Abstract
Over the past decade, there has been a rising interest in Achromobacter sp., an emerging opportunistic pathogen responsible for nosocomial and cystic fibrosis lung infections. Species of this genus are ubiquitous in the environment, can outcompete resident microbiota, and are resistant to commonly used disinfectants as well as antibiotics. Nevertheless, the Achromobacter genus suffers from difficulties in diagnosis, unresolved taxonomy and limited understanding of how it adapts to the cystic fibrosis lung, not to mention other host environments. The goals of this first genus-wide comparative genomics study were to clarify the taxonomy of this genus and identify genomic features associated with pathogenicity and host adaptation. This was done with a widely applicable approach based on pan-genome analysis. First, using all publicly available genomes, a combination of phylogenetic analysis based on 1,780 conserved genes with average nucleotide identity and accessory genome composition allowed the identification of a largely clinical lineage composed of Achromobacter xylosoxidans, Achromobacter insuavis, Achromobacter dolens, and Achromobacter ruhlandii. Within this lineage, we identified 35 positively selected genes involved in metabolism, regulation and efflux-mediated antibiotic resistance. Second, resistome analysis showed that this clinical lineage carried additional antibiotic resistance genes compared with other isolates. Finally, we identified putative mobile elements that contribute 53% of the genus’s resistome and support horizontal gene transfer between Achromobacter and other ecologically similar genera. This study provides strong phylogenetic and pan-genomic bases to motivate further research on Achromobacter, and contributes to the understanding of opportunistic pathogen evolution.
Keywords: comparative genomics, phylogenomics, antibiotic resistance, horizontal gene transfer, mobilome
Introduction
Achromobacter sp. is a nonfermenting Gram-negative bacterium, part of the Burkholderiales order, and considered an emerging opportunistic pathogen in the context of cystic fibrosis (CF) lung infections (Davies and Rubin 2007; Emerson et al. 2010; Jakobsen et al. 2013). Although it is not closely related to Pseudomonas aeruginosa, one of the most common causes of lung infection among CF patients (Filkins and O’Toole 2016), the two organisms are similar in many respects. Both carry relatively large genomes (6–7 Mb) and are ubiquitous in the environment, which is considered as the main source of infection (Ridderberg et al. 2011; Amoureux et al. 2013a; Kupfahl et al. 2015), along with documented cases of cross-infection (Hansen et al. 2013; Cools et al. 2015, 2016). Moreover, like P. aeruginosa, Achromobacter sp. can outcompete resident microbiota (Talbot and Flight 2016), establish persistent chronic infections associated with inflammation (Hansen et al. 2010; Lambiase et al. 2011), produce biofilm, resist common disinfectants (Jeukens et al. 2015; Günther et al. 2016) and readily acquire antibiotic resistance (Trancassini et al. 2014).
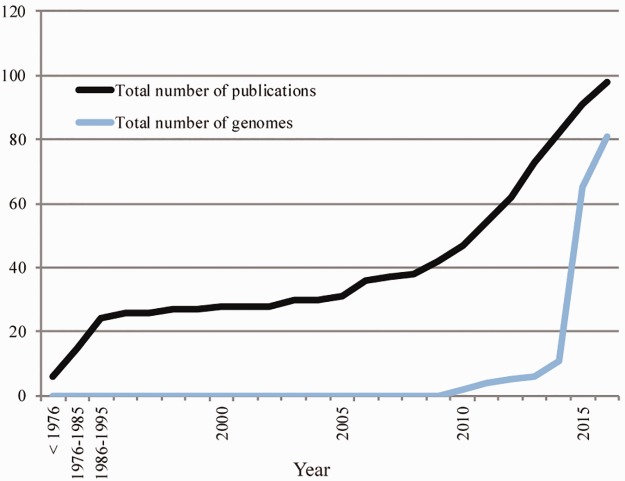
While its prevalence among CF patients is estimated to be as high as 10–20% (Lambiase et al. 2011; Trancassini et al. 2014), Achromobacter species can also cause infections such as bacteremia, pneumonia, meningitis, urinary tract infections, and nosocomial infections (reviewed in Abbott and Peleg 2015). This may be the reason why, after a decade of stagnation, research focusing on clinical Achromobacter isolates was revived ∼10 years ago (fig. 1). This increase in research interest was potentiated by the advent of next-generation sequencing technologies (Vincent et al. 2016) and the explosion in the number of sequenced bacterial genomes that followed (Land et al. 2015). As a result, most of the genomes available for the genus today are of clinical origin, while a few were sequenced in the context of bioremediation research. Despite this renewed interest, the Achromobacter genus suffers from a complex unresolved taxonomy (Vandamme et al. 2013), rare comparative genomics studies (Li et al. 2013; Ridderberg et al. 2015), and very limited understanding of how it adapts to the CF lung (Dupont et al. 2015), let alone other host environments.
Fig. 1.—
Achromobacter research interest from a clinical perspective. As of June 3, 2016. The number of publications is based on a literature search using Web of Science (www.webofknowledge.com; last accessed June 3, 2016) with Title=“Achromobacter” and Topic=“clinical.” The number of genomes is based on the content of NCBI.
In order to identify the genetic characteristics that allow some isolates to thrive in opportunistic infections, we combined all available Achromobacter genomes with four new genome sequences to perform the first large-scale comparative genomics study for this genus. More specifically, the goals were to (1) resolve the taxonomic uncertainties of the genus, and (2) identify virulence-associated genes, antimicrobial resistance genes, and other genomic features associated with pathogenicity and host adaptation. To achieve these goals, we developed an approach based on pan-genome analysis that could be applied to gain significant knowledge on any microbial species or genus.
Materials and Methods
DNA Preparation, Sequencing, and Assembly
In the context of an international Pseudomonas genome project (Freschi et al. 2015), we sequenced four Achromobacter genomes (table 1), one of which was already published: A. xylosoxidans CF304 (Jeukens et al. 2015). Bacterial colonies were isolated on Difco™ Pseudomonas Isolation Agar (BD, Sparks, MD). Genomic DNA was extracted from overnight cultures using the DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany). Genomic DNA (500 ng) was mechanically fragmented for 40 s using a Covaris M220 (Covaris, Woburn, MA) with default settings. Fragmented DNA was transferred to a PCR tube and library synthesis was performed with the Kapa Hyperprep kit (Kapa biosystems, Wilmington, MA) according to manufacturer’s instructions. TruSeq HT adapters (Illumina, SanDiego, CA) were used to barcode the libraries, which were each sequenced in 1/48 of an Illumina MiSeq 300 bp paired-end run at the Plateforme d’Analyses Génomiques of the Institut de Biologie Intégrative et des Systèmes (Laval University, Quebec, Canada). Each data set was assembled de novo with the A5 pipeline version A5-miseq 20140521 (Tritt et al. 2012).
Table 1.
New Achromobacter Genome Assemblies
| Isolate Namea | Source | City | Country | Size (Mb) | Contigs | Median Coverageb | N50 | Accession |
|---|---|---|---|---|---|---|---|---|
| A. xylosoxidans CF304c | cf, sputum | Montreal | Canada | 6.3 | 29 | 61 | 586493 | NZ_LFHA00000000 |
| A. xylosoxidans 4124363476 | cf, throat | Rouyn-Noranda | Canada | 6.8 | 42 | 26 | 503116 | MJMP00000000 |
| A. xylosoxidans AUS488 | bronchiectasis, sputum | Brisbane | Australia | 7.1 | 65 | 20 | 257078 | MJMN00000000 |
| A. xylosoxidans U400 | cf patient | Hobart | Australia | 6.5 | 61 | 47 | 275436 | MJMO00000000 |
The phylogenetic analysis presented in figure 2 enabled species identification.
Median depth of coverage in a de novo assembly with A5-MiSeq (Tritt et al. 2012).
This genome was previously published (Jeukens et al. 2015).
Public Data Selection
All genomes of the genus Achromobacter that had 200 scaffolds or less were downloaded from RefSeq (in June 2016). The 200-scaffold cut-off was selected as a compromise to ensure an assembly quality similar to our own (<100 scaffolds), without having to eliminate too many genomes. A preliminary phylogenetic analysis showed that four genomes were extremely distant from the others: Achromobacter sp. ATCC8750 (NZ_CYTB01000000), Achromobacter sp. 2789STDY5608636 (NZ_CYTV00000000), Achromobacter sp. ATCC13047 (NZ_CYTA00000000) and Achromobacter sp. ATCC35328 (NZ_CYUC00000000). BLAST searches revealed that they respectively corresponded to Alcaligenes, Bordetella, Enterobacter and Escherichia coli isolates. Therefore, these genomes were excluded from this study. The final set of publicly available genomes used with accession numbers and source are provided in supplementary file 1, Supplementary Material online.
