Abstract
ATG14 binding to BECN/Beclin homologs is essential for autophagy, a critical catabolic homeostasis pathway. Here, we show that the α‐helical, coiled‐coil domain (CCD) of BECN2, a recently identified mammalian BECN1 paralog, forms an antiparallel, curved homodimer with seven pairs of nonideal packing interactions, while the BECN2 CCD and ATG14 CCD form a parallel, curved heterodimer stabilized by multiple, conserved polar interactions. Compared to BECN1, the BECN2 CCD forms a weaker homodimer, but binds more tightly to the ATG14 CCD. Mutation of nonideal BECN2 interface residues to more ideal pairs improves homodimer self‐association and thermal stability. Unlike BECN1, all BECN2 CCD mutants bind ATG14, although more weakly than wild type. Thus, polar BECN2 CCD interface residues result in a metastable homodimer, facilitating dissociation, but enable better interactions with polar ATG14 residues stabilizing the BECN2:ATG14 heterodimer. These structure‐based mechanistic differences in BECN1 and BECN2 homodimerization and heterodimerization likely dictate competitive ATG14 recruitment.
Keywords: autophagy, BECN2, coiled‐coil domain, ATG14, BECN2:ATG14 heterodimer
Short abstract
Introduction
Autophagy is a catabolic pathway essential for organismal homeostasis in all eukaryotes. Autophagy enables the degradation of cytoplasmic components that are surplus, defective, or damaged, facilitating nutrient recycling.1, 2, 3 Autophagy is involved in many physiological and pathophysiological processes, including antiaging mechanisms, development and differentiation, elimination of microorganisms, and immunity.4, 5, 6, 7, 8 Autophagy is a highly regulated process executed by autophagy‐related effectors, many of which are called ATG proteins. The first committed step of autophagy is autophagosome nucleation in which macromolecular assemblies selected for degradation are surrounded by isolation membranes. Autophagosome nucleation is executed by a protein complex whose core includes the class III phosphatidylinositol‐3‐kinase (PI3KC3); the PI3KC3 regulatory Ser/Thr kinase, p150; a BECN (or Beclin) homolog;1, 9 and either ATG14 or UVRAG.
BECN1 (or Beclin 1) was among the first mammalian autophagy proteins to be identified.10 A recently identified, 431‐residue paralog of human BECN1, BECN2 (or Beclin 2), which shares 57% sequence identity with BECN1, is also involved in autophagy and in ligand‐induced endolysosomal degradation of several cellular GPCRs. BECN2 heterozygous knockout mice have defective autophagy, increased levels of brain cannabinoid 1 receptor, elevated food intake, obesity and insulin resistance.9 Furthermore, BECN2 regulates endolysosomal degradation and oncogenic signaling of the Kaposi's Sarcoma‐associated herpesvirus encoded GPCRs.11
BECN1 domain architecture12 consists of an intrinsically disordered region (IDR),13, 14 a flexible helical domain (FHD),15, 16 a coiled‐coil domain (CCD),17, 18 and a β‐α repeat autophagy‐specific domain (BARAD).19, 20 BECN homologs in higher organisms contain a BCL2 homology 3 domain (BH3D) within the IDR that enables binding of and regulation by the antiautophagic and antiapoptotic BCL2 proteins,21, 22, 23, 24, 25 but this is not a common feature of all homologs.12 The BECN1 CCD is required and sufficient for binding other CCD‐containing autophagy proteins such as ATG14 and UVRAG.26, 27, 28 BECN1:ATG14 and BECN1:UVRAG heterodimers serve as scaffolds for recruiting PIK3C3 and p150 to form two different PIK3C3 core complexes, Complex I and Complex II respectively.16, 28, 29 The interaction of BECN1 with ATG14 is important for autophagy as BECN1 CCD mutations that disrupt the interaction of BECN1 with ATG14 impair autophagy.18
Like BECN1, BECN2 coimmunoprecipitates (CoIPs) with each of the proteins in the PI3KC3 core complexes: PI3KC3, p150, ATG14, and UVRAG, as well as with AMBRA1,9 which upregulates autophagy,30 and with BCL2 homologs, which down‐regulate autophagy.31 However, there are clear differences in the interactions of BECN1 and BECN2. BECN2 binds AMBRA1 more strongly than does BECN1.9 Furthermore, unlike BECN1, the BECN2:BCL2 interaction is not disrupted upon starvation. Lastly, BECN2 does not CoIP with Rubicon,9 a negative regulator of autophagy.27 Thus, although BECN2 and BECN1 are broadly similar in sequence and function, differences in their sequence, detailed structure, interactions and mechanism likely result in their distinct cellular roles. A structure‐based understanding of these differences will help elucidate the mechanistic bases for the different biological functions of BECN paralogs and consequently, the reason that mammals have two paralogs of this protein.
