Abstract
Salmonella enterica subsp. enterica serovar Newport has been associated with various foodborne outbreaks in humans and animals. Phylogenetically, serovar Newport is one of several Salmonella serovars that are polyphyletic. To understand more about the polyphyletic nature of this serovar, six food, environment, and human isolates from different Newport lineages were selected for genome comparison analyses. Whole genome comparisons demonstrated that heterogeneity mostly occurred in the prophage regions. Lineage-specific characteristics were also present in the Salmonella pathogenicity islands and fimbrial operons.
Keywords: Salmonella Newport, lineage divergence, complete genome
Introduction
Salmonellosis is a major global public health concern. Of all foodborne diseases, diarrheal and invasive infections due to nontyphoidal Salmonella enterica have resulted in the highest medical burden, causing an average of 4.07 million Disability Adjusted Life Years (DALYs) between 1990 and 2012 (Kirk et al. 2015). Salmonella enterica subsp. enterica serovar Newport has been among the top three Salmonella serovars associated with foodborne outbreaks in the United States since 1970 (Centers for Disease Control and Prevention 2016). S. Newport has been found associated with a wide variety of food commodities including, but not limited to, ground beef, tomatoes, cucumbers, chicken, alfalfa sprouts, and soft cheese. (Angelo et al. 2015; Centers for Disease Control and Prevention 2010; Greene et al. 2008; Schneider et al. 2011).
According to multilocus enzyme electrophoresis (MLEE) (Beltran et al. 1988) and multilocus sequence typing (MLST) (Sangal et al. 2010), S. Newport is polyphyletic, showing a high level of genomic diversity. Newport-I is rarer than the other two lineages, and mostly specific to Europe. Newport-II and Newport-III are common in both Europe and North America, and associated with a variety of hosts. In contrast to Newport-I and Newport-II, most lineage III Newport strains are pansusceptible to antibiotics (Sangal et al. 2010). In the same report, Sangal et al. suggested that Newport-II and Newport-III may have arisen from a single lineage that has now differentiated or frequent recombination events occurred between the two lineages due to niche sharing and may merge via recombination in the future. However, Cao et al. (2013) found that Newport lineage II and III diverged early in the serovar evolution and have evolved largely independently. To further characterize Newport strains from different lineages, we closed the genomes of six S. Newport strains from three lineages.
Materials and Methods
Salmonella enterica subsp. Enterica Serovar Newport Isolates
Six S. Newport strains were obtained from the stock culture collection of the Division of Microbiology, Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration, College Park, MD: CFSAN000825 (environmental isolate from pond water, VA), CFSAN000836 (environmental isolate, VA), CFSAN001660 (environmental isolate from creek water, VA), CFSAN000871 (stool isolate, WA), CFSAN003890 (almond kernel isolate, CA), and CFSAN003887 (almond kernel isolate, CA).
DNA Extraction and Genome Sequencing
All the isolates were cultured in Trypticase soy broth (Becton, Dickinson, Franklin Lakes, NJ, USA) overnight at 37 °C. The genomic DNA was isolated from the overnight cultures using the DNeasy blood and tissue kit (Qiagen, Inc., Valencia, CA, USA). The DNA was sequenced using the Pacific Biosciences (PacBio) RS II sequencing platform, as previously reported (Hoffmann et al. 2015). Genomic DNA was sheared into approximately 20-kb fragments using g-TUBE (Covaris, Inc., Woburn, MA, USA). The library was prepared based on the 20-kb PacBio sample preparation protocol and sequenced using P6/C4 chemistry on four single-molecule real-time (SMRT) cells with a 240-min collection time. The continuous long-read data were de novo assembled using the PacBio hierarchical genome assembly process (HGAP version 3.0) with default parameters in SMRT Analysis v2.3.0, including consensus polishing with Quiver (Chin et al. 2013).
Genomic Analyses
Annotations of assemblies were processed using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) and subsequently deposited at DDBJ/EMBL/GenBank. Genomes were aligned with MAUVE aligner version 2.3.1 using the progressive algorithm with default setting (Darling et al. 2010). Genome comparisons were undertaken using the BLAST Ring Image Generator (BRIG) (Alikhan et al. 2011). Salmonella Pathogenicity Islands (SPIs) and fimbriae were identified and annotated using Artemis (Rutherford et al. 2000). Putative genomic islands containing prophage sequences and antibiotic resistance gene were identified using PHASTER, a new version of PHAST (Arndt et al. 2016) and ResFinder (Zankari et al. 2012), respectively. Single nucleotide polymorphism (SNP) phylogeny was constructed based on the pan SNP matrix generated by kSNP3.0 (Gardner et al. 2015).
Results and Discussion
Whole Genome Comparison
The closed S. Newport genomes were sequenced with 100 to 900× coverage. A final consensus was reached with predicted accuracy at 100% for each complete genome. The average size of the completed genomes of Newport strains was 4.8 Mb. Assembly and annotation results are shown in table 1. Annotation revealed between 4,359 and 4,719 coding sequence (CDS) features. Among these isolates, CFSAN003890 has the largest genome size of 4,719 CDS. CFSAN003890 and CFSAN001660 carried a 130 and 68 kb plasmid, respectively.
