Abstract
Organizing heterologous biosyntheses inside bacterial cells can alleviate common problems owing to toxicity, poor kinetic performance, and cofactor imbalances. A subcellular organelle known as a bacterial microcompartment, such as the 1,2‐propanediol utilization microcompartment of Salmonella, is a promising chassis for this strategy. Here we demonstrate de novo design of the N‐terminal signal sequences used to direct cargo to these microcompartment organelles. We expand the native repertoire of signal sequences using rational and library‐based approaches and show that a canonical leucine‐zipper motif can function as a signal sequence for microcompartment localization. Our strategy can be applied to generate new signal sequences localizing arbitrary cargo proteins to the 1,2‐propanediol utilization microcompartments.
Keywords: Salmonella, bacterial microcompartments, signal sequence, nanoreactor
Introduction
Bacterial microcompartments (MCPs) present an attractive scaffold to spatially organize biosynthetic pathways while simultaneously controlling small molecule transport to and from the catalytic enzymes.1, 2, 3, 4, 5 Bound by a protein monolayer, these subcellular bacterial organelles encapsulate enzymes by means of the interaction of N‐terminal signal sequences with the C‐termini of shell proteins.6, 7, 8, 9 Native microcompartment systems are known to metabolize 1,2‐propanediol, ethanolamine, and plant saccharides.10, 11, 12 The organelles are thought to function by sequestering toxic intermediates,13 providing a private pool of cofactors,14, 15 and enhancing pathway flux.16 Synthetic biologists and metabolic engineers hope to manipulate microcompartment systems to gain these benefits for heterologous pathways of interest. A central aspect of these engineering efforts is the controlled encapsulation of heterologous cargo in the microcompartment lumen.17, 18, 19
Recent experimental and bioinformatic studies have shown that the interaction between the shell and the encapsulated cargo is mediated by a common hydrophobic motif across diverse MCP systems, including the 1,2‐propanediol utilization (Pdu) and ethanolamine utilization (Eut) MCPs.20, 21 Here we demonstrate that this common motif can form the basis of both rational and library‐based strategies to generate Pdu MCP signal sequences de novo, including signal sequences bearing a canonical leucine zipper motif. Our results suggest that an amphipathic alpha‐helical motif, lacking bulky residues that may disrupt helix‐helix interactions, is a key attribute of these signal sequences. Our approach contributes another key part to a toolkit for controlling enzyme loading to a custom subcellular nanoreactor inside a bacterial cell.6, 7, 18, 20, 21
Results
Previous experimental studies in our laboratory, as well as bioinformatic studies of the bacterial MCP operons of diverse bacteria, have shown that localization of proteins to the Pdu MCPs is mediated by a common amphipathic alpha‐helical motif.20, 21 We hypothesized that if we designed an N‐terminal signal sequence bearing this motif, it would also function to localize heterologous proteins to the Pdu MCP. We therefore examined the various signal sequences that we previously found to be Pdu‐localized and developed a consensus de novo signal sequence bearing this motif, but distinct from the known signal sequences [Fig. 1(A)]. We tested the function of this signal sequence, denoted rational, by constructing a fusion of this signal sequence to gfp along with a C‐terminal ssrA cytosolic degradation tag, and examining cells expressing Pdu MCPs along with the Rational‐GFP–SsrA fusion protein.22 The Rational‐GFP–SsrA fusion is degraded in the cytosol by the ClpXP protease, but is rescued by encapsulation in the Pdu MCP. Punctate fluorescence was observed in these cultures [Fig. 2(A)], indicating that Rational functions as a Pdu MCP signal sequence, a finding we confirmed by purifying Pdu MCPs containing Rational‐GFP‐SsrA and analyzing the samples by electrophoretic separation and western blotting against GFP [Fig. 2(B)]. MCP morphology was confirmed to be normal by SDS‐PAGE and transmission electron microscopy (Supporting Information Fig. S1).
Figure 1.