Pan-Genome and Phylogenetic Analyses
With the goal of obtaining a robust core genome phylogeny, that is based on conserved regions among genomes, we used the tool SaturnV (https://github.com/ejfresch/saturnV) with the “strictest” algorithm. Genes were considered as orthologous when they shared at least 50% identity over 85% of their length. Orthologous genes for which there was no ambiguous relationship (e.g. putative paralogous genes) were aligned (codon alignments) using PRANK version 150803 (Loytynoja 2014). Sites with no genetic variation were removed with BMGE version 1.02 (Criscuolo and Gribaldo 2010). The resulting alignments were concatenated into a matrix of 595,807 sites and the best-fit evolutionary model was chosen using jModelTest version 2.1.7 (Darriba et al. 2012).
The phylogenetic analysis was performed by maximum-likelihood using RAxML version 8.1.17 (Stamatakis 2014) under the model GTR + Γ4 with 1000 rapid bootstraps. The resulting tree was visualized and midpoint rooted using FigTree version 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/; last accessed June 15, 2016). Average nucleotide identity (ANI) analysis between all genome pairs was performed using pyani (https://github.com/widdowquinn/pyani; last accessed June 15, 2016) with the MUMmer algorithm (Kurtz et al. 2004). Species identification was supported by comparisons with molecular typing data (Jolley 2016).
Genes under Selection
The second goal of this study was to identify genomic features associated with pathogenicity. After the phylogenetic analysis demonstrated that clinical isolates tended to cluster together, we sought to identify genes that were under positive selection (dN/dS > 1) in these clusters specifically. Previously generated orthologous gene alignments were analyzed with HyPhy version 2.2.4 (Pond et al. 2005) using the adaptive branch-site random effects likelihood (aBSREL) method (Smith et al. 2015). A gene was considered under positive selection for a given node or tip in the tree if the P value, after correction for multiple testing with the Holm–Bonferroni method, was <0.05.
Accessory Genome Diversity and Association Analyses
The core genome phylogeny does not take into account information from the accessory genome, that is the genes that are not conserved among isolates. In order to determine whether this information was consistent with the core genome phylogeny, we used a binary matrix of presence/absence for all flexible genes, that is nonunique accessory genes. Genes that are unique to a given genome are uninformative and were excluded from the following analyses. We first assessed flexible gene diversity and relatedness among genomes with a principal component analysis (PCA) and hierarchical clustering using R package ade4 (Dray and Dufour 2007). We then used a discriminant analysis of principal components (DAPC) using R package adegenet (Jombart 2008) to identify genes that are most discriminant between isolates of clinical and nonclinical origin. The goal of this analysis was mainly to find genes that are beneficial in the context of opportunistic infection. To specifically look at gene presence/absence in the context of virulence, we also extracted all annotated virulence genes for Achromobacter genomes on PATRIC (Wattam et al. 2014).
Open/Closed Pan-Genome
In order to calculate how the pan-genome size increases as a function of the number of genomes sampled, 10 sets of the 29 A. xylosoxidans genomes in which the order of the genomes was randomly assigned were generated. For each set, the first n genomes (1 ≤ n ≤ 29) were selected and the number of genes in the pan-genome was calculated using the output table generated by SaturnV, using an R script. To calculate how many new genes are discovered while adding a new genome to a data set of n−1 genomes (1 ≤ n ≤ 29), the same 10 sets were used by subtracting the pan-genome size of n genomes from that of n−1 genomes.
Mobilome Analysis
With the aim of detecting horizontal gene transfer events between Achromobacter isolates and other prokaryotes, we developed two approaches instead of relying on existing tools, because they generally require raw sequencing data, which was not available for our complete data set.
The first approach to investigate the mobilome directly targeted plasmid elements. All protein sequences coded by each genome (according to the annotation) were compared with the NCBI database of proteins found on plasmids (ftp://ftp.ncbi.nlm.nih.gov/refseq/release/plasmid/; last accessed November 1, 2016). The software used to perform the sequence comparisons was Usearch v8.1.1861 (Edgar 2010), with a threshold of 50% protein sequence identity. Modules of five or more contiguous proteins found on the same strand were identified. For each module, the list of unique species where the proteins of the module were found was determined using the annotation present on the NCBI database of proteins found on plasmids. With this information, a network that connects Achromobacter to other genera was drawn using Cytoscape 3.3 (Shannon et al. 2003). Another network representing how the modules are shared among Achromobacter isolates was also drawn using Cytoscape.
The second approach was based on finding regions of high nucleotide identity between Achromobacter genomes and genomes from other genera. More specifically, MegaBLAST (BLAST Tools version 2.4.0, using default parameters) (Altschul et al. 1990) searches were conducted for each Achromobacter genome against the NCBI nucleotide database (nt, downloaded on September 9, 2016) while excluding the Achromobacter genus. BLAST results were filtered to select alignments of at least 5 kb in length and 95% identity. Redundancy was removed for each BLAST result using the longest alignment as a reference and removing smaller alignments that were included in this reference. Two hundred sequences ranging from 5,026 to 121,784 bp were thus extracted. In this work, we refer to these sequences as putative mobile genetic elements (MGEs). Putative MGEs were annotated with the RAST webserver (http://rast.nmpdr.org/; last accessed November 1, 2016, supplementary file 2, Supplementary Material online). A nonredundant table of MGE proteins along with their functional category was generated with the SEED-Viewer environment available in RAST (supplementary files 3 and 4, Supplementary Material online). We generated a final, nonredundant list of putative MGEs, such that two MGEs were considered the same element if they aligned with each other with >95% identity and no more than 5% variation in length at both ends, always retaining the longest element as the reference. This step reduced the number of sequences to 158 elements (supplementary file 2, Supplementary Material online, column “contig_id,” non-redundant_database = Yes). MegaBLAST searches were conducted once again with the nonredundant list of putative MGEs against the NCBI nucleotide database (nt, downloaded September 9, 2016) while excluding the Achromobacter genus, and the BLASTn results were visualized using MEGAN5 (Huson et al. 2007).
Resistome Analysis
Antimicrobial resistance (AMR) genes were identified in all genomes based on the Comprehensive Antibiotic Resistance Database (CARD) (McArthur et al. 2013). This was done using the command-line version of the Resistance Gene Identifier (RGI) software, version 3.0.9 (McArthur et al. 2013). This software is based on BLASTp search against the CARD and uses curated e-value cut-offs to determine the presence of genes.
Results
Genome Sequencing and Public Data Selection
For comparative genomics analyses, all genomes of the genus Achromobacter were downloaded from RefSeq. Four genomes were excluded because they were incorrectly classified on NCBI (see Methods). The final set of 55 publicly available genomes used with accession numbers and isolation source are provided in supplementary file 1, Supplementary Material online. We also sequenced four Achromobacter genomes, one of which was released prior to this study (Jeukens et al. 2015). Newly available genomes have been deposited on NCBI as part of BioProject PRJNA343957. Table 1 contains descriptions and accession numbers for these four genomes, along with assembly statistics.