Here, we present the 2.3 Å X‐ray crystal structure of the human BECN2 CCD homodimer. This structure demonstrates that the BECN2 CCD has a nonideal packing interface. We use structure‐based mutagenesis, circular dichroism (CD) and isothermal titration calorimetry (ITC) to show that interface mutations that improve or diminish homodimerization, stabilize or destabilize homodimer structure respectively. We have also solved the X‐ray crystal structure of the N187L BECN2 CCD mutant, verifying how it homodimerizes better than wild‐type (WT) BECN2. Furthermore, we use ITC to show that these mutations also impact ATG14 binding. An ab initio bead model of the BECN2:ATG14 CCD heterodimer generated by small angle X‐ray scattering (SAXS) demonstrates that the BECN2:ATG14 heterodimer has a parallel arrangement of CCDs. Additionally, a computational model of the heterodimer that has optimal packing and fits the SAXS data well suggests that the heterodimer is stabilized by conserved polar interface interactions and only a few ideal hydrophobic interactions. Our results indicate that, compared to the BECN1 CCD, the BECN2 CCD forms a weaker homodimer but a stronger heterodimer with the ATG14 CCD. Thus, this study provides insights into the structural and thermodynamic features that dictate BECN2 homodimerization and heterodimerization, as well as into the competitive recruitment of ATG14 by different BECN homologs.
Results
BECN2 domain architecture
BECN1 and BECN2 likely have similar architecture based on their sequence similarity (56.6% overall sequence identity for human paralogs) and predicted BECN2 secondary structure. BECN2 domain architecture (Fig. S1) likely consists of an IDR comprising residues 1–121, a FHD comprising residues 122–152, a CCD comprising residues 158–250, and a BARAD comprising residues 251–431. The most variable region of BECN1 and BECN2 is the IDR. Within the BECN2 IDR, residues 88–111 are equivalent to the BECN1 BH3D. However, the highly conserved BECN1 BH3D residue, L116, that is critical for binding to various BCL2 proteins,21, 24, 32 is not conserved in BECN2. The FHD‐CCD‐BARAD region of BECN1 and BECN2 is highly conserved, sharing 60.3% identity, with the CCD sharing 53.8% sequence identity (Fig. S1).
The BECN2 CCD forms a curved, antiparallel homodimer with multiple nonideal interface interactions
We have solved the 2.3 Å X‐ray crystal structure of the human BECN2 CCD (Table 1). The BECN2 CCD forms an antiparallel coiled‐coil homodimer in which two α‐helices coil around each other in a left‐handed twist [Fig. 1(A,B)]. Each crystallographic asymmetric unit has two CCD homodimers: one comprising chain A and chain B and another comprising chain C and chain D. The AB and CD homodimers superimpose with an RMSD of 1.49 Å over 160 Cα atoms. As the two dimer pairs are almost identical, subsequent discussion focuses on the AB dimer, unless mentioned otherwise.
Table 1.
