described in the next paragraph. The 95% limit of detection (LOD) for the Altona PCR was determined to be the equivalent of 0.015 PFU/ml of serum (95% CI, 0.004 to 0.982 PFU/ml). The 95% LODs for the E and prM gene targets of the reference PCR were determined to be 0.128 PFU/ml (95% CI, 0.042 to 4.645 PFU/ml) and 0.061 PFU/ml (95% CI, 0.018 to 2.323 PFU/ml), respectively. The 95% LOD of the NS5 PCR was determined to be 0.213 PFU/ml; there were insufficient replicates to generate a 95% CI.
Assay reproducibility.
Excellent inter- and intra-assay reproducibility was documented down to 0.005 PFU/ml, with all replicates being detected at this concentration on all 3 days of testing.
DISCUSSION
To our knowledge, this is the first study comparing the commercial Altona Diagnostics RealStar Zika virus rRT-PCR to the CDC-designed dual-target ZIKV rRT-PCR reference assay with a large number of clinical specimens.
After a large number of clinical serum specimens were tested, the Altona PCR showed a sensitivity and a specificity of 91% and 92%, respectively, compared to the reference PCR. It is unclear whether the discrepant results between the 2 PCR assays represent false-positive or -negative results of the reference PCR or the Altona PCR. Even though the NS5 PCR was not used to resolve discrepant results, as recommended by the U.S. FDA (17), the 100% positive and negative percent agreement of this assay with the Altona PCR may suggest that Altona PCR-negative, reference PCR-positive specimens likely represent specimens false positive by the reference PCR rather than a true lack of sensitivity of the Altona assay. Similarly, all Altona PCR-positive, reference PCR-equivocal specimens were positive by the NS5 PCR, suggesting that they represent specimens false negative by the reference PCR.
According to the manufacturer, the possibility of cross-reactivity of the Altona assay with Usutu virus, another flavivirus, cannot be ruled out due to sequence homology with the target region used for the detection of ZIKV RNA (18); even when the Altona PCR was performed on only 4 serum specimens, it did not show any cross-reactivity with specimens confirmed to be positive for DENV RNA by a verified PCR assay in use in PHOL, which provides additional data on the specificity of the assay (19). Although the number of urine specimens available for this study was small, the Altona PCR performed as well as the reference PCR with this specimen type. Moreover, the fact that the Altona PCR detected ZIKV RNA in 100% of the urine specimens that were spiked with ZIKV RNA provides additional data on the sensitivity of the Altona PCR with urine specimens. Together, these data confirm the reliability of the Altona RealStar ZIKV PCR kit and allow the safe implementation of the assay to expedite testing of clinical blood and urine specimens.
Our data also confirm that viremia is of short duration in most patients. In our study, 89.6% of patients whose serum was PCR positive had blood collected during the first 5 days of illness. This finding is consistent with the findings of previous studies showing that viremia is transient (7, 8, 20). The positivity rate was somewhat stable from day 0 to day 4 and then rapidly decreased, which correlates with data recently published by Bingham et al. (8). Interestingly, four serum specimens collected after day 5 were Altona PCR positive and reference PCR equivocal or negative.
