LETTER
A previously published one-step real-time quantitative reverse transcription-PCR (qRT-PCR) targeting the group A rotavirus (RVA) NSP3 gene is a sensitive technique for detection of RVA (1). This assay used the GeneAmp EZ recombinant thermostable Thermus thermophilus (rTth) RNA PCR kit (Applied Biosystems, Inc., Foster City, CA, USA), which was discontinued in July 2014. Thus, a replacement kit was needed to perform the assay.
Here, we report evaluation of a one-step RT-PCR master mix kit (EMD Millipore Corporation, Billerica, MA, USA) for detection of the RVA NSP3 gene. RNA extraction with an MS2 bacteriophage RNA (ZeptoMetrix, Buffalo, NY, USA) internal process control (IPC) (1–3) was conducted as described previously. Each 25-μl reaction mixture contained 7.75 μl nuclease-free water, 12.5 μl 2× one-step RT-PCR master mix (rTth DNA polymerase, antibody, buffer, deoxynucleotides), 1.25 μl 50 mM manganese(II) acetate [Mn(OAc)2], the NSP3 (400 nM) and MS2 (400 nM) forward and reverse oligonucleotide primers and probes (NSP3-FAM, MS2-Texas Red, with the probes at 100 nM), and 2 μl of undenatured RNA extract. NSP3 and MS2 primer/probe sequences were published previously (1–4). Assay modifications for use of the EMD Millipore kit were as follows: NSP3 primer and probe concentrations were reduced to 400 nM and 100 nM, respectively; thermal cycling conditions were changed to 40 cycles of 15 s at 95°C and 1 min at 60°C; and the assay was run on an Applied Biosystems 7500 Fast real-time PCR system in fast mode instead of standard mode. Establishment of the limit of detection and efficiency of the EMD Millipore-based assay was as described previously (1), except that the calculation to determine the double-stranded RNA (dsRNA) copy number per reaction was amended to use RNA template volume rather than the ratio of RNA template volume to overall reaction volume (see Fig. S1 in the supplemental material).
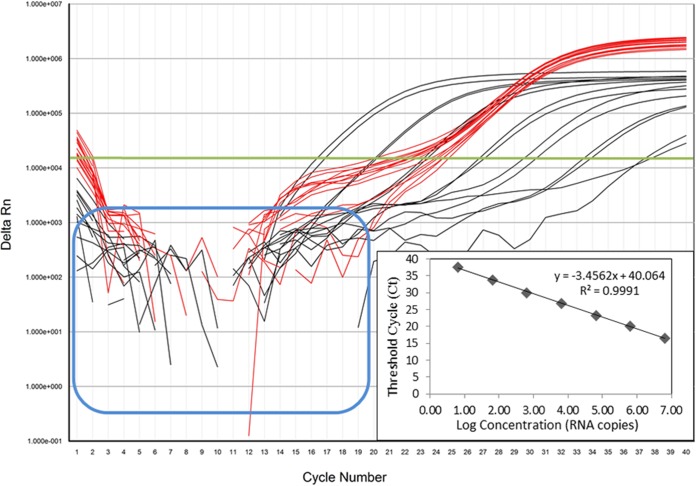
Amplification curves of 10-fold dilutions of RVA strain Wa dsRNA transcript spiked with MS2 RNA showed that the dynamic range of the EMD Millipore-based assay spanned 6.5 × 106 to 6.5 copies/reaction, with threshold cycle (CT) values ranging from 16.6 to 37.6. Approximately 3.3 cycles separated the CT values of each dilution, with an R2 value of 0.9991 and an assay efficiency of 94.69% (Fig. 1). The MS2 IPC was detected across the dilution series and in the NSP3-negative controls, and no amplified product was detected in the no-template controls (NTCs) (Fig. 1). Using the previously published calculations, the dynamic range for the GeneAmp-based assay spanned 9.0 × 105 copies to 0.9 copy per reaction, with CT values ranging from 17.0 to 37.4, an R2 value of 0.9985, and an assay efficiency of 99.38% (1). The change in assay efficiency using the EMD Millipore-based assay may be attributed to reduced primer/probe concentrations, running the assay in fast mode instead of standard mode, and/or differing polymerase efficiencies between kits.
FIG 1.