Phylogenetic Reconstruction
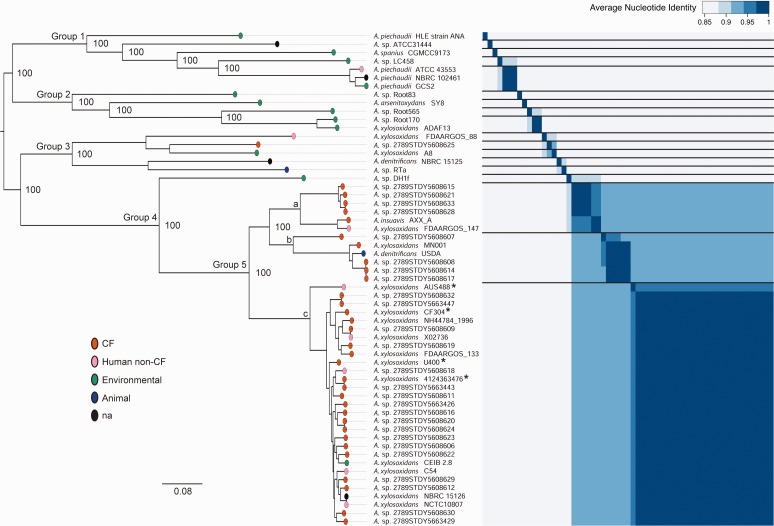
Pan-genome analysis on 59 Achromobacter genomes revealed that the gene inventory of this genus contained a total of 26,365 genes: 12,272 were unique to specific genomes, 12,252 were shared among two or more genomes (“flexible genes”), and 1,841 represented the conserved core genome (31.6% of an average genome). The first goal of this study was to provide a strong phylogenetic basis for the classification of this group. To achieve this, we generated a maximum likelihood tree based on the 1,780 conserved genes that had no ambiguous paralogous relationships. We also computed the average nucleotide identity (ANI) between genome pairs. ANI is a measure of genetic similarity that is used to define boundaries between different species (Konstantinidis and Tiedje 2005), such that a value of 94% or more is indicative of two genomes being part of the same species. The phylogenetic tree and ANI data were combined (fig. 2) to obtain a detailed overview of the Achromobacter phylogeny along with a cross-validation of the two methods. The tree itself was highly robust, with 100% bootstrap values on all basal nodes, and well supported by the ANI results. Of the five labelled groups, group 5 was the most abundant in our data set. According to molecular typing (Jolley 2016), the largest cluster (c) at the bottom of group 5, which included all four of our newly sequenced genomes, corresponded to A. xylosoxidans (nrdA 765 types 14, 36 and 42). It was very closely related to two other clusters, which comprised A. insuavis (cluster a, nrdA 765 types 45 and 51), A. dolens (cluster b, isolate A. sp. 2789STDY5608607, nrdA 765 type 90) and A. ruhlandii (cluster b, 5 isolates, nrdA 765 types 20 and 23). As for the remainder of the tree, including groups 1–3 as well as A. sp. DH1f from group 4, both ANI and branch length suggested that almost every genome was its own species. By labeling genomes based on isolation source, we found that group 5 included 95% of clinical isolates from CF and non-CF human infections, while this frequency dropped to 20% for the rest of the tree (Fisher’s exact test: P < 0.0001). Hence, understanding the genomic characteristics that differentiate this group from the rest of the Achromobacter genus became a central aspect of this study.
Fig. 2.—
Achromobacter core genome phylogeny. Midpoint rooted maximum likelihood tree of 59 Achromobacter genomes based on 595,807 core SNPs and computed with RAxML. Results from 1,000 bootstraps are indicated for basal nodes. Colored ovals represent the isolation source. The bottom scale represents substitutions per site. Genomes presented in this paper are identified with an asterisk. The heat map represents an all-against-all matrix of average nucleotide identity (ANI) between pairs of isolates. Horizontal lines were included to separate clusters where ANI ≥94%, which indicates that genomes are part of the same species (Konstantinidis and Tiedje 2005). According to molecular typing (Jolley 2016): cluster a is A. insuavis (six isolates, nrdA 765 types 45 and 51), cluster b is A. dolens (A. sp. 2789STDY5608607, nrdA 765 type 90) and A. ruhlandii (five isolates, nrdA 765 types 20 and 23), and cluster c is A. xylosoxidans (29 isolates, nrdA 765 types 14, 36 and 42). Labels for groups 1–5 were arbitrarily included for reference in the main text.
Positively Selected Genes in the Core Genome
The second goal of this study was to identify genomic features associated with pathogenic success. Considering the apparent phylogenetic clustering of clinical isolates, we sought to identify genes under positive selection that were specific to group 5. All genes that had a nonsynonymous to synonymous substitution rate ratio (dN/dS) significantly >1, that is under positive selective pressures, for any given node or tip in the tree are listed in supplementary file 5 (supported by supplementary file 6, Supplementary Material online), whereas those that were specific to group 5 and/or group 4 (group 5 plus its closest relative) are summarized in table 2. These 35 genes included 15 metabolic genes, 8 genes related to regulation (for transcription, translation or post-translational modification), and 3 genes implicated in antibiotic resistance, all of which encode efflux pump components. Moreover, two positively selected genes were involved in nitrogen metabolism, one directly and the other indirectly, by being implicated in the synthesis of molybdenum cofactor (MoCo), which is essential for molybdoenzymes such as nitrate reductase.
Table 2.
Genes under Positive Selection in Groups 4 and 5 of the Achromobacter Genus
| Node(s)a | Annotationb | KEGG Pathway(s)c | COG Functional Categoryd |
|---|---|---|---|
| Group 5 | Indole-3-glycerol phosphate synthase | KEGG:00400 Phenylalanine, tyrosine and tryptophan biosynthesis | Amino acid transport and metabolism |
| Group 5 | Sulfate/thiosulfate import ATP-binding protein CysA | na | Amino acid transport and metabolism |
| Group 5 | High-affinity branched-chain amino acid transport system permease protein LivN | na | Amino acid transport and metabolism |
| Group 5 | Phospho-2-dehydro-3-deoxyheptonate aldolase, Phe-sensitive | KEGG:00400 Phenylalanine, tyrosine and tryptophan biosynthesis | Amino acid transport and metabolism |
| Group 5, group 4 | aromatic amino acid exporter | na | Amino acid transport and metabolism |
| a*, b*, c*, group 5, group 4 | putative D, D-dipeptide transport system permease protein DdpC | na | Amino acid transport and metabolism, Inorganic ion transport and metabolism |
| a*, group 5 | Sialic acid TRAP transporter permease protein SiaT | na | Carbohydrate transport and metabolism |
| Group 5 | GlcNAc-PI de-N-acetylase | na | Carbohydrate transport and metabolism |
| Group 5 | Prolipoprotein diacylglyceryl transferase | KEGG:00601 Glycosphingolipid biosynthesis—lacto and neolacto series KEGG:00604 Glycosphingolipid biosynthesis—ganglio series | Cell wall/membrane/envelope biogenesis |
| Group 5 | Molybdopterin-synthase adenylyltransferase | na | Coenzyme transport and metabolism |
| Group 5 | Energy-coupling factor transporter transmembrane protein BioN | na | Coenzyme transport and metabolism |
| c | Multidrug export protein EmrA | na | Defense mechanismse |
| c*, group 5 | Multidrug resistance protein MexB | na | Defense mechanismse |
| Group 5 | Macrolide export protein MacA | na | Defense mechanismse, Cell wall/membrane/envelope biogenesis |
| c† | Cytochrome bo(3) ubiquinol oxidase subunit 3 | KEGG:00965 Betalain biosynthesis | Energy production and conversion |
| b*, c | Succinate dehydrogenase hydrophobic membrane anchor subunit | na | Energy production and conversion |
| c, group 4 | Nitrite reductase (NADH) small subunit | KEGG:00910 Nitrogen metabolism | Inorganic ion transport and metabolism, Secondary metabolites biosynthesis, transport and catabolism |
| Group 5 | 2,3-dehydroadipyl-CoA hydratase | na | Lipid transport and metabolism |
| Group 5 | 3-oxoadipate CoA-transferase subunit B | na | Lipid transport and metabolism |
| c | NAD-dependent protein deacetylase | na | Post-translational modification, protein turnover, chaperones |
| Group 5 | Protease HtpX | na | Post-translational modification, protein turnover, chaperones |
| Group 5 | putative 3′-5′ exonuclease related to the exonuclease domain of PolB | na | Replication, recombination and repair |
| Group 5 | Isocitrate dehydrogenase kinase/phosphatase | KEGG:00051 Fructose and mannose metabolism KEGG:00053 Ascorbate and aldarate metabolism KEGG:00565 Ether lipid metabolism KEGG:00600 Sphingolipid metabolism KEGG:00730 Thiamine metabolism KEGG:00740 Riboflavin metabolism KEGG:00760 Nicotinate and nicotinamide metabolism | Signal transduction mechanisms |
| Group 5 | hypothetical protein | na | Signal transduction mechanisms |
| Group 5, group 4 | hypothetical protein | na | Signal transduction mechanisms |
| Group 5, group 4 | Transcriptional regulatory protein QseB | na | Signal transduction mechanisms, Transcription |
| c, group 4 | Transcriptional activator FeaR | na | Transcription |
| Group 5 | HTH-type transcriptional activator CmpR | na | Transcription |
| Group 5 | Valine–tRNA ligase | KEGG:00970 Aminoacyl-tRNA biosynthesis | Translation, ribosomal structure and biogenesis |
| a*, group 5, group 4 | 7-cyano-7-deazaguanine synthase | KEGG:00790 Folate biosynthesis | Translation, ribosomal structure and biogenesis |
| c*, group 5 | Cysteine–tRNA ligase | KEGG:00970 Aminoacyl-tRNA biosynthesis | Translation, ribosomal structure and biogenesis |
| Group 5 | hypothetical protein | na | na |
| Group 5 | Hydroxyacylglutathione hydrolase | na | na |
| Group 5 | putative hydrolase | na | na |
| Group 5, group 4 | hypothetical protein | na | na |
Node(s) in the phylogenetic tree where positive selection (dN/dS > 1) was detected. a: cluster formed by A. insuavis, b: cluster formed by A. ruhlandii and dolens, c: cluster formed by A. xylosoxidans, group 5: cluster formed by a + b + c, group 4: group 5 plus closest relative A. sp DH1f, * positive selection on internal node or isolate, not on whole group, † positive selection on whole group in addition to internal nodes and isolates.