Only a small subset of our specimens were urine. This is related to the fact that more than half of the specimens were collected before CDC and Canada's National Microbiology Laboratory (NML) recommended PCR testing of urine, in addition to blood (21). In our study, almost a third of positive urine specimens were detected >10 days after symptom onset, which confirms previous data showing that virus shedding in urine continues after the resolution of viremia (8, 20, 22). Bingham et al. showed that the urine of 82% of patients with confirmed ZIKV disease tested positive after 5 days from symptom onset, whereas the blood of none of the patients tested positive when it was collected after 5 days from symptom onset (8). Because of the noninvasive character and prolonged shedding of ZIKV in the urine, ZIKV PCR testing of urine may provide better sensitivity during the acute phase of the illness (days 0 to 5 following symptom onset); this is most likely related to the more transient character of the viremia than the viruria. Among 55 patients with ZIKV disease who had both serum and urine collected during the first 5 days following symptom onset, the rates of positivity were 56% and 95%, respectively (8). Another study with sequential paired urine and blood specimens also showed that urine was more likely to be positive than blood during the acute phase of illness (20). In our study, only 4 patients with paired serum and urine specimens had ZIKV detected in either blood or urine during the first 5 days following symptom onset: urine was positive in 4/4 patients, whereas blood was positive in 2/4 patients. Despite the very small numbers, this tends to support previous data showing the better sensitivity of urine than blood. However, a recent study with paired specimens collected between day 0 and day 5 after symptom onset reported a positivity rate of 75% in plasma and 61% in urine (23), which may indicate that plasma is a better specimen than serum or urine during the acute phase of the illness. This should be confirmed in other studies. Interestingly, Lustig et al. recently showed in a small subset of patients that whole blood remained positive for ZIKV RNA longer than serum (24). The few data on PCR testing of saliva suggest that it is more sensitive than blood but less sensitive than urine (8, 25). As the ideal specimen for ZIKV PCR testing is not defined yet, CDC currently recommends that RT-PCR be performed on urine and blood specimens collected ≤14 days since symptom onset for all symptomatic patients (26, 27). Moreover, asymptomatic pregnant women should also be tested by blood and urine RT-PCR within 14 days of the last potential ZIKV exposure and if they are positive for ZIKV IgM antibodies by serology within 2 to 12 weeks of exposure (27).
The rate of reference PCR-equivocal specimens was very high in our cohort (>25% of serum specimens). To our knowledge, this has not been previously reported. This is most likely explained by a technical issue at PHOL that we were unable to identify.
The EUA of commercial assays, such as the Altona Diagnostics RealStar Zika virus rRT-PCR test kit, can be of great value in assisting with the response to emerging pathogens. Our data confirm the utility and reliability of the Altona PCR, which can be implemented by laboratories wishing to expedite implementation of ZIKV RT-PCR testing with reduced verification requirements compared with those that would be needed for a laboratory-developed test.
MATERIALS AND METHODS
Definitions.
The following definitions were used for analysis: the reference PCR was the dual-target ZIKV rRT-PCR reference assay designed by the U.S. CDC, and the Altona PCR was the Altona Diagnostics RealStar ZIKV RT-PCR test kit (version 1.0; Altona Diagnostics GmbH, Hamburg, Germany).
Study setting.
The study was conducted at PHOL, Ontario's reference microbiology laboratory, where all molecular testing for ZIKV is performed for the province, together with the support of Canada's National Microbiology Laboratory (NML). Every clinical specimen that was submitted to PHOL for ZIKV testing between 8 March and 21 July 2016 and that met the criteria for rRT-PCR testing was tested using the reference PCR, and the results were reported for clinical purposes.
Clinical specimens.
All specimen types submitted during the study period were included in the study (serum, urine, placenta, cord blood, cerebrospinal fluid [CSF], nasopharyngeal swab, and tissue). Due to the limited number of positive urine specimens, 14 previously tested ZIKV-negative urine specimens were spiked with a commercial ZIKV control (Vircell Technologies, Granada, Spain) quantified to have a stock ZIKV RNA concentration of 100 copies/μl. Each 250-μl aliquot of urine was spiked with 10 μl of the control for a concentration of 4,000 spiked copies/ml of specimen; the spiked urine aliquots were then processed per the standard operating procedures for the Altona PCR.
Reference PCR.