Summary of X‐ray Diffraction Data Collection and Refinement Statistics
| Wild type | N187L | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.97920 | 0.97918 |
| Data range (Å) | 40.33–2.19 | 72.09–2.52 |
| Space group | I2 | C2 |
| Unit cell parameters | A = 95.46 Å; b = 44.32 Å; c = 97.96 Å; β = 96.77° | a = 127.97 Å; b = 44.71 Å; c = 94.90 Å; β = 130.57° |
| Average mosaicity (°) | 0.77 | 0.18 |
| Unique reflections | 17,947 (1589) | 13,749 (1362) |
| Avèrage multiplicity | 3.4 (3.3) | 3.7 (3.4) |
| Completeness (%) | 84.6 (87.0) | 97.1 (86.9) |
| CC1/2 | 0.997 (0.473) | 0.991 (0.761) |
| R meas a | 0.11 (0.76) | 0.26 (1.43) |
| I/σI | 7.7 (2.6) | 6.1 (1.1) |
| Refinement | ||
| Model | PDB ID: 5K7B | PDB ID: 5K9L |
| Monomer A | 87 residues | 89 residues |
| Monomer B | 91 residues | 90 residues |
| Monomer C | 86 residues | 89 residues |
| Monomer D | 88 residues | 89 residues |
| Water molecules | 139 | 60 |
| Data range (Å) | 40.15–2.30 | 72.09–2.52 |
| R work a (%) | 22.1 | 23.5 |
| R free b, c (%) | 23.6 | 27.8 |
| Average B‐values (Å2) | 43.5 | 47.6 |
| Macromolecules | 43.7 | 47.8 |
| Water | 37.7 | 37.6 |
| RMSDs from target values | ||
| Bond lengths (Å) | 0.022 | 0.018 |
| Bond Angles (°) | 1.55 | 1.38 |
| Ramachandran favored (%) | 98.5 | 96.8 |
| Ramachandran outliers (%) | 0 | 0 |
Values in parentheses pertain to the outermost shell of data.
R meas = ∑hkl(n/n − 1)1/2∑h , i|Ihkl , i − ⟨Ihkkl⟩|/∑hkl∑h , iIhkl , i.
R factor = ∑h|F obs − |F calc|/∑h|F obs|.
Test set for R free consisted of 5% of data.
Figure 1.
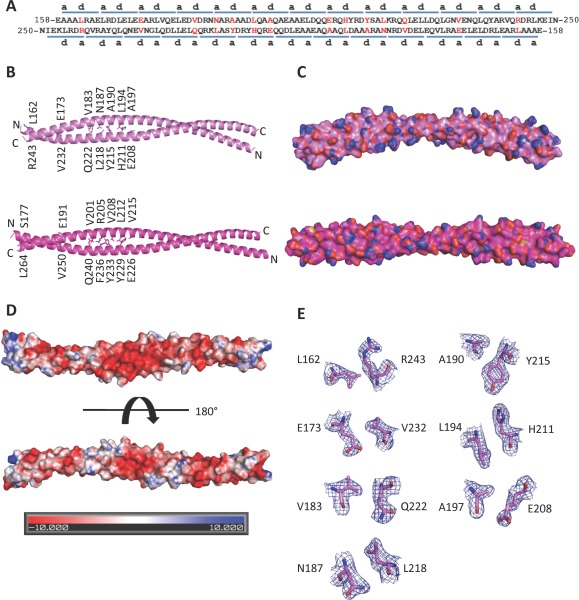
The BECN2 CCD. (A) Paired residues (red) of the two antiparallel monomers of the BECN2 CCD homodimer. The blue bars above or below the sequence indicate the heptad repeats of the BECN2 CCD. The letters a and d indicate the residues at the a and d positions of each heptad repeats, respectively. The nonideal interface pairs are colored red. (B) The X‐ray crystal structures of the BECN2 CCD (violet ‐ top) and the BECN1 CCD (magenta ‐ bottom) antiparallel homodimers. The side chains of nonideal interface residues are displayed in stick with atoms colored by type—oxygen red, nitrogen blue, and carbon violet (BECN2 CCD) or magenta (BECN1 CCD). (C) Surface representation, colored as in (B) of the BECN2 CCD homodimer demonstrates that it is a curved molecule with a radius of 15 nm in contrast to the straight rod‐like BECN1 CCD. (D) Electrostatic surface of the BECN2 CCD homodimer generated by APBS.40 The top panel shows the convex face and the bottom panel shows the concave face. (E) Atomic details of nonideal packing residues, color‐coded as in (B). The blue mesh represents the 2Fo–Fc electron density maps contoured at 1σ above the mean. This and other molecular figures were made using PyMOL (An interactive view is available in the electronic version of the article).41, 42
The average surface area of the BECN2 CCD buried upon homodimerization is 2070 ± 37 Å, constituting ∼24% of the total surface per monomer. The surface area buried in the human BECN1 CCD homodimer interface is 2504 Å2, which constitutes 27% of the total surface area per monomer.18 The BECN2 CCD homodimer forms a curved coiled‐coil with a 15 nm radius of curvature, in contrast to the BECN1 CCD homodimer, which is not curved [Fig. 1(C)]. Like BECN1,18 the BECN2 surface CCD is mostly negatively charged as indicated by its electrostatic surface potential [Fig. 1(D)], especially at the center of the convex face [Fig. 1(D), top panel].