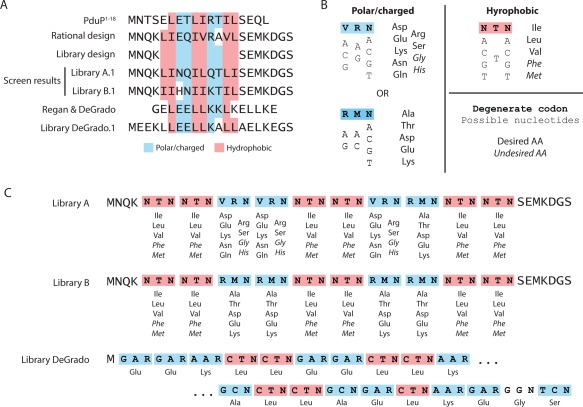
Library and degenerate codon designs to generate Pdu signal sequences. (A) Multiple sequence alignment of PduP1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, rational signal sequence design, library hits A.1 and B.1, Regan and DeGrado alpha helical design, and library hit DeGrado.1; (B) degenerate codons used to generate polar/charged or hydrophobic amino acids, and corresponding encoded amino acids; and (C) detailed library designs for Library A, Library B, and Library DeGrado. Degenerate codons are specified according to the Nomenclature for Incompletely Specified Bases in Nucleic Acid Sequences.
Figure 2.
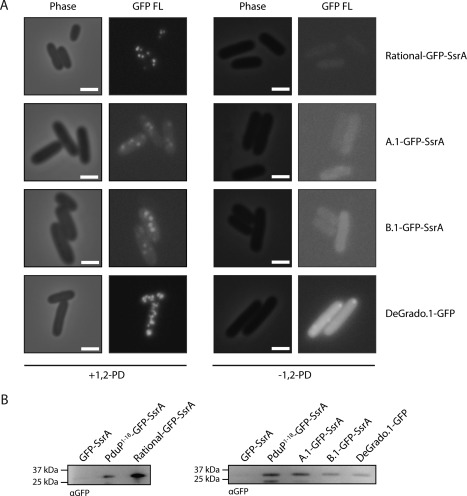
De novo signal sequences localize GFP to Pdu microcompartments. (A) Phase contrast and fluorescence microscopy of S. enterica expressing Pdu MCPs and signal sequence–GFP fusions as indicated. Scale bars indicate 1 μm. (B) Anti‐GFP western blot of Pdu MCPs purified from S. enterica expressing Pdu MCPs and signal sequence–GFP fusions as indicated.
Having shown that a new signal sequence could be created based on simple design rules, we then set out to demonstrate the creation of multiple new Pdu MCP signal sequences by library‐based approaches. To this end, we designed two signal‐sequence libraries based on degenerate codons encoding amino acids with the appropriate hydrophobic or polar/charged chemical properties, as shown [Fig. 1(C)]. In each case, a framework from the above rational design was retained at the N‐ and C‐termini of the 20‐amino acid signal sequence; 10 residues were randomized between these constant regions. One library (A) used a more general degenerate codon (VRN) for three of the four polar/charged positions; the other (B) used the more restrictive RMN codon at all these positions. Both used the NTN codon to encode hydrophobic amino acids [Fig. 1(B)]. The libraries were ordered as oligonucleotide primers designed to amplify the gfp gene. We generated amplicon pools of each library of potential signal sequences fused to gfp and a C‐terminal ssrA cytosolic degradation tag, assembled these amplicons into an appropriate vector using GoldenGate assembly,23 and transformed the plasmids directly into S. enterica for screening. The theoretical size of each library is greater than 1010, and we observed ∼104−105 S. enterica transformants per 10 μL GoldenGate assembly reaction. We confirmed the diversity of each pool of signal sequence variants by Sanger sequencing of a small selection of colonies. The designs of Libraries A and B are shown in Figure 1(C).