Amplification curves of 10-fold dilutions of the RVA strain Wa dsRNA transcript spiked with MS2 bacteriophage RNA (from 6.5 × 106 to 6.5 copies per reaction), obtained with the EMD Millipore NSP3 qRT-PCR assay. Using the threshold for delta Rn (the normalized reporter value [Rn] of the reaction minus the Rn of the baseline signal) (green line), black curves show the 10-fold dilutions of the NSP3 gene transcript and red curves show MS2 IPC amplification. The graph showing the CT value versus the log copy number was fitted with a regression line, and the slope for calculation of efficiency was obtained from the regression line. The fluorescent signals from RVA-negative samples and no-template controls are indicated by the blue outline.
The EMD Millipore-based assay was compared to the GeneAmp-based assay using previously frozen clinical stool samples (n = 111; 50 samples were positive and 61 negative as determined by previous testing for RVA antigen and/or RNA) run in duplicate to measure agreement (5) between the two kits (222 total qRT-PCRs) (Tables S1 and S2). Positive percent agreement (PPA), negative percent agreement (NPA), and overall percent agreement (OPA) of the EMD Millipore-based assay were 100.00%, 83.05%, and 90.99% (5), respectively, when the GeneAmp-based assay was used as the gold-standard assay (Table S2). Poor NPA can be attributed to the use of a less-sensitive gold-standard assay, introducing test bias (1). Increased NSP3 gene detection by the EMD Millipore-based assay can be attributed to differences in enzyme activity, which were confirmed through discrepant analysis. Three of seven samples, where both replicates showed NSP3 qRT-PCR amplification using the EMD Millipore-based assay but were not detected using the GeneAmp-based assay (Tables S1 and S2), were confirmed as RVA positive using a nested RT-PCR assay for detection of RVA (6). Comparatively, the EMD Millipore-based assay showed significantly lower CT values (t test [paired], P < 0.0001; the average difference was 0.815) (Table S1), along with an increase of 9.01% in RVA detection (Tables S1 to S2). A qualitative assessment of precision for the EMD Millipore-based assay in which three operators tested 20 samples (10 positive samples and 10 negative samples, as determined by previous testing for RVA RNA) in triplicate on different days showed that all 90 replicates for the RVA-negative samples were negative and that 86 of 90 replicates for the RVA-positive samples were positive. The four false-negative results for detection of RVA were for low-titer samples likely just under the established limit of detection. The precision of the EMD Millipore-based assay was calculated to be 97.78%. Despite differences from analysis of assay standards and the NPA observed through parallel-kit comparison, evaluation of the EMD Millipore one-step master mix kit suggests that it is a suitable alternative for the GeneAmp EZ rTth RNA PCR kit used in the original rotavirus NSP3 gene qRT-PCR assay.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, Atlanta, GA.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the Centers for Disease Control and Prevention or the U.S. Department of Health and Human Services.
We thank Slavica Mijatovic-Rustempasic for her assistance with the study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00126-17.
REFERENCES
- 1.Mijatovic-Rustempasic S, Tam KI, Kerin TK, Lewis JM, Gautam R, Quaye O, Gentsch JR, Bowen MD. 2013. Sensitive and specific quantitative detection of rotavirus A by one-step real-time reverse transcription-PCR assay without antecedent double-stranded-RNA denaturation. J Clin Microbiol 51:3047–3054. doi: 10.1128/JCM.01192-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rolfe KJ, Parmar S, Mururi D, Wreghitt TG, Jalal H, Zhang H, Curran MD. 2007. An internally controlled, one-step, real-time RT-PCR assay for norovirus detection and genogrouping. J Clin Virol 39:318–321. doi: 10.1016/j.jcv.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Gautam R, Mijatovic-Rustempasic S, Esona MD, Tam KI, Osbourne Q, Bowen MD. 2016. One-step multiplex real-time RT-PCR assay for detecting and genotyping wild-type group A rotavirus strains and vaccine strains (Rotarix® and RotaTeq®) in stool samples. PeerJ 4:e1560. doi: 10.7717/peerj.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freeman MM, Kerin TK, Hull J, McCaustland K, Gentsch J. 2008. Enhancement of detection and quantification of rotavirus in stool using a modified real-time RT-PCR assay. J Med Virol 80:1489–1496. doi: 10.1002/jmv.21228. [DOI] [PubMed] [Google Scholar]
- 5.National Committee for Clinical Laboratory Standards. 2002. User protocol for evaluation of qualitative test performance; approved guideline. NCCLS document EP12-A. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 6.Mijatovic-Rustempasic S, Esona MD, Williams AL, Bowen MD. 2016. Sensitive and specific nested PCR assay for detection of rotavirus A in samples with a low viral load. J Virol Methods 236:41–46. doi: 10.1016/j.jviromet.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.