Annotation with Prokka.
Annotation with PATRIC.
Annotation with SaturnV.
EmrA: no match in the Comprehensive Antibiotic Resistance Database (CARD), MacA: match with CARD’s macA, MexB: match with CARD’s mexW.
Pan-Genome and Accessory Genome Diversity
A large part of the pan-genome resides in the accessory genome, that is genes that are not conserved among isolates. Comparing the presence/absence of accessory genes among genomes allowed us to produce a global portrait of genome architecture variability. More specifically, a principal component analysis (PCA) based on presence/absence of the 12,252 flexible genes showed that accessory gene content was consistent with the classification that emerged from the core genome phylogeny. In figure 3a, Axis 1 (24% explained variance) separated species of group 4, formed by A. xylosoxidans, A. ruhlandii, A. dolens, A. insuavis, and isolate A. sp. DH1f from other species. Axis 2 (8% explained variance), on the other hand, discriminated A. xylosoxidans from other members of group 4. Representing the same analysis, but with labeling based on clinical or nonclinical isolation source, showed that accessory genome content is more consistent with evolutionary history than with sampling source (fig. 3b).
Fig. 3.—
Principal component analysis of accessory gene content in Achromobacter. PCA analyses based on the presence/absence of 12,252 nonunique accessory (flexible) genes. Inertia ellipses are used to represent groups of genomes. Percentage of the variance explained by principal components 1 and 2 are shown on axes 1 and 2, respectively. (a) The 59 genomes are labelled based on previous core genome phylogeny. (b) Genomes are colored according to their isolation source (clinical: CF and human non-CF, nonclinical: environmental and animal). Four genomes had unknown sources, hence this panel represents 55 genomes instead of 59.
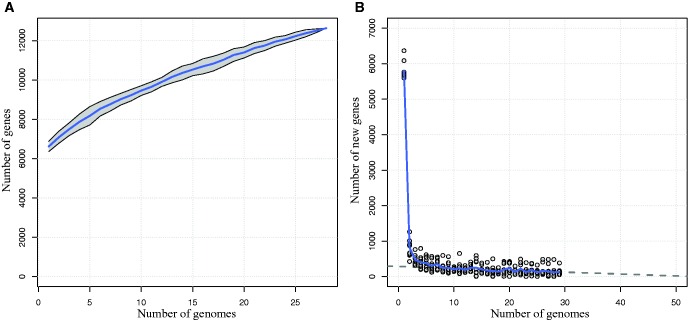
By studying how pan-genome size increases as a function of the number of genomes sampled, it is possible to estimate our knowledge of a species’ genetic repertoire as well as to get some insight on its lifestyle. Using A. xylosoxidans, which had the largest data set, we first constructed a rarefaction curve (fig. 4a) showing that with 28 sequenced genomes, new genes were still being discovered when adding an additional genome. This pattern is the signature of an open pan-genome, in opposition to a closed pan-genome, where pan-genome size stabilizes after just a few sequenced genomes (Tettelin et al. 2008). A large genome size coupled to an open pan-genome typically characterize species living in a community with high levels of lateral gene transfer (Rouli et al. 2015). From these results, it was possible to estimate that, on an average, 133 new genes were discovered by adding a genome to a data set of n−1 isolates, where n is the number of isolates (1 ≤ n ≤ 29, fig. 4b). We were also able to estimate that at least 50 high-quality genomes would be required to obtain a better survey of this species’ genetic repertoire.
Fig. 4.—
Pan-genome size and new gene discovery in the pan-genome of Achromobacter xylosoxidans. (a) Twenty-nine genomes were randomly sampled 10 times and the number of genes present in the pan-genome was calculated for the first n genomes of each sample. The curve thus represents the mean of 10 independent observations. The grey area around this curve represents the standard deviation from the mean. (b) For each of the 10 samples described earlier, the number of new genes found in the pan-genome when an additional genome was added to the previous n−1 genomes (1≤n≤29) was plotted. The curve represents the mean of the 10 observations. Grey dots represent individual observations. The dashed line represents a linear fit to the nearly linear part of the curve (n≥10).
To investigate the accessory genome in the context of pathogenicity, we compiled genes associated with virulence for each phylogenetic lineage (table 3). For most virulence factor categories, namely the type III secretion system (T3SS), endotoxins and proteases, associated genes were more common, but not exclusive to group 5.
Table 3.
Virulence-Associated Genes in Achromobacter Species
| Virulence Genesa |
Speciesb |
|||
|---|---|---|---|---|
| Other (18) | A. insuavis (6) | A. ruhlandii/dolens (6) | A. xylosoxidans (26) | |
| Endotoxin | ||||
| Glutamate–UDP-2-acetamido-2-deoxy-D-ribohex-3-uluronic acid aminotransferase (PLP cofactor) (EC 2.6.1.98) | 2 | 2 | 1 | 14 |
| Putative oxidoreductase | 2 | 2 | 1 | 14 |
| UDP-2-acetamido-3-amino-2,3-dideoxy-D-glucuronic acid acetyltransferase (EC 2.3.1.201) | 2 | 2 | 1 | 14 |
| Invasion | ||||
| Chemotaxis regulator—transmits chemoreceptor signals to flagelllar motor components CheY | 14 | 6 | 6 | 26 |
| Flagellar motor switch protein FliN | 2 | |||
| Protease, serine protease | ||||
| Mobile element protein | 3 | 1 | 13 | |
| Transposon Tn21 resolvase | 3 | |||
| TnpA transposase | 2 | |||
| Integron integrase IntI1 | 2 | 1 | ||
| Resolvase family recombinase | 1 | |||
| Streptomycin 3''-O-adenylyltransferase (EC 2.7.7.47), Spectinomycin 9-O-adenylyltransferase | 1 | |||
| Secretion, invasion, motility | ||||
| Flagellar basal-body rod protein FlgC | 1 | |||
| Secretion, Type III secretion system | ||||
| Type III secretion cytoplasmic ATP synthase (EC 3.6.3.14, YscN,SpaL,MxiB,HrcN,EscN) | 5 | 6 | 6 | 22 |
| Type III secretion inner membrane channel protein (LcrD,HrcV,EscV,SsaV) | 2 | 6 | 6 | 19 |
| Type III secretion inner membrane protein (YscS,homologous to flagellar export components) | 5 | 6 | 6 | 19 |
| Type III secretion inner membrane protein (YscR,SpaR,HrcR,EscR,homologous to flagellar export components) | 3 | |||
| Secretion, Type III secretion system, Invasion | ||||
| Nicotinamidase family protein YcaC | 7 | 1 | ||
| Serum resistance | ||||
| Inner membrane protein YihY, formerly thought to be RNase BN | 6 | 6 | 6 | 26 |
| Other virulence factors | ||||
| 5-Enolpyruvylshikimate-3-phosphate synthase (EC 2.5.1.19) | 7 | 6 | 6 | 26 |
| Argininosuccinate synthase (EC 6.3.4.5) | 17 | 6 | 6 | 26 |
| Lipid A biosynthesis lauroyl acyltransferase (EC 2.3.1.241) | 5 | 6 | 6 | 26 |
| Translation elongation factor Tu | 17 | 6 | 6 | 26 |
| Imidazole glycerol phosphate synthase cyclase subunit (EC 4.1.3.-) | 18 | 6 | 5 | 25 |
| RNA-binding protein Hfq | 18 | 6 | 6 | 25 |
| LSU ribosomal protein L36p, LSU ribosomal protein L36p, zinc-dependent | 13 | 5 | 6 | 24 |
| RecA protein | 18 | 4 | 5 | 22 |
| MotA/TolQ/ExbB proton channel family protein | 8 | 4 | 2 | 7 |
| RNA polymerase sigma factor RpoE | 1 | |||
| Chorismate synthase (EC 4.2.3.5) | 1 | |||
Virulence-associated genes were extracted from PATRIC (Wattam et al. 2014), using a combination of databases Victors and VFDB.