We then examined 48 clones from each library for punctate fluorescence upon Pdu MCP expression and isolated the plasmids encoding the functional signal sequences. A functional signal sequence was discovered from each small screening set, denoted Library A.1 and Library B.1, respectively [Fig. 1(A)]. Interestingly, the Library B.1 sequence contained a nucleotide not encoded by the degenerate primer (presumably as a result of an error during PCR amplification), and encoded a His residue at the seventh position. The coding sequences of these hits were reconstituted by PCR, retransformed in S. enterica, and tested for encapsulation both by fluorescence microscopy and by purification of Pdu MCPs followed by electrophoretic separation and western blotting against GFP (Fig. 2). The morphology of the purified MCPs was confirmed by SDS‐PAGE and TEM (Supporting Information Fig. S1). These measurements confirmed the initial microscopy results from the earlier screen. We also identified the amino acid sequences of 20 randomly chosen nonfunctional library members (Supporting Information Fig. S2; 10 members from each library).
We next attempted to abstract further from the native consensus motif and generate a signal sequence based on the amphipathic alpha‐helical peptides designed de novo by DeGrado and coworkers.24, 25 We therefore designed a library of degenerate signal sequences encoding a canonical leucine‐zipper peptide [Fig. 1(A)]. We retained the framework used in the above libraries, using Leu for the hydrophobic positions, and using Glu, Lys, or Ala as appropriate to both match the signal sequence motif and mimic the alpha helical design of Regan and DeGrado [Fig. 1(C)]. Degenerate primers encoding these amino acids using a variety of codons were used to amplify gfp‐ssrA, and these various signal sequences were screened as above for the ability to target GFP encapsulation in the Pdu MCP. A screen of 48 such degenerate coding sequences revealed a hit, which was once again recapitulated and confirmed for GFP encapsulation by fluorescence microscopy and western blotting of purified MCPs against GFP (Fig. 2). Some minor polar aggregation of the fusion protein was observed in the absence of 1,2‐PD, but the +1,2‐PD condition reveals predominant localization of the tagged GFP to the more numerous microcompartments. We again confirmed normal MCP morphology using SDS‐PAGE and TEM (Supporting Information Fig. S1).
Discussion
In addition to the several heterologous signal sequences shown previously to localize proteins to the Pdu MCP,21 we demonstrate here that new Pdu signal sequences can be designed both rationally and using library‐based approaches. A functional signal sequence was designed rationally, without optimization, by examining signal sequences from nature already known to encapsulate cargo in the Pdu MCP. This encouraged us to pursue the generation of further signal sequences de novo.
A library‐based approach also yielded functional signal sequences. The absence of Phe and Met residues in the library‐derived signal sequences is notable, since both can be encoded by the NTN degenerate codon. Each NTN codon has a 19% chance of encoding one of these bulky residues; therefore, neglecting other selective pressures such as mRNA secondary structure, the chances of the two library hits excluding both of these residues from a total of 12 NTN sites by chance are approximately 12 to one against. In contrast, examination of various nonfunctional library members reveals that 40% of the nonfunctional peptides contained the bulky residues Phe or Met (Supporting Information Fig. S2). Another 30% encoded nonsense mutations, 15% were Ala‐ or Gly‐rich (residues which may disfavor the formation of amphipathic helices), and the remaining 15% (three sequences) lacked any obvious defect. The possibility of selection against Phe/Met in the signal sequence suggests that the helix–helix interaction between the signal sequences and the C‐termini of the PduA and PduJ shell proteins may disfavor such bulky residues, consistent with our current structural understanding of such interactions.6, 17 Furthermore, the peak expression levels of the three functional library‐derived signal sequences were higher than those of the majority of the nonfunctional but expression‐competent library members (Supporting Information Fig. S3). Two of the three mysteriously nonfunctional signal sequences (i.e., those without evident defects) exhibited low expression levels, and may simply have lacked favorable codon choices for expression in S. enterica. The third exhibited expression comparable to one of the functional signal sequences, and therefore may violate an as‐yet unknown criterion for proper signal sequence function. Observed peak expression levels are in general agreement with translation rates in Salmonella as predicted by the RBS calculator,26 so future library generation strategies may wish to take predicted expression level into account during library design.