The reference PCR was performed as previously described by Lanciotti et al. (7). Total nucleic acid was extracted from serum and urine specimens using a NucliSENS easyMAG automated platform (bioMérieux, St. Lauren, Quebec, Canada; amount of matrix extracted, 250 μl; elution volume, 25 μl). PCR was performed on an Applied Biosystems 7900HT Fast real-time PCR system (Thermo Fisher Scientific Inc., Waltham, MA, USA). The positive control (the ZIKV [African strain] RNA-positive control) was supplied by the NML, and the GADPH (glyceraldehyde-3-phosphate dehydrogenase) control was used as an extraction/housekeeping control. On the basis of CDC interpretation criteria, a specimen was considered positive if both primer sets showed amplification with cycle threshold (CT) values of ≤38.5, whereas a specimen was considered equivocal if only one primer set showed amplification (at any CT value) or if both showed amplification but at least one had a CT value of >38.5 (7, 10). To evaluate positivity related to the interval from symptom onset to specimen collection, the reference PCR was used, as more specimens had been tested using this method than the Altona PCR.
NS5 gene PCR and sequencing.
All reference PCR-positive and -equivocal specimens underwent an endpoint PCR targeting 191 bp of the ZIKV NS5 gene, regardless of the Altona PCR results, on the basis of a previously published protocol modified by NML (28). Bidirectional Sanger sequencing of the PCR product was completed with the same primer set using a BigDye Terminator (version 3.1) cycle sequencing kit and an ABI Prism 3730XL genetic analyzer (Applied Biosystems). Sequencing was performed on all early specimens and subsequently on later specimens only if a band was visualized on the endpoint PCR. Sequences were analyzed using Vector NTI Advance software (Life Technologies, CA) and aligned with those of the reference strains.
Altona PCR.
All serum and urine specimens testing positive or equivocal using the reference PCR, as well as a convenience sample of specimens testing negative using the reference PCR, were retested using the Altona PCR, which targets the NS1 gene (personal communication with the manufacturer), according to the manufacturer's instructions (18). The PCR was performed at PHOL on an Applied Biosystems 7500 real-time PCR system (Thermo Fisher Scientific Inc., Waltham, MA, USA). Extraction was performed as described above for the reference PCR; the positive control and the extraction control were provided by the manufacturer. The assay runs 45 cycles; any specimen for which the sigmoidal curve crosses the set fluorescence threshold is considered positive. There is no indeterminate range for this assay.
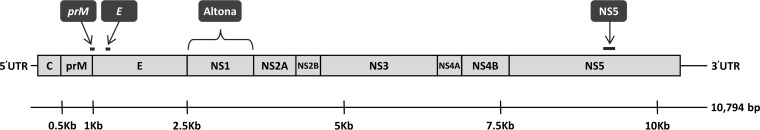
The gene locations targeted by the reference PCR, Altona PCR, and NS5 PCR are shown in Fig. 4.
FIG 4.
ZIKV genome and locations targeted by the ZIKV reference PCR, Altona PCR, and NS5 gene PCR. ZIKV, Zika virus; reference PCR, 2-step dual-target real-time reverse transcription-PCR, designed by the U.S. Centers for Disease Control and Prevention, targeting the Zika virus envelope (E) and premembrane (prM) genes; NS, nonstructural protein gene; C, capsid protein gene. The schematic representation depicts the coding and untranslated regions (UTRs) of the ZIKV genome. The three structural proteins C, prM, and E and the nonstructural (NS) proteins NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 are specified. The assay targets are indicated above. Briefly, the reference PCR dual-target assay amplifies a 75-bp region spanning the prM-E boundary (Zika1) and a 76-bp portion of the E gene (Zika2), the NS5 PCR targets a 191-bp internal region of the NS5 gene, and the Altona PCR targets the NS1 gene (personal communication with the manufacturer). The viral schematic and genomic locations are based on Zika virus reference strain MR-766 (GenBank accession no. NC_012532.1).
Analytical sensitivity, specificity, and LOD.