Species were determined based on previous core genome phylogeny, total number of genomes per species is indicated between parentheses.
Resistome and Mobilome
In order to further characterize clinically relevant genomic features in Achromobacter, we identified genes associated with antibiotic resistance (fig. 5). Species of group 5 carried more resistance genes than other species (Fisher’s exact test P = 0.0003), namely for resistance against aminoglycosides (six additional genes), beta-lactams (six additional genes), chloramphenicol (three additional genes) and sulfonamides (two additional genes).
Fig. 5.—
Antimicrobial resistance (AMR) genes among Achromobacter genomes. Gene presence was determined using the RGI-CARD (McArthur et al. 2013). Genomes are ordered based on the phylogenetic tree (fig. 2), and vertical lines represent putative species boundaries. AMR genes are grouped by antibiotic family or function. Green: perfect match to a gene in the CARD, red: similar to a gene in the CARD, according to curated cut-offs, black: no match in the CARD, gyrA and rpoB mutants: specific variants conferring resistance, * Genomes presented in this paper, † Genes with a significant difference in frequency between group 5 (four labelled species) and the other isolates (Fisher’s exact test P<0.05).
Some of these results clarified observations from previous gene-specific, PCR-based studies. For instance, figure 5 shows that OXA-114a, which has been used to diagnose A. xylosoxidans infection (Turton et al. 2011) and encodes a constitutive β-lactamase (Doi et al. 2008), was present in A. xylosoxidans, whereas A. ruhlandii carried OXA-258 (Papalia et al. 2013), and A. insuavis as well as A. dolens carried OXA-243. Analysis of the genomic context of these three sequences confirmed that they corresponded to the same locus, while OXA-2 was a different gene. Isolates outside group 5 did not have an OXA gene, except for closest relative A. sp. DH1f, which shared OXA-114a with A. xylosoxidans. Previous PCR-based studies, mostly because they were misguided by the incorrect classification of the genus, present different results regarding allelic variants and the presence of OXA genes in Achromobacter species, and should be treated with caution (Amoureux et al. 2013b; Traglia et al. 2013, 2014). Figure 5 also contains two previously described efflux protein complexes of the resistance–nodulation–division (RND) family. AxyABM (Bador et al. 2011), which is implicated in the efflux of β-lactams, fluoroquinolones and chloramphenicol, but not aminoglycosides, corresponds to MexAD–OprM in figure 5 and was present in all genomes of this study. The AxyXY–OprZ complex (Bador et al. 2013), which confers broad spectrum resistance, including intrinsic resistance to aminoglycosides and a possible role in acquired resistance as well, corresponds to MexC–AcrF–OprA in figure 5. While MexC and AcrF were present in all genomes, OprA was absent in many species outside group 4. Still, a previous PCR-based study suggested that the entire AxyXY–OprZ operon was absent in aminoglycoside sensitive Achromobacter species (Bador et al. 2016), which is inconsistent with our results.
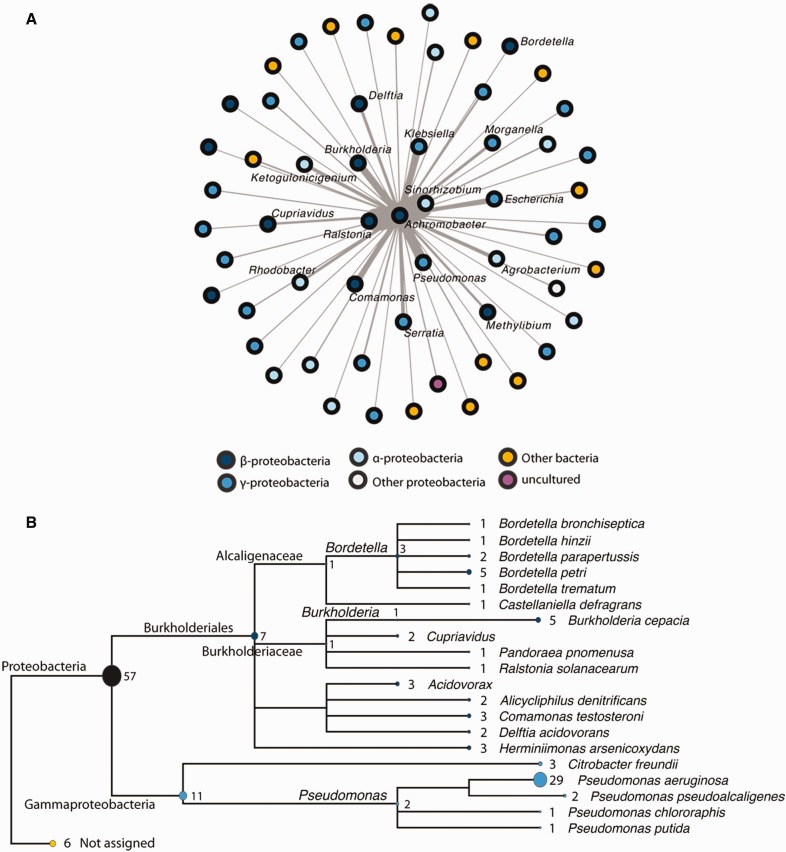
Some of the results from figure 5 suggested horizontal acquisition of resistance. For instance, among the six perfect matches to sequences in the Comprehensive Antibiotic Resistance Database (CARD), ANT(2'')-Ia, APH(6)-Id, sul1 and sul2 were annotated as plasmid or integron encoded, and present in multiple bacterial genera. Considering that lateral gene transfer is a major mechanism in the evolution of resistance, pathogenicity and virulence in bacteria (Ochman et al. 2000), we decided to investigate mobile genetic material in a more formal manner using two independent approaches. First, we searched for modules of contiguous genes (≥ 5 genes) located in the same strand that matched entries of the NCBI plasmid database. This allowed us to identify 466 modules that are shared among isolates of the Achromobacter genus (supplementary file 7, Supplementary Material online). These modules have a large variance in their length (up to 44 genes) and their distribution within the genus (supplementary file 8, Supplementary Material online). We then sought to investigate with which bacterial genera these modules were exchanged. The most likely candidates were plant growth promoting Sinorhizobium (Hayat et al. 2010), as well as Ralstonia, Pseudomonas and Burkholderia, which, like Achromobacter, are soil organisms that are also responsible for opportunistic infections, namely in CF (Lipuma 2015) (fig. 6a). To validate these results, we performed an orthogonal analysis based on the search for stretches of at least 5 kb with high nucleotide identity (≥ 95%) that are shared between Achromobacter and other genera. This method allowed the identification of 158 putative mobile elements (supplementary file 2, Supplementary Material online, non-redundant_database = Yes), which are represented here using a tree of the taxa with which Achromobacter is likely to have exchanged these elements (fig. 6b). The taxonomic content of this tree has much in common with figure 6a (where outermost taxa names were removed for clarity), hence the two independent methods used to identify putative mobile genetic elements can validate and complement one another.
Fig. 6.—
Horizontal gene transfer between Achromobacter and other bacterial species. (a) Network showing the modules (five or more contiguous genes that match entries of the NCBI plasmid database and are present on the same strand) shared between Achromobacter isolates and other bacterial taxa. The color of each node reflects classification (grey to blue: different groups of proteobacteria; yellow: other bacterial species; violet: uncultured bacteria; outermost taxa labels were removed for clarity). A force-directed layout was used to draw the network so that the more a node is close to Achromobacter, the more it shares the same modules). (b) Tree showing the species with which putative mobile elements were exchanged. This analysis is based on a BLASTn search of Achromobacter DNA sequences (>5 kb) matching those of other bacterial species (>95% identity). MEGAN5 (Huson et al. 2007) was used to represent the BLAST results in the form of a tree. The ellipses’ width and the numbers next to them represent the number of sequences shared with each species/genus. The colours reflect classification, like in panel (a).
RAST annotation of the protein sequences within putative mobile elements revealed that 27% of them participated in resistance to antibiotics and toxic compounds (supplementary files 3 and 4, Supplementary Material online). Of these 27%, most proteins were implicated in heavy metal resistance rather than antibiotic resistance, which reflects the overall genomic content of Achromobacter with respect to these functions (Jakobsen et al. 2013). As a final analysis, we were interested in pinpointing the genes of the resistome that were also part of putative mobile elements. From the plasmid database approach, we found that 22 modules contained at least one AMR gene (supplementary file 7, Supplementary Material online), while from the more general second approach, we identified 29 putative mobile elements with at least one AMR gene (supplementary file 2, Supplementary Material online). If both analyses were combined, a total of 26 different AMR genes out of the 49 represented in figure 5 were part of the putative mobilome.