Despite the vast theoretical library size of each degenerate primer (>1010 theoretically possible members), a screen of only 48 library members yielded a hit in each case. This implies a greater than 97% chance that at least one functional signal sequence will be found if 200 of these library members are tested, and a greater than 99% chance if at least 500 library members are tested. Therefore, it is likely that the motifs demonstrated here will generate functional signal sequences even if a relatively small number (on the order of 100) of fusions are tested. Strikingly, these probabilities suggest that approximately 107 possible signal sequences exist and can be discovered based on these libraries. This is encouraging to those wishing to generate signal sequences to encapsulate proteins, such as enzymes, the encapsulation of which is more laborious to ascertain than that of our model protein, GFP.
Taking the idea of an amphipathic alpha‐helical motif to its logical extreme, we finally sought to design a Pdu signal sequence based on the self‐assembling helical bundles created by DeGrado and colleagues24, 25 (Fig. 1). We used another small screen (48 members) to find just such a signal sequence. This implies that the structural constraints for Pdu signal sequence function are very simple: an alpha‐helical peptide containing only Glu, Leu, Ala, and Lys was found to have signal sequence function, implying that the chemical character of individual residues is not as important to encapsulation as the secondary structure of the critical N‐terminal region. The notion that proper alpha‐helical register is crucial is supported by our previous finding that the correction of a single‐amino‐acid “typo” in a putative signal sequence from Clostridium kluyveri restores Pdu signal sequence function.21
It is important to note that the experiments described herein (and other similar studies of N‐terminal Pdu signal sequences) have been conducted using episomal cargo fusion proteins. It remains to be seen whether the trends in encapsulation efficiency and other phenotypes observed for episomal cargo will persist if cargo is expressed in a chromosomal context. Moreover, the expression levels of our de novo signal sequence fusions to GFP varied somewhat (Supporting Information Fig. S3), and this necessitated the removal of the SsrA degradation tag in the case of the DeGrado.1 sequence. A comparison of the behavior of A.1, B.1, and Degrado.1 fusions to GFP with and without the SsrA tag suggests that both approaches should be tested for each new fusion protein to be encapsulated (Supporting Information Fig. S4). Some signal sequences may require the removal of the degradation tag to facilitate effective encapsulation.
Herein, we demonstrate the facile discovery of new Pdu signal sequences by screening small libraries. These libraries are generated based on an amphipathic motif easily encoded by degenerate codons. An important attribute may be the omission of bulky residues which disfavor tight helix‐helix packing between the cargo and container binding helices, but further investigation will be required to confirm this criterion. The technique we demonstrate here could readily be applied to any protein of interest for which encapsulation can be determined, and could be extended in future studies to tune expression and encapsulation in detail. Crucially, the hit rate is high given the theoretical library complexity, indicating that signal sequences could be designed even for proteins requiring labor‐intensive procedures to determine encapsulation.
Materials and Methods
Bacterial strains and plasmids
Plasmid assembly was conducted using E. coli DH10B unless otherwise noted. Microcompartment experiments were conducted in Salmonella enterica serovar Typhimurium LT2. Strains and plasmids used in this study can be found in Supporting Information Table S1.
Molecular biology methods
Molecular biology reagents were procured from New England Biolabs. Plasmids were assembled by the GoldenGate protocol23 and transformed into E. coli DH10B (see Supporting Information Table S2). ORFs of interest were generated as single dsDNA cassettes by PCR using the Phusion polymerase and ligated into a pBAD p15A vector also encoding the chloramphenicol resistance gene cat. Primers used in this study can be found in Table S3. Each primer generates a 5' extension to the gfp‐ssrA gene encoding the N‐terminal signal sequence of interest.
Library generation and screening
Plasmids were assembled by the GoldenGate protocol,23 then 1 μL of the GoldenGate reaction mixture was electroporated with 40 μL of electrocompetent S. enterica LT2. Transformants were selected on solid LB‐agar medium with 34 μg/mL chloramphenicol. Screening was conducted by inoculating 48 library transformants into 5 mL of liquid LB medium with 34 μg/mL chloramphenicol. Cultures were grown at 30°C for 24 h, then subcultured 1:1000 into 5 mL NCE medium with 55 mM 1,2‐propanediol and 17 μg/mL chloramphenicol. The LB overnight cultures were then sedimented at 3,000×g and the pellets stored at −80°C. At OD600∼0.4, expression of gfp fusions in the NCE cultures was induced with 0.02% w/v arabinose. Cultures were grown for a further 5.5 h, then examined by epifluorescence microscopy. The overnight cultures corresponding to positive samples, as determined by microscopy, were retrieved from storage at −80°C, the corresponding plasmids recovered, and the plasmids sequenced by Sanger sequencing.