Positions a and d of heptad amino‐acid repeats (a–b–c–d–e–f–g) of classical CCDs are occupied by hydrophobic residues that enable formation of a well‐packed hydrophobic interface that holds the two chains together.33 The BECN2 CCD homodimer interface consists of 26 pairs of interacting residues (or 13 pairs of heptad repeats), comprising 13 unique pairs related by molecular twofold symmetry, similar to the BECN1 CCD homodimer.18 Within the 13 pairs of heptad repeats, the first heptad (residues 159–165) of one CCD pack against the thirteenth heptad (residues 243–249) of the partner CCD [Fig. 1(A)]. Within each pair of heptad repeats, residues at the a and d positions of one repeat interact with residues at the d and a positions, respectively, of the partner heptad. However, unlike classical CCD dimers, the BECN2 CCD has only six unique pairs of acceptable packing interactions: A159–L246, L166–A239, L169–L236, L176–L229, L180–L225, and A201–L204, which includes three ideal leucine pairs. Notably, these ideal packing interactions are conserved in the BECN CCD.18
The BECN2 CCD homodimer has seven unique pairs of nonideal packing interactions, wherein either the a position or the d position is occupied by either a charged or a bulky polar residue [Fig. 1(A,B,E)]. This includes four pairs where one partner is a charged residue (L162–R243, E173–V232, L194–H211, and A197–E208) and three pairs where one partner is a polar residue (V183–Q222, N187–L218, and A190–Y215). Equivalent pairs in BECN1 also have similar nonideal pairings that include charged or polar residues, except for the terminal L162–R243 pairs, which are ideal leucine pairs in BECN1.18 Thus, six pairs of nonideal packing interactions are conserved between BECN1 and BECN2.
Besides the hydrophobic interactions at the interface, the BECN2 CCD homodimer is stabilized by two unique pairs of inter‐chain polar interactions: Q222 hydrogen bonds D184 of the partner helix, and R240 ion pairs with E170 of the partner helix. In comparison, the BECN1 CCD has three unique pairs of interchain polar interactions.18 Of these, BECN1 Q240 is equivalent to BECN2 Q222, but other BECN1 residues contributing to polar interactions are not located at equivalent positions in BECN2. Thus, surface polar interactions stabilize both the BECN1 and BECN2 CCD homodimers, but unlike the interface residues, these interactions are not well conserved between these paralogs.
Nonideal packing differentially impacts BECN2 CCD homodimerization
Our ITC experiments indicate that the WT BECN2 CCD homodimerizes weakly, with a self‐dissociation constant (K d) of 138 µM, which is driven by entropy, not enthalpy (Table 2), consistent with the structural evidence that the majority of CCD homodimer interface interactions are nonideal. BECN2 CCD homodimerization is approximately threefold weaker than BECN1 CCD homodimerization with a K d of 48 µM,18 consistent with structural differences in the interactions stabilizing the BECN2 and BECN1 CCD homodimers discussed above.
Table 2.
Summary of the Self‐Dissociation Constants of Wild‐Type and Mutant BECN2 CCD
| BECN2 CCD | K d (μM) | dH (kJ/mol) | dS (J/K mol) |
|---|---|---|---|
| Wild type | 138 ± 6 | 120.7 ± 7.4 | 492.7 ± 25.4 |
| E173L | 5180 ± 940 | 205.6 ± 12.4 | 757.2 ± 41.6 |
| N187L | 0.04 ± 0.01 | −877.8 ± 89.9 | −2904.7 ± 311.4 |
| A190L + Y215L | 104 ± 9 | −85.4 ± 1.4 | −220.0 ± 4.2 |
| A197L + E208L | Not detectable | ||
| H211L | 13.3 ± 2.2 | 168.2 ± 1.1 | 677.4 ± 5.1 |
| Q222L | 4800 ± 750 | 98.5 ± 9.1 | 386.3 ± 32.6 |
| R243L | 326 ± 4 | 165.3 ± 7.6 | 640.6 ± 26.5 |
The impact of nonideal packing on BECN2 CCD dimer stability was investigated by mutating the seven pairs of residues involved in nonideal packing to more ideal leucine–leucine or leucine–valine pairs to create five single mutants and two double mutants wherein both partners within the imperfect pair were altered (Table 2). Relative to the WT CCD homodimer, the mutants were expected to have stronger hydrophobic interface interactions, leading to more stable homodimers. Preliminary evidence of this was provided by the improved purification yields of four of the BECN2 mutants, N187L, A190L + Y215L, A197L + E208L, and H211L, which increased two‐ to fivefold relative to WT, with the highest yield observed for the A197L + E208L mutant (Table SI).