Discussion
Phylogeny and Species Boundaries in Achromobacter
According to sequence typing database PubMLST (Jolley 2016), which is by far the most comprehensive molecular database for this organism (Spilker et al. 2013), there are over 18 Achromobacter species, only a subset of which have been fully genome sequenced. This study provides genome-wide evidence to support the classification of group 5, that is A. xylosoxidans, A. ruhlandii, A. dolens and A. insuavis. However, with respect to ANI and accessory genome content, A. ruhlandii and A. dolens should be part of the same species. Moreover, because species of group 5 are within the 93–95% ANI “grey zone” (Konstantinidis and Tiedje 2005), it would be equally appropriate to classify them as subspecies. Achromobacter xylosoxidans is the most prevalent species of the genus among infected CF patients (Spilker et al. 2013; Coward et al. 2016) as well as nonrespiratory clinical samples (Amoureux et al. 2016), followed in frequency by the three other species of group 5. This explains the over-representation of these species in our data set, as there is a clear sampling bias favoring clinical isolates in both typing and sequence databases. Still, the prevalence of A. xylosoxidans, A. ruhlandii, A. dolens and A. insuavis among clinical isolates combined to their phylogenetic clustering suggest that they are better adapted to cause opportunistic infections. If this is the case, Achromobacter evolution would be reminiscent of that of its closest relative, Bordetella (Li et al. 2013), a largely pathogenic genus involved in human respiratory infections (Melvin et al. 2014) that evolved from a metabolically versatile, environmental ancestor (Gross et al. 2008; Zelazny et al. 2013; Linz et al. 2016). As for the 18 genomes outside of group 5, our results suggest that they represent almost as many different species. More genome sequences would be required to confidently support taxonomic inference, but it is reasonable to assume that group 1 comprises A. spanius and A. piechaudii, and that A. arsenitoxydans is part of group 2.
Mechanisms of Adaptation to the Host
A central goal of this study was to identify genomic features implicated in pathogenicity, or more broadly speaking, in adaptation to the human host environment. Based on the evolutionary tree of the Achromobacter genus, this goal became intimately linked to understanding the evolution of group 5, which we propose to call “the clinical lineage.” In looking at both the core and the accessory genome, we found that this lineage of Achromobacter is likely to share adaptive mechanisms with other biological systems.
Identifying positively selected genes in the clinical lineage led us to find similarities with a study on within-host evolution of Achromobacter spp. in CF patients, where most mutated genes were involved in general metabolism, and some were related to virulence and antimicrobial resistance (Ridderberg et al. 2015). Metabolic genes are often identified in screening approaches aimed at finding genes implicated in virulence, but, even though metabolism and virulence are known to be intimately linked, understanding of detailed mechanisms is extremely limited (Rohmer et al. 2011; Fuchs et al. 2012). Metabolism is key to adapt to host conditions and effectively compete against resident microbiota (Rohmer et al. 2011; Olive and Sassetti 2016), hence it may not be surprising that almost half of positively selected genes in the clinical lineage were implicated in metabolic processes. This analysis also led to the identification of three antibiotic resistance genes, which will be discussed later on.
Although Achromobacter and Pseudomonas are distantly related bacteria, both are obviously well equipped to thrive in the CF lung environment. We found two possible common features between them, the first one pertaining to the type III secretion system (T3SS), which pathogenic bacteria use to inject effectors into host cells (Coburn et al. 2007). A previous comparative genomics analysis of Achromobacter using only three genomes, one from a CF patient and two from environmental isolates, showed that some virulence genes, mostly T3SS related, were only present in the CF isolate (Li et al. 2013). Considering that we had a larger data set, coupled to a better understanding of the evolutionary history of the genus, we compiled virulence gene presence per phylogenetic lineage. As suspected based on our observation that accessory genome content is largely influenced by evolutionary history, patterns of gene presence/absence are not as clear cut as was previously suggested. T3SS genes were more common in the clinical lineage, but they were not systematically present, and some of the other species carried them as well. The T3SS anciently evolved from the flagellum machinery (Abby and Rocha 2012) and is highly conserved among Bordetella and Achromobacter species. However, it may be absent in some isolates, which are generally, but not exclusively, nonpathogenic (Li et al. 2013). Therefore, this virulence factor may have been lost during adaptation to environmental niches, or selected against in certain cases of chronic lung infection (Ridderberg et al. 2015). This last mechanism is reminiscent of adaptation to the CF lung for P. aeruginosa, where virulence factor loss is thought to be advantageous to evade host defenses (Nguyen and Singh 2006; Smith et al. 2006).
The second potential mechanism that we identified relates to survival with limited oxygen. It was demonstrated that the CF mucus is oxygen depleted, prompting P. aeruginosa to use denitrification for energy production (Schobert and Jahn 2010). Moreover, there is evidence that molybdenum uptake, upon which denitrification depends, is essential for anaerobic proliferation and influences virulence in this pathogen (Pederick et al. 2014; Perinet et al. 2016). Considering that most of the genomes of clinical origin used here are from CF patients, it was interesting to find two positively selected genes involved in nitrogen metabolism, suggesting that Achromobacter and P. aeruginosa may share this adaptive mechanism to the CF lung environment.
Flexible and Mobile Genomic Diversity
We have shown that accessory genome content generally matches evolutionary history more than it does ecological niche. These results are consistent with a study on opportunistic pathogen P. aeruginosa, where no correlation was found between genome content and infection type or environmental source (Wolfgang et al. 2003). Nevertheless, using a discriminant analysis of principal components (DAPC), we attempted to identify genes that were specific to isolates of clinical or nonclinical origin. The 28 most discriminant genes identified (based on loading > 0.001, i.e. above background noise) are listed in supplementary file 9, Supplementary Material online. It is noteworthy that many of them are consecutive (locus tags separated by increments of 5), and are likely part of prophages or genomic islands. Especially among genes that were more common among clinical isolates, we identified multiple hypothetical proteins, highlighting the fact that genes with unknown function may play very important roles. Among characteristic genes of nonclinical isolates, this analysis revealed an arsenic efflux pump protein. This is interesting, considering that arsenic resistance is a staple of contaminated soil isolate A. arsenitoxidans SY8 (Li et al. 2012), although this result is influenced by the presence of multiple contaminated site isolates in our data set.
Compiling the number of genes found as a function of the number of genomes available showed that each new A. xylosoxidans genome results in gene discovery, which corresponds to the concept of an open pan-genome. It was suggested that this type of pan-genome reflects a need for high adaptability in the face of diverse environmental conditions, which may translate into high levels of lateral gene transfer among organisms (Tettelin et al. 2008). This result motivated a systematic search for mobile genetic material in the Achromobacter genus. Using two independent approaches, we found putative mobile elements that support past exchange of genetic material between Achromobacter and bacterial genera that share the same ecological setting, without assumptions on the direction of this exchange. Considering that many of these other genera are also environmental organisms capable of causing opportunistic infections, determining whether gene transfer in the soil has predominated over gene transfer in host environments is far from being trivial, although it is probably reasonable to assume that this mechanism has played an important role in both cases. It is now widely recognized that horizontal gene transfer is key to rapid adaptation in the contexts of infectious disease, plant symbiosis and bioremediation (Frost et al. 2005), all of which are relevant to the ecology of Achromobacter.
While they yielded a similar taxonomic breakdown of gene exchange, results of the two approaches used here to find evidence of horizontal transfer shared 338 proteins, representing only 6.6% of the plasmid-related proteins and 19.4% of the putative mobile element proteins. This is due to the fact that the first approach uses a less stringent identity cut-off, thus allowing the identification of more distantly related elements, while the second approach, with a 95% sequence identity cut-off, has a clear bias in favor of more recent transfer events. Thus, the two methods are complementary in this respect. It is also important to note that these results likely include false positives in the form of conserved regions inherited from a common ancestor, for example between Achromobacter and Bordetella. Within their overlapping results, there were 9 AMR genes: aadA25, acrF, ceoB, cmlA1, golS, mexQ, mexT, sul1 and sul2. Five of them (aadA25, cmlA1, golS, sul1 and sul2) were infrequent among Achromobacter isolates, which is what would be expected in cases of acquired resistance (Hu et al. 2015).