Culture methods
Cultures were grown in LB–Miller medium at 37°C with 225 rpm orbital shaking in 24‐well blocks unless otherwise noted. Samples expressing microcompartments were grown in No‐Carbon E (NCE) medium supplemented with 0.4% succinate and 55 mM 1,2‐propandediol.
Microcompartment purification
Pdu microcompartments were purified from 200 mL cultures of S. enterica LT2 by sedimentation at 21,000xg as described previously.21
Epifluorescence microscopy
1 μL of culture (OD600∼2) was mounted on a glass slide for epifluorescence microscopy. Bacteria were viewed using a Nikon Ni‐U upright microscope with a 100× 1.45 n.a. plan apochromat objective. Images were captured using an Andor Clara‐Lite digital camera and Nikon NIS Elements software. Fluorescence images were collected using a C‐FL Endow GFP HYQ bandpass filter. All images intended for comparison were adjusted identically for contrast in Adobe Photoshop software.
Transmission electron microscopy
Samples were placed on 400 mesh formvar coated copper grids with a carbon film after glow discharge treatment. 10 μL of purified MCPs at a concentration of approximately 0.1 mg/mL was placed on grids for 2 min. The grids were washed three times with deionized water before fixation. 10 μL of 2% glutaraldehyde in water was placed on grids for 1 min. Grids were then washed an additional three times with deionized water. Finally, samples were stained in 1.6% aqueous uranyl acetate for 1 min. Samples were imaged using a Hitachi HT‐7700 transmission electron microscope.
SDS‐PAGE and Western blotting
SDS‐PAGE was carried out by the methods of Laemmli.27 Discontinuous separating gels containing equal volumes of 12.5%/17.5% acrylamide polymer were used to resolve the microcompartment constituents. Protein gels were stained nonspecifically using the method of Studier.28 Transfer of proteins from denaturing gels to PVDF membranes was carried out using Bio‐Rad wet transfer equipment according to the manufacturer's directions. Western blotting against GFP was performed using a Clontech mouse anti‐GFP primary antibody diluted 1:2000 in 50 mM Tris, 150 mM NaCl, pH 7.6 with 0.05% Tween 20 (TBST) with 1% (w/v) dry milk and then with a Thermo HRP‐conjugated goat anti‐mouse secondary antibody diluted 1:1000 in TBST. Samples were imaged using the Thermo SuperSignal West Femto substrate.
Supplementary material
The supplementary material contains four supplementary figures and three supplementary tables. The supplementary figures illustrate the morphology of purified MCPs (Supporting Information Fig. S1), the coding sequences of nonfunctional N‐terminal peptides (Supporting Information Fig. S2), the expression levels of various library members (Supporting Information Fig. S3), and the behavior of signal sequence‐GFP fusions with and without the SsrA degradation tag (Supporting Information Fig. S4). The supplementary tables describe the bacterial strains (Supporting Information Table S1), plasmids (Supporting Information Table S2), and primers (Supporting Information Table S3) used in this study.
Supporting information
Supporting Information
Acknowledgments
We thank the Tullman‐Ercek Lab for helpful discussions.