The sensitivity and specificity of the Altona PCR were determined by comparison to the reference PCR; reference PCR-equivocal specimens were excluded from the sensitivity and specificity analysis. The cross-reactivity of the Altona PCR was determined by testing a small set of specimens positive for DENV RNA. Analyses of the 95% limit of detection (LOD) of all 3 assays (reference PCR, Altona PCR, and NS5 PCR) were performed using total nucleic acid extracted from an inactivated ZIKV culture suspension provided by the NML for use as a positive PCR control. The stock suspension obtained from the NML was provided at 5 × 106 PFU/ml. Extraction of the intact virus suspension was performed on the easyMAG platform per the standard ZIKV testing protocols at PHOL, generating a neat total nucleic acid preparation containing the equivalent of 5 × 106 PFU/ml (assuming full recovery of viral RNA during extraction for the purposes of the 95% LOD estimate). Serial 10-fold dilutions of the initial undiluted ZIKV total nucleic acid preparation (5 × 106 PFU/ml) were prepared using nuclease-free water. The dilutions ranged from 10−1 to 10−12. Each replicate dilution was tested using the Altona PCR and the reference PCR, and the mean CT value for each dilution in the series was calculated. The 95% LOD of the NS5 PCR was also assessed using the generation of a PCR band and confirmation by Sanger sequencing as successful detection of each replicate at each dilution. The 95% LOD was calculated using Probit regression (IBM SPSS Statistics for Windows, version 24.0, released in 2010; IBM Corp., Armonk, NY).
Assay reproducibility.
Reproducibility studies were performed over three sequential days. The reproducibility panel consisted of serial 10-fold dilutions of extracted ZIKV total nucleic acid tested in triplicate on each testing day. The dilution range was determined by the 95% LOD studies and included serial dilutions of between 5 × 10−6 and 5 × 10−10 PFU/ml, which were immediately above and below the 95% LOD, respectively.
ACKNOWLEDGMENTS
Ernesto Lombos, Elaine Tang, Stephen Perusini, Sandeep Nagra, Christine Frantz, and Jonathan B. Gubbay designed the study. Ernesto Lombos, Elaine Tang, Stephen Perusini, Alireza Eshaghi, and Kristina Dimitrova performed the laboratory analyses. Alireza Eshaghi and Stephen Perusini contributed to the initial verification of the assay. Arnaud G. L'Huillier, Ernesto Lombos, Romy Olsha, Erik Kristjanson, and Jonathan B. Gubbay analyzed the data. Arnaud G. L'Huillier, Ernesto Lombos, and Jonathan B. Gubbay wrote the manuscript. All authors reviewed and agreed to the final version of the manuscript.
This study was supported by Public Health Ontario.
Public Health Ontario had no role in study design, data collection, or results interpretation.
Jonathan B. Gubbay has received research grants from GlaxoSmithKline Inc. and Hoffman-La Roche Ltd. to study antiviral resistance in influenza virus and from Pfizer Inc. to conduct microbiological surveillance of Streptococcus pneumoniae. The other authors have no conflict of interest to disclose.
REFERENCES
- 1.Kuno G, Chang GJ. 2007. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol 152:687–696. doi: 10.1007/s00705-006-0903-z. [DOI] [PubMed] [Google Scholar]
- 2.Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS, Pretrick M, Marfel M, Holzbauer S, Dubray C, Guillaumot L, Griggs A, Bel M, Lambert AJ, Laven J, Kosoy O, Panella A, Biggerstaff BJ, Fischer M, Hayes EB. 2009. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 3.Cao-Lormeau VM, Roche C, Teissier A, Robin E, Berry AL, Mallet HP, Sall AA, Musso D. 2014. Zika virus, French Polynesia, South Pacific, 2013. Emerg Infect Dis 20:1085–1086. doi: 10.3201/eid2006.140138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao-Lormeau VM, Musso D. 2014. Emerging arboviruses in the Pacific. Lancet 384:1571–1572. doi: 10.1016/S0140-6736(14)61977-2. [DOI] [PubMed] [Google Scholar]