Antibiotic Resistance
It has been known for some time that Achromobacter species have innate resistance against multiple antibiotics, namely cephalosporins (beta-lactam), aztreonam (beta-lactam), and aminoglycosides (Glupczynski et al. 1988; Saiman et al. 2001; Almuzara et al. 2010), which include antibiotics relevant to CF lung infection treatment (Tom et al. 2016). Most likely due to the selective pressures that they have undergone, our results show that isolates of the clinical lineage carry more resistance genes than other isolates, namely for resistance against aminoglycosides, beta-lactams, chloramphenicol and sulfonamides. These additional genes presumably contribute to acquired resistance. There are certain limitations to the approach used here to identify genes of the resistome. First, EmrA, while it was annotated as an AMR gene and found to be under positive selective pressures, was not detected in the analysis presented in figure 5. Second, even if a gene is present and reasonably similar to a known AMR gene, there is no guarantee that it is expressed, or even functional. These issues highlight the need for tools that are not database dependant, especially when it comes to organisms that remain to be well described like Achromobacter. Unfortunately, identifying the genetic basis of a trait de novo requires a large data set of both genotypes and phenotypes (Bradley et al. 2015), which is simply not available at the moment for this organism.
Three genes encoding efflux pump components were under positive selection in the clinical lineage: (1) EmrA, a periplasmic fusion protein part of a major facilitator superfamily multidrug export complex (2) MacA, the periplasmic fusion part of an ABC efflux pump that exports macrolides, and (3) MexW, the multidrug transporter of a RND-type efflux pump. The periplasmic fusion protein is essential to anchor the inner membrane transporter and the outer membrane channel in tripartite efflux systems; it can even play a regulatory role (Lin et al. 2009; Modali and Zgurskaya 2011). Although efflux pumps are anciently evolved systems, substrate changes have been observed relatively frequently in prokaryotes (Saier et al. 1998) and show a tendency to favor loss of specificity, which translates into multi-resistance (Lewis 1994; Vargiu et al. 2016). Moreover, efflux pumps do not exclusively export antimicrobials, and studies on multiple pathogens suggest that they are implicated in bacterial virulence as well (Alcalde-Rico et al. 2016). Hence, efflux pump components, which are generally part of the core genome, represent potent targets for adaptation to a pathogenic lifestyle.
Conclusion
With this study, we first provide a strong phylogenetic basis upon which further research efforts on Achromobacter classification, identification and evolution can build. Second, we show that pathogenicity and host adaptation in this opportunistic pathogen can rest both on elements of the conserved core genome, such as metabolic, regulatory and efflux components, as well as accessory elements, such as virulence-associated and antibiotic resistance genes, which may be horizontally acquired from other bacteria adapted to the same ecological niches. Third, we propose a general strategy to efficiently analyze and gain insight on any microbe’s pan-genome. As emerging pathogens and drug resistance are increasingly recognized as serious healthcare issues (http://www.who.int/antimicrobial-resistance/en/; last accessed October 30, 2016), this work contributes to the critical understanding of how versatile environmental microbes evolve into serious threats to human health.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by Cystic Fibrosis Canada [grant number 2610]. We would like to thank Dao Nguyen, Clara Popa, Timothy Kidd, Scott Bell, Iain Lamont and David Reid for providing the four isolates sequenced for this study. We also acknowledge the Natural Sciences and Engineering Research Council of Canada (NSERC) for an Alexander Graham Bell Canada Graduate Scholarship to A.T.V. and a grant to S.J.C. S.J.C. is a research scholar of the Fonds de la Recherche du Québec-Santé (FRQS).
Literature Cited
- Abbott IJ, Peleg AY. 2015. Stenotrophomonas, Achromobacter, and nonmelioid Burkholderia species: antimicrobial resistance and therapeutic strategies. Semin Respir Crit Care Med. 36:99–110. [DOI] [PubMed] [Google Scholar]
- Abby SS, Rocha EPC. 2012. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 8:e1002983.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcalde-Rico M, Hernando-Amado S, Blanco P, Martínez JL. 2016. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front Microbiol. 7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almuzara M, et al. 2010. In vitro susceptibility of Achromobacter spp. isolates: comparison of disk diffusion, Etest and agar dilution methods. Int J Antimicrob Agents 35:68–71. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Amoureux L, et al. 2013a. Detection of Achromobacter xylosoxidans in hospital, domestic, and outdoor environmental samples and comparison with human clinical isolates. Appl Environ Microbiol. 79:7142–7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoureux L, et al. 2013b. Epidemiology and resistance of Achromobacter xylosoxidans from cystic fibrosis patients in Dijon, Burgundy: first French data. J Cyst Fibros. 12:170–176. [DOI] [PubMed] [Google Scholar]
- Amoureux L, et al. 2016. Achromobacter xylosoxidans is the predominant Achromobacter species isolated from diverse non-respiratory samples. Epidemiol Infect. 18:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bador J, Amoureux L, Blanc E, Neuwirth C. 2013. Innate aminoglycoside resistance of Achromobacter xylosoxidans is due to AxyXY-OprZ, an RND-type multidrug efflux pump. Antimicrob Agents Chemother. 57:603–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bador J, et al. 2011. First description of an RND-type multidrug efflux pump in Achromobacter xylosoxidans, AxyABM. Antimicrob Agents Chemother. 55:4912–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bador J, et al. 2016. Distribution of innate efflux-mediated aminoglycoside resistance among different Achromobacter species. N Microbes N Infect. 10:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley P, et al. 2015. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun. 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn B, Sekirov I, Finlay BB. 2007. Type III secretion systems and disease. Clin Microbiol Rev. 20:535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools P, et al. 2015. A single clone of Achromobacter xylosoxidans colonizes Belgian cystic fibrosis patients from different centres. J Cystic Fibros. 14(Suppl 1):S76. [Google Scholar]
- Cools P, et al. 2016. Epidemic Achromobacter xylosoxidans strain among Belgian cystic fibrosis patients and review of literature. BMC Microbiol. 16:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward A, et al. 2016. Use of nrdA gene sequence clustering to estimate the prevalence of different Achromobacter species among cystic fibrosis patients in the UK. J Cystic Fibros. 15:479–485. [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772–772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JC, Rubin BK. 2007. Emerging and unusual Gram-negative infections in cystic fibrosis. Semin Respir Crit Care Med. 28:312–321. [DOI] [PubMed] [Google Scholar]
- Doi Y, Poirel L, Paterson DL, Nordmann P. 2008. Characterization of a naturally occurring class D β-lactamase from Achromobacter xylosoxidans. Antimicrob Agents Chemother. 52:1952–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray S, Dufour A-B. 2007. The ade4 package: implementing the duality diagram for ecologists. J Stat Soft. 22:20. [Google Scholar]
- Dupont C, et al. 2015. Intrapatient diversity of Achromobacter spp. involved in chronic colonization of cystic fibrosis airways. Infect Genet Evol. 32:214–223. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- Emerson J, Mcnamara S, Buccat AM, Worrell K, Burns JL. 2010. Changes in cystic fibrosis sputum microbiology in the United States between 1995 and 2008. Pediatr Pulmonol. 45:363–370. [DOI] [PubMed] [Google Scholar]
- Filkins LM, O’Toole GA. 2016. Cystic fibrosis lung infections: polymicrobial, complex, and hard to treat. PLoS Pathog. 11:e1005258.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freschi L, et al. 2015. Clinical utilization of genomics data produced by the international Pseudomonas aeruginosa consortium. Front Microbiol. 6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 3:722–732. [DOI] [PubMed] [Google Scholar]
- Fuchs TM, Eisenreich W, Heesemann J, Goebel W. 2012. Metabolic adaptation of human pathogenic and related nonpathogenic bacteria to extra- and intracellular habitats. FEMS Microbiol Rev. 36:435–462. [DOI] [PubMed] [Google Scholar]
- Glupczynski Y, Hansen W, Freney J, Yourassowsky E. 1988. In vitro susceptibility of Alcaligenes denitrificans subsp. xylosoxidans to 24 antimicrobial agents. Antimicrob Agents Chemother. 32:276–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross R, et al. 2008. The missing link: Bordetella petrii is endowed with both the metabolic versatility of environmental bacteria and virulence traits of pathogenic Bordetellae. BMC Genomics 9:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther F, Merle U, Frank U, Gaida MM, Mutters NT. 2016. Pseudobacteremia outbreak of biofilm-forming Achromobacter xylosoxidans – environmental transmission. BMC Infect Dis. 16:584.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CR, et al. 2010. Inflammation in Achromobacter xylosoxidans infected cystic fibrosis patients. J Cyst Fibros. 9:51–58. [DOI] [PubMed] [Google Scholar]
- Hansen CR, Pressler T, Ridderberg W, Johansen HK, Skov M. 2013. Achromobacter species in cystic fibrosis: cross-infection caused by indirect patient-to-patient contact. J Cystic Fibros. 12:609–615. [DOI] [PubMed] [Google Scholar]
- Hayat R, Ali S, Amara U, Khalid R, Ahmed I. 2010. Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol. 60:579–598. [Google Scholar]
- Hu Y, et al. 2015. Genomic insights into intrinsic and acquired drug resistance mechanisms in Achromobacter xylosoxidans. Antimicrob Agents Chemother. 59:1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res. 17:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen TH, et al. 2013. Complete genome sequence of the cystic fibrosis pathogen Achromobacter xylosoxidans NH44784-1996 complies with important pathogenic phenotypes. PLoS One 8:e68484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeukens J, Freschi L, Kukavica-Ibrulj I, Nguyen D, Levesque RC. 2015. Draft genome sequence of triclosan-resistant cystic fibrosis isolate Achromobacter xylosoxidans CF304. Genome Announc. 3:0086500815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA. 2016. Achromobacter MLST website. Available from: http://pubmlst.org/achromobacter/.