References
- 1. Kim E, Tullman‐Ercek D (2012) Engineering nanoscale protein compartments for synthetic organelles. Curr Opin Biotechnol 4:627–632. [DOI] [PubMed] [Google Scholar]
- 2. Kerfeld CA, Erbilgin O (2015) Bacterial microcompartments and the modular construction of microbial metabolism. Trends Microbiol 23:22–34. [DOI] [PubMed] [Google Scholar]
- 3. Chowdhury C, Sinha S, Chun S, Yeates TO, Bobik TA (2014) Diverse bacterial microcompartment organelles. Microbiol Mol Biol Rev 78:438–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giessen TW, Silver PA (2015) Encapsulation as a strategy for the design of biological compartmentalization. J Mol Biol 428:916–927. [DOI] [PubMed] [Google Scholar]
- 5. Chessher A, Breitling R, Takano E (2015) Bacterial microcompartments: biomaterials for synthetic biology‐based compartmentalization strategies. ACS Biomater Sci Eng 1:345–351. [DOI] [PubMed] [Google Scholar]
- 6. Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA (2010) Short N‐terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci USA 107:7509–7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fan C, Bobik TA (2011) The N‐terminal region of the medium subunit (PduD) packages adenosylcobalamin‐dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J Bacteriol 193:5623–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fan C, Cheng S, Sinha S, Bobik TA (2012) Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc Natl Acad Sci USA 109:14995–15000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt‐Dannert C (2012) Engineered protein nano‐compartments for targeted enzyme localization. PloS One 7:e33342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC (1999) The propanediol utilization (pdu) operon of Salmonella enterica Serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12‐dependent 1,2‐propanediol degradation. J Bacteriol 181:5967–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kofoid E, Rappleye C, Stojiljkovic I, Roth J (1999) The 17‐gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol 181:5317–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Erbilgin O, McDonald KL, Kerfeld CA (2014) Characterization of a planctomycetal organelle: A novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl Environ Microbiol 80:2193–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sampson EM, Bobik TA (2008) Microcompartments for B12‐dependent 1,2‐propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol 190:2966–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng S, Bobik TA (2012) The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica . PLoS ONE 10:e47144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huseby DL, Roth JR (2013) Evidence that a metabolic microcompartment contains and recycles private cofactor pools. J Bacteriol 195:2864–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jakobson CM, Slininger MF, Tullman‐Ercek D, Mangan NM (2016) A systems‐level model reveals that 1,2‐propanediol utilization microcompartments enhance pathway flux through intermediate sequestration. bioRxiv 69542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawrence AD, Frank S, Newnham S, Lee MJ, Brown IR, Xue W‐F, Rowe ML, Mulvihill DP, Prentice MB, Howard MJ, Warren MJ (2014) Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth Biol 3:454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jakobson CM, Chen Y, Slininger MF, Valdivia E, Kim EY, Tullman‐Ercek D (2016) Tuning the catalytic activity of subcellular nanoreactors. J Mol Biol 428:2989–2996. [DOI] [PubMed] [Google Scholar]
- 19. Wagner HJ, Capitain CC, Richter K, Nessling M, Mampel J (2017) Engineering bacterial microcompartments with heterologous enzyme cargos. Eng Life Sci 17:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aussignargues C, Paasch BC, Gonzalez‐Esquer R, Erbilgin O, Kerfeld CA (2015) Bacterial microcompartment assembly: The key role of encapsulation peptides. Commun Integr Biol 8:e1039755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jakobson CM, Kim EY, Slininger MF, Chien A, Tullman‐Ercek D (2015) Localization of proteins to the 1,2‐propanediol utilization microcompartment by non‐native signal sequences is mediated by a common hydrophobic motif. J Biol Chem 290:24519–24533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim E, Tullman‐Ercek D (2014) A rapid flow cytometry assay for the relative quantification of protein encapsulation into bacterial microcompartments. Biotechnol J 9:348–354. [DOI] [PubMed] [Google Scholar]
- 23. Engler C, Kandzia R, Marillonnet S (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3:e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DeGrado WF, Regan L, Ho SP (1987) The design of a four‐helix bundle protein. Cold Spring Harb Symp Quant Biol 52:521–526. [DOI] [PubMed] [Google Scholar]
- 25. Regan L, DeGrado WF (1988) Characterization of a helical protein designed from first principles. Science 241:976–978. [DOI] [PubMed] [Google Scholar]
- 26. Salis HM, Mirsky EA, Voigt CA (2009) Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol 27:946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 28. Studier FW (2005) Protein production by auto‐induction in high density shaking cultures. Protein Expr Purif 41:207–234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