Table 1.
Assembly and Annotation Statistics for Salmonella enterica Serovar Newport Strains
| Isolate | DNA | Coverage | Contig | Length (bp) | CDS | tRNA | rRNA | ncRNA | Pseudogene |
|---|---|---|---|---|---|---|---|---|---|
| CFSAN000825 | chromosome | 110 | 1 | 4841916 | 4,495 | 85 | 7 | 21 | 87 |
| CFSAN000836 | chromosome | 900 | 1 | 4883715 | 4,563 | 86 | 7 | 18 | 66 |
| CFSAN001660 | chromosome | 100 | 1 | 4833991 | 4,611 | 84 | 7 | 15 | 49 |
| CFSAN001660 | plasmid | 100 | 1 | 67730 | 68 | 2 | |||
| CFSAN003890 | chromosome | 600 | 1 | 4898059 | 4,719 | 88 | 7 | 16 | 79 |
| CFSAN003890 | plasmid | 600 | 1 | 130039 | 139 | 6 | |||
| CFSAN000871 | chromosome | 600 | 1 | 4712541 | 4,368 | 83 | 7 | 15 | 50 |
| CFSAN003887 | chromosome | 500 | 1 | 4746969 | 4,359 | 84 | 7 | 16 | 60 |
The phylogeny of these six isolates, in the context of the phylogenetic diversity for serovar Newport, was determined combining with 59 genomes from other clade A2 S. enterica serovars (Timme et al. 2013) (supplementary fig. S1, Supplementary Material online). Briefly, the pan SNP matrix was generated by kSNP3.0 using 65 draft or complete genomes with the optimum kmer size of 19, which was determined by Kchooser provided in kSNP3.0 (Gardner et al. 2015). The phylogenetic tree was inferred using FastTree 2.1.7 (Price et al. 2009) with generalized time-reversible (GTR) models of nucleotide evolution, 2 rate categories of sites, and Gamma likelihoods. The overall tree topology is concordant with that shown previously (Sangal et al. 2010; Timme et al. 2013) with three distinctive phylogenetic lineages.
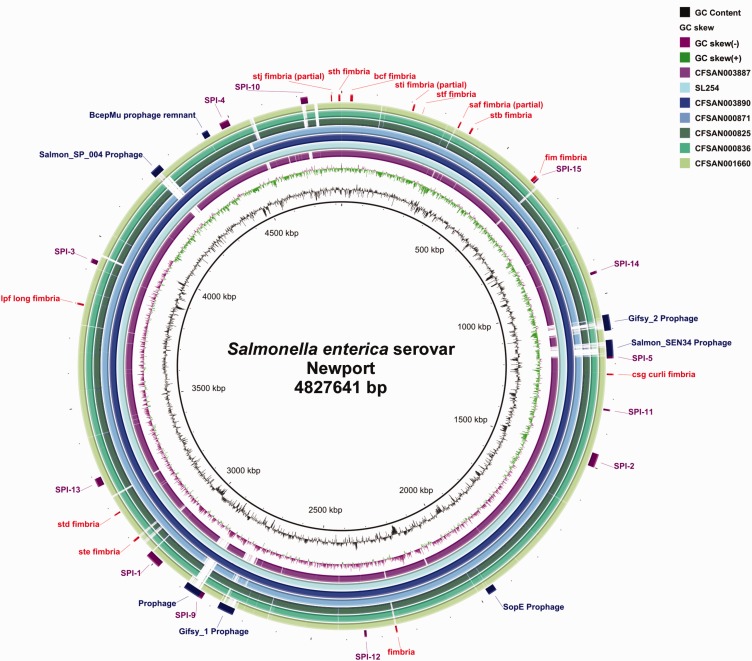
An all-by-all BLAST comparison of the six Newport strains with S. Newport SL254 (CP001113) was done using BRIG (fig. 1). Integrated bacteriophages represent major loci of genetic diversity in these Newport genomes. Varied numbers of prophage regions were identified by PHASTER. Genomes from Newport strains CFSAN003887 and CFSAN000871 only carried one prophage which was named ΦSN20 and ΦSN19, respectively, after their position in the genome. CFSAN003890 genome contained the most prophages (eight), and the rest of the Newport genomes each harbored five to six prophages. All of the prophages displayed a mosaic of genes from related bacteriophages such as Gifsy-1, Gifsy-2, Fels-1, Fels-2, SJ46, SEN34, ST64B, and prophage elements from other bacteria genera. Detailed information on size, location, number of genes and the G + C content are listed in supplementary table S1, Supplementary Material online. To benefit the host, some prophages carry extra DNA that is necessary for phage function like genes encoding enterohemolysin, virulence proteins, PagK, secretion protein HlyD, type III translocated protein SseI, and arsenic resistance protein. The phage-encoded extra functions might allow the host bacterium to increase its fitness within its current ecological niche or even to conquer new niches. It is also interesting to note that all the Newport genomes in this study contained ΦSN44 (supplementary table S1, Supplementary Material online), a prophage remnant, which might shed light to the evolution of serovar Newport. Comparisons using BRIG revealed some interesting differences in Salmonella pathogenicity islands (SPIs) and fimbriae between the genomes. All six Newport genomes contained 12 common SPIs (SPIs-1 to 6, 9, 11, 12, 13, 14, and 15) (fig. 1). SPI-10 was only found in lineage II Newport (CFSAN003890 and CFSAN000871). Fimbriae are another type of virulence factor and can be used at different critical times during infection. Whole genome sequence analysis revealed eight intact putative fimbrial operons shared by all Newport genomes here (sth, bcf, stf, stb, fim, csg, std, and lpf), and three partial operons as well (stj, sti, and saf). Notably, one fimbrial operon, ste, is unique to lineage I and II Newport, while another fimbrial operon was found unique to lineage II Newport (fig. 1). The unique fimbrial adhesion systems or repertoire may explain, in part, some of the niche difference observations.