We used ITC to quantify self‐dissociation constants of mutant BECN2 CCDs. We found that three of the seven BECN2 mutants associate more tightly as homodimers, consistent with the expectation of improved stability (Table 2). Among these three mutants, we found that the N187L mutant homodimerizes most tightly, with a K d of 0.04 µM, which is ∼3500‐fold tighter than the WT BECN2 CCD. Homodimerization of the H211L mutant is ∼10‐fold tighter compared to the WT BECN2 CCD, while that of the A190L + Y215L mutant is only 1.3‐fold tighter than WT.
No self‐dissociation of the A197L + E208L mutant was detected, suggesting that either there is no detectable homodimer dissociation, the heat generated during dissociation is too small to be detected by ITC, or this mutant does not homodimerize and hence does not self‐dissociate. The dramatically improved purification yield for this mutant (Table SI) suggests that the first possibility is the most likely and the A197L + E208L mutant forms a very stable homodimer.
In contrast to the mutants discussed earlier, and contrary to our initial expectations, the remaining three mutants, R243L, E173L, and Q222L, homodimerize more weakly than the WT BECN2 CCD, with the latter two forming substantially weaker homodimers (Table 2). In the WT BECN2 CCD homodimer, the Cα, Cβ, and Cδ atoms constituting the aliphatic part of R243 pack against a Cδ atom of L162 of the partner helix, while the charged R243 guanidinium protrudes out of the homodimer interface. Analysis of modeled backbone‐dependent leucine side chain rotamers of the R243L mutation indicates that L162 and L166 of the partner helix would sterically clash in 65% of leucine conformations, thereby weakening homodimerization. Similarly, in the WT BECN2 CCD homodimer, the E173 Cβ and Cγ pack against the L229 Cδ and V232 Cγ of the partner helix, while the E173 carboxylate is solvent exposed. Mutation of E173 to leucine causes steric clashes with L229 or V232 of the partner in all backbone‐dependent rotamer conformations, explaining the significantly weaker homodimerization of the E173L mutant compared to WT. Finally, in the WT BECN2 CCD homodimer, the Q222 Cα and Cβ pack against the V183 Cγ atom of the partner helix, while the Q222 Cγ packs with the partner helix L180 Cδ. Furthermore, the Q222 side chain amide hydrogen bonds the D184 carboxylate of the partner helix, while the Q222 side chain carbonyl oxygen makes an intrachain hydrogen bond to the K219 amino group. Q222 mutation to leucine would cause loss of these hydrogen bonds and also, in all rotamer conformations, cause steric conflicts with V183, D184, or L180 of the partner helix. Thus, like the E173L mutant, the Q222L mutant would form a much weaker homodimer compared to WT.
Nonideal packing impacts thermal stability of the BECN2 CCD
We used CD to assess the secondary structure of each WT and mutant BECN2 CCD. The CD spectra recorded for each protein show two minima at wavelength 208 and 222 nm, indicative of α‐helical conformation (Fig. 2), consistent with expectations from the crystal structure. We find (Table SII) that all the mutants, except the Q222L mutant, have increased helicity and decreased coil content. In general, mutants that homodimerize better have increased helicity and decreased disorder, with the most significant increase in helicity observed in the N187L, which also had the tightest measured homodimerization. The Q222L mutant, which has barely detectable homodimerization, has helicity similar to WT. Thus, none of the mutations adversely impacted CCD secondary structure.
Figure 2.
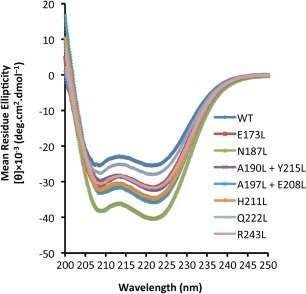
CD spectra of the BECN2 CCD WT and mutants at 4°C.