- 5.Tognarelli J, Ulloa S, Villagra E, Lagos J, Aguayo C, Fasce R, Parra B, Mora J, Becerra N, Lagos N, Vera L, Olivares B, Vilches M, Fernandez J. 2016. A report on the outbreak of Zika virus on Easter Island, South Pacific, 2014. Arch Virol 161:665–668. doi: 10.1007/s00705-015-2695-5. [DOI] [PubMed] [Google Scholar]
- 6.Campos GS, Bandeira AC, Sardi SI. 2015. Zika virus outbreak, Bahia, Brazil. Emerg Infect Dis 21:1885–1886. doi: 10.3201/eid2110.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bingham AM, Cone M, Mock V, Heberlein-Larson L, Stanek D, Blackmore C, Likos A. 2016. Comparison of test results for Zika virus RNA in urine, serum, and saliva specimens from persons with travel-associated Zika virus disease—Florida, 2016. MMWR Morb Mortal Wkly Rep 65:475–478. doi: 10.15585/mmwr.mm6518e2. [DOI] [PubMed] [Google Scholar]
- 9.Rabe IB, Staples JE, Villanueva J, Hummel KB, Johnson JA, Rose L, Hills S, Wasley A, Fischer M, Powers AM. 2016. Interim guidance for interpretation of Zika virus antibody test results. MMWR Morb Mortal Wkly Rep 65:543–546. doi: 10.15585/mmwr.mm6521e1. [DOI] [PubMed] [Google Scholar]
- 10.Waggoner JJ, Pinsky BA. 2016. Zika virus: diagnostics for an emerging pandemic threat. J Clin Microbiol 54:860–867. doi: 10.1128/JCM.00279-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barzon L, Pacenti M, Franchin E, Lavezzo E, Trevisan M, Sgarabotto D, Palu G. 2016. Infection dynamics in a traveller with persistent shedding of Zika virus RNA in semen for six months after returning from Haiti to Italy, January 2016. Euro Surveill 21(32):pii=30316. doi: 10.2807/1560-7917.ES.2016.21.32.30316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicastri E, Castilletti C, Liuzzi G, Iannetta M, Capobianchi MR, Ippolito G. 2016. Persistent detection of Zika virus RNA in semen for six months after symptom onset in a traveller returning from Haiti to Italy, February 2016. Euro Surveill 21(32):pii=30314. doi: 10.2807/1560-7917.ES.2016.21.32.30314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waggoner JJ, Gresh L, Mohamed-Hadley A, Ballesteros G, Davila MJ, Tellez Y, Sahoo MK, Balmaseda A, Harris E, Pinsky BA. 2016. Single-reaction multiplex reverse transcription PCR for detection of Zika, chikungunya, and dengue viruses. Emerg Infect Dis 22:1295–1297. doi: 10.3201/eid2207.160326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waggoner JJ, Gresh L, Vargas MJ, Ballesteros G, Tellez Y, Soda KJ, Sahoo MK, Nunez A, Balmaseda A, Harris E, Pinsky BA. 2016. Viremia and clinical presentation in Nicaraguan patients infected with Zika virus, chikungunya virus, and dengue virus. Clin Infect Dis 63:1584–1590. doi: 10.1093/cid/ciw589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pabbaraju K, Wong S, Gill K, Fonseca K, Tipples GA, Tellier R. 2016. Simultaneous detection of Zika, chikungunya and dengue viruses by a multiplex real-time RT-PCR assay. J Clin Virol 83:66–71. doi: 10.1016/j.jcv.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 16.U.S. Food and Drug Administration. 2016. Emergency use authorization: Altona Diagnostics RealStar® Zika virus RT-PCR kit. U.S. Food and Drug Administration, Silver Spring, MD; https://www.fda.gov/downloads/MedicalDevices/Safety/EmergencySituations/UCM501023.pdf Accessed 2 March 2017. [Google Scholar]
- 17.U.S. Food and Drug Administration. 2007. Statistical guidance on reporting results from studies evaluating diagnostic tests. U.S. Food and Drug Administration, Silver Spring, MD: https://www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm071148.htm Accessed 2 March 2017. [Google Scholar]
- 18.Altona Diagnostics GmbH. 2015. RealStar® Zika virus RT-PCR kit 1.0. Altona Diagnostics GmbH, Hamburg, Germany. http://www.fda.gov/downloads/MedicalDevices/Safety/EmergencySituations/UCM501027.pdf Accessed 2 March 2017.