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci. 102:2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfahl C, Walther M, Wendt C, Von Baum H. 2015. Identical Achromobacter strain in reusable surface disinfection tissue dispensers and a clinical isolate. Infect Control Hosp Epidemiol. 36:1362–1364. [DOI] [PubMed] [Google Scholar]
- Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambiase A, et al. 2011. Achromobacter xylosoxidans respiratory tract infection in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 30:973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land M, et al. 2015. Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics 15:141–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. 1994. Multidrug-resistance pumps in bacteria – variations on a theme. Trends Biochem Sci. 19:119–123. [DOI] [PubMed] [Google Scholar]
- Li X, et al. 2012. Genome sequence of the highly efficient arsenite-oxidizing bacterium Achromobacter arsenitoxydans SY8. J Bacteriol. 194:1243–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Hu Y, Gong J, Zhang L, Wang G. 2013. Comparative genome characterization of Achromobacter members reveals potential genetic determinants facilitating the adaptation to a pathogenic lifestyle. Appl Microbiol Biotechnol. 97:6413–6425. [DOI] [PubMed] [Google Scholar]
- Lin HT, et al. 2009. MacB ABC transporter is a dimer whose ATPase activity and macrolide-binding capacity are regulated by the membrane fusion protein MacA. J Biol Chem. 284:1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linz B, et al. 2016. Acquisition and loss of virulence-associated factors during genome evolution and speciation in three clades of Bordetella species. BMC Genomics 17:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipuma JJ. 2015. Assessing airway microbiota in cystic fibrosis: what more should be done? J Clin Microbiol. 53:2006–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A. 2014. Phylogeny-aware alignment with PRANK. Methods Mol Biol. 1079:155–170. [DOI] [PubMed] [Google Scholar]
- Mcarthur AG, et al. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 57:3348–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melvin JA, Scheller EV, Miller JF, Cotter PA. 2014. Bordetella pertussis pathogenesis: current and future challenges. Nat Rev Microbiol. 12:274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modali SD, Zgurskaya HI. 2011. The periplasmic membrane proximal domain of MacA acts as a switch in stimulation of ATP hydrolysis by MacB transporter. Mol Microbiol. 81:937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Singh PK. 2006. Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc Natl Acad Sci U S A. 103:8305–8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. [DOI] [PubMed] [Google Scholar]
- Olive AJ, Sassetti CM. 2016. Metabolic crosstalk between host and pathogen: sensing, adapting and competing. Nat Rev Microbiol. 14:221–234. [DOI] [PubMed] [Google Scholar]
- Papalia M, et al. 2013. OXA-258 from Achromobacter ruhlandii: a species-specific marker. J Clin Microbiol. 51:1602–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederick VG, et al. 2014. Acquisition and role of molybdate in Pseudomonas aeruginosa. Appl Environ Microbiol. 80:6843–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perinet S, et al. 2016. Molybdate transporter ModABC is important for Pseudomonas aeruginosa chronic lung infection. BMC Res Notes 9:016–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. [DOI] [PubMed] [Google Scholar]
- Ridderberg W, Bendstrup KE, Olesen HV, Jensen-Fangel S, Nørskov-Lauritsen N. 2011. Marked increase in incidence of Achromobacter xylosoxidans infections caused by sporadic acquisition from the environment. J Cyst Fibros. 10:466–469. [DOI] [PubMed] [Google Scholar]
- Ridderberg W, Nielsen SM, Nørskov-Lauritsen N. 2015. Genetic adaptation of Achromobacter sp. during persistence in the lungs of cystic fibrosis patients. PLoS One 10:e0136790.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L, Hocquet D, Miller SI. 2011. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouli L, Merhej V, Fournier PE, Raoult D. 2015. The bacterial pangenome as a new tool for analysing pathogenic bacteria. N Microbes N Infect. 7:72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, et al. 1998. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 12:265–274. [DOI] [PubMed] [Google Scholar]
- Saiman L, et al. 2001. Identification and antimicrobial susceptibility of Alcaligenes xylosoxidans isolated from patients with cystic fibrosis. J Clin Microbiol. 39:3942–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobert M, Jahn D. 2010. Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int J Med Microbiol. 300:549–556. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EE, et al. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci. 103:8487–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, et al. 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol. 32:1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilker T, Vandamme P, Lipuma JJ. 2013. Identification and distribution of Achromobacter species in cystic fibrosis. J Cystic Fibros. 12:298–301. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot NP, Flight WG. 2016. Severe Achromobacter xylosoxidans infection and loss of sputum bacterial diversity in an adult patient with cystic fibrosis. Paediatr Respir Rev. 20:27–29. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 11:472–477. [DOI] [PubMed] [Google Scholar]
- Tom SK, Yau YCW, Beaudoin T, Lipuma JJ, Waters V. 2016. Effect of high-dose antimicrobials on biofilm growth of Achromobacter species isolated from cystic fibrosis patients. Antimicrob Agents Chemother. 60:650–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traglia G, et al. 2014. Presence of OXA-type enzymes in Achromobacter insuavis and A. dolens. Curr Microbiol. 69:501–506. [DOI] [PubMed] [Google Scholar]
- Traglia GM, et al. 2013. Distribution of allelic variants of the chromosomal gene bla OXA-114-like in Achromobacter xylosoxidans clinical isolates. Curr Microbiol. 67:596–600. [DOI] [PubMed] [Google Scholar]
- Trancassini M, et al. 2014. Outbreak of Achromobacter xylosoxidans in an Italian Cystic fibrosis centre: genome variability, biofilm production, antibiotic resistance, and motility in isolated strains. Front Microbiol. 5:8. doi:10.3389/fmicb.2014.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritt A, Eisen JA, Facciotti MT, Darling AE. 2012. An integrated pipeline for de novo assembly of microbial genomes. PLoS One 7:e42304.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turton JF, et al. 2011. Identification of Achromobacter xylosoxidans by detection of the blaOXA-114-like gene intrinsic in this species. Diagn Microbiol Infect Dis. 70:408–411. [DOI] [PubMed] [Google Scholar]
- Vandamme P, et al. 2013. Classification of Achromobacter genogroups 2, 5, 7 and 14 as Achromobacter insuavis sp. nov., Achromobacter aegrifaciens sp. nov., Achromobacter anxifer sp. nov. and Achromobacter dolens sp. nov., respectively. Syst Appl Microbiol. 36:474–482. [DOI] [PubMed] [Google Scholar]
- Vargiu AV, Pos KM, Poole K, Nikaido H. 2016. Editorial: bad bugs in the XXIst century: resistance mediated by multi-drug efflux pumps in Gram-negative bacteria. Front Microbiol. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent AT, Derome N, Boyle B, Culley AI, Charette SJ. 2016. Next-generation sequencing (NGS) in the microbiological world: how to make the most of your money. J Microbiol Methods 16:30031–30038. [DOI] [PubMed] [Google Scholar]
- Wattam AR, et al. 2014. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42:D581–D591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang MC, et al. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc Natl Acad Sci. 100:8484–8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny AM, et al. 2013. Adaptability and persistence of the emerging pathogen Bordetella petrii. PLoS One 8:e65102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.