Fig. 1.—
Genome comparisons of six Salmonella Newport isolates against Sl254. Legend showing color gradient for % similarity. The prophage regions, Salmonella pathogenicity islands (SPIs), and fimbrial operons from SL254 genome are annotated in Artemis and marked in blue, purple, and red, respectively.
Plasmids
S. Newport strains CFSAN001660 and CFSAN003890 each contained one self-transmissible plasmid of about 67 kb (pCFSAN001660) and 130 kb (pCFSAN003890), respectively. pCFSAN001660 is an IncFII plasmid, harboring virulence genes saf fimbrial operon and type IVb pili (T4bP) (supplementary fig. S2a, Supplementary Material online), which are involved not only in mediating adhesion to host cells but also performing complex functions such as force-driven contraction. Newport strain CFSAN003890 contained an IncA/C2 plasmid carrying antimicrobial resistance genes that confer resistance to aminoglycosides (aadA2, strB, strA), cephalosporins (blaCMY-2), chloramphenicol (floR), sulphonamides (sul1, sul2), tetracyclines (tetA), and trimethoprim (dfrA12) (supplementary fig. S2b, Supplementary Material online). Comparative sequence analysis of pCFSAN003890 with pSN254, plasmid from the same Newport lineage strain SL254, showed a highly conserved, largely syntenic plasmid backbone of about 120 kb except for the dfrA12-aad2-sul1 resistance cassette which was acquired elsewhere (supplementary fig. S2b, Supplementary Material online).
Conclusion
Salmonellosis caused by Salmonella enterica serovar Newport remains a significant global challenge. Given the polyphyletic nature of this serovar and its niche adaptation, it remains vital to explore the genome repertoire of different Newport lineages. Here, we compared the genomes of Newport strains from different lineages. Despite overall similarity between genomes, an astonishing number of lineage-specific differences were noted at loci related to virulence and niche colonization.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Literature Cited
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo KM, et al. 2015. Outbreak of Salmonella Newport Infections Linked to Cucumbers: United States, 2014. Morbid Mortal Wkly Rep. 64:144–147. [PMC free article] [PubMed] [Google Scholar]
- Arndt D, et al. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44:W16–W21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran P, et al. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc Natl Acad Sci U S A. 85:7753–7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, et al. 2013. Phylogenetics and differentiation of Salmonella Newport lineages by whole genome sequencing. PLoS One 8:e55687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multistate Outbreak of Human Salmonella Newport Infections Linked to Raw Alfalfa Sprouts (Final Update) [Internet]. Centers for Disease Control and Prevention; 2010. Available from: https://www.cdc.gov/salmonella/2010/newport-alfalfa-sprout-6-29-10.html
- Centers for Disease Control and Prevention. 2016. National enteric disease surveillance: Salmonella annual report, 2013 In: Disease NCfEaZI, editor. Atlanta: Centers for Disease Control and Prevention. [Google Scholar]
- Chin CS, et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. [DOI] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. [DOI] [PubMed] [Google Scholar]
- Greene SK, et al. 2008. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol Infect. 136:157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, et al. 2015. Complete genome sequence of Salmonella enterica subsp. enterica Serovar Agona 460004 2-1, associated with a multistate outbreak in the United States. Genome Announc. 3:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk MD, et al. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 26:1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. [DOI] [PubMed] [Google Scholar]
- Sangal V, et al. 2010. Evolution and population structure of Salmonella enterica serovar Newport. J Bacteriol. 192:6465–6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JL, et al. 2011. Multistate outbreak of multidrug-resistant Salmonella newport infections associated with ground beef, October to December 2007. J Food Prot. 74:1315–1319. [DOI] [PubMed] [Google Scholar]
- Timme RE, et al. 2013. Phylogenetic diversity of the enteric pathogen Salmonella enterica subsp. enterica inferred from genome-wide reference-free SNP characters. Genome Biol Evol. 5:2109–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E, et al. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 67:2640–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.