- 19.Pongsiri P, Praianantathavorn K, Theamboonlers A, Payungporn S, Poovorawan Y. 2012. Multiplex real-time RT-PCR for detecting chikungunya virus and dengue virus. Asian Pac J Trop Med 5:342–346. doi: 10.1016/S1995-7645(12)60055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gourinat AC, O'Connor O, Calvez E, Goarant C, Dupont-Rouzeyrol M. 2015. Detection of Zika virus in urine. Emerg Infect Dis 21:84–86. doi: 10.3201/eid2101.140894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.U.S. Centers for Disease Control and Prevention. 2016. Interim guidance for Zika virus testing of urine—United States, 2016. MMWR Morb Mortal Wkly Rep 65:474. [DOI] [PubMed] [Google Scholar]
- 22.Campos RDM, Cirne-Santos C, Meira GL, Santos LL, de Meneses MD, Friedrich J, Jansen S, Ribeiro MS, da Cruz IC, Schmidt-Chanasit J, Ferreira DF. 2016. Prolonged detection of Zika virus RNA in urine samples during the ongoing Zika virus epidemic in Brazil. J Clin Virol 77:69–70. doi: 10.1016/j.jcv.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Pessoa R, Patriota JV, de Souza MDL, Abd El Wahed A, Sanabani SS. 2016. Detection of Zika virus in Brazilian patients during the first five days of infection—urine versus plasma. Euro Surveill 21(30):pii=30302. doi: 10.2807/1560-7917.ES.2016.21.30.30302. [DOI] [PubMed] [Google Scholar]
- 24.Lustig Y, Mendelson E, Paran N, Melamed S, Schwartz E. 2016. Detection of Zika virus RNA in whole blood of imported Zika virus disease cases up to 2 months after symptom onset, Israel, December 2015 to April 2016. Euro Surveill 21(26):pii=30269. doi: 10.2807/1560-7917.ES.2016.21.26.30269. [DOI] [PubMed] [Google Scholar]
- 25.Bonaldo MC, Ribeiro IP, Lima NS, Dos Santos AA, Menezes LS, da Cruz SO, de Mello IS, Furtado ND, de Moura EE, Damasceno L, da Silva KA, de Castro MG, Gerber AL, de Almeida LG, Lourenco-de-Oliveira R, Vasconcelos AT, Brasil P. 2016. Isolation of infective Zika virus from urine and saliva of patients in Brazil. PLoS Negl Trop Dis 10:e0004816. doi: 10.1371/journal.pntd.0004816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.U.S. Centers for Disease Control and Prevention. 2016. Guidance for U.S. laboratories testing for Zika virus infection. U.S. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 27.Oduyebo T, Igbinosa I, Petersen EE, Polen KN, Pillai SK, Ailes EC, Villanueva JM, Newsome K, Fischer M, Gupta PM, Powers AM, Lampe M, Hills S, Arnold KE, Rose LE, Shapiro-Mendoza CK, Beard CB, Munoz JL, Rao CY, Meaney-Delman D, Jamieson DJ, Honein MA. 2016. Update: interim guidance for health care providers caring for pregnant women with possible Zika virus exposure—United States, July 2016. MMWR Morb Mortal Wkly Rep 65:739–744. doi: 10.15585/mmwr.mm6529e1. [DOI] [PubMed] [Google Scholar]
- 28.Balm MN, Lee CK, Lee HK, Chiu L, Koay ES, Tang JW. 2012. A diagnostic polymerase chain reaction assay for Zika virus. J Med Virol 84:1501–1505. doi: 10.1002/jmv.23241. [DOI] [PubMed] [Google Scholar]