CCDs are unique in terms of protein folding because their tertiary structures, and often their secondary structure as well, are coupled to their oligomerization.34 Therefore, we assessed and compared structural stability of the WT and seven mutant BECN2 CCD constructs using thermal denaturation coupled to CD measurements at 222 nm (Fig. S2) to quantify T m (Table 3). The relatively low T m of 18.1°C (Table 3) determined from the single transition melting curve of the WT BECN2 CCD indicates that the BECN2 CCD is a metastable homodimer.
Table 3.
Summary of Tms of WT and Mutant BECN2 CCDs
| Beclin 2 CCD | T m (°C) |
|---|---|
| Wild type | 18.1 ± 0.7 |
| E173L | NDa |
| N187L | 42.6 ± 0.1 |
| A190L + Y215L | 27.2 ± 0.1 |
| A197L + E208L | 40.3 ± 0.3 |
| H211L | 30.4 ± 0.2 |
| Q222L | NDa |
| R243L | 27.3 ± 0.5 |
ND: Not determined.
The T m of these mutants could not be obtained because the melting process is not cooperative.
All the mutants, except the two that form significantly weaker homodimers, also have single transition melting curves (Fig. S2), and strikingly, each of these has a T m significantly higher than WT (Table 3). The N187L and A197L + E208L mutants have the highest melting temperatures, a striking 22–24°C higher than WT, and 10–15°C higher than the other mutants. Notably, the N187L BECN2 mutant has the tightest measured homodimerization, about ∼3500‐fold lower self‐dissociation than WT, while no self‐dissociation was detected for the A197L + E208L mutant. The dramatically improved thermal stability, combined with significantly increased helicity, of the A197L + E208L mutant confirms that the nondetectable self‐dissociation by ITC is the result of strong homodimerization, rather than lack of dimerization or low heat of dissociation. The H211L mutant, which homodimerizes ∼10‐fold tighter than the WT BECN2 CCD, also shows an improvement of 12°C in thermal stability while the A190L + Y215L and R243L mutants, which have homodimerization affinities comparable to WT, display a smaller ∼9°C increase in thermal stability.
Melting temperatures of the remaining two mutants, E173L and Q222L, could not be quantified because their melting curves are not cooperative (Fig. S2). Significantly, these two mutations also result in the weakest homodimerization, which likely leads to reduced stability of CCD secondary and tertiary structure. In sum, structural stability of BECN2 CCD mutants (Table 3) appears correlated with their ability to form CCD homodimers (Table 2).
The N187L BECN2 CCD X‐ray crystal structure reveals why it homodimerizes better than WT
To understand how the N187L single mutation impacts BECN2 CCD homodimer structure, we determined the 2.5 Å X‐ray crystal structure of N187L BECN2 CCD mutant [Fig. 3(A)]. Crystallographic data collection and refinement statistics are summarized in Table 1. The N187L BECN2 CCD crystallized in a different space group than the WT (Table 1), but like the WT, the asymmetric unit contains the following two pairs of antiparallel homodimers: AB and CD. The N187L BECN2 CCD AB homodimer superimposes on the CD homodimer with an RMSD of 1.49 Å over 160 Cα atoms, indicating that the two homodimers are very similar. The BECN2 N187L dimers superimpose very well on the WT dimers, with an RMSD of 1.08 ± 0.58 Å over 160 Cα atoms [Fig. 3(A)]. Thus, the N187L mutant and WT BECN2 CCD have very similar structures.
Figure 3.
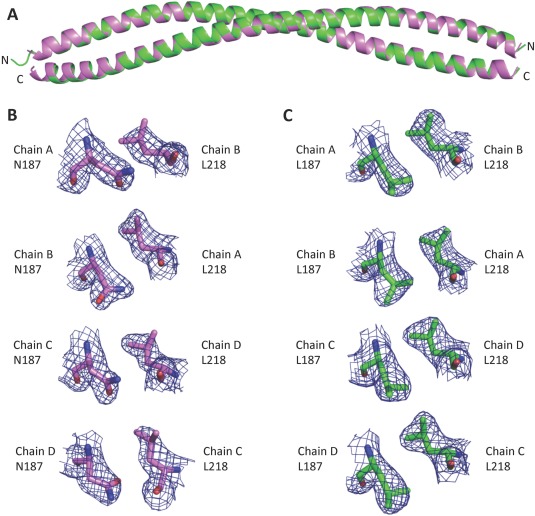
Superposition of WT and N187L BECN2 CCD structures. (A) Superposition of the N187L mutant (green) and WT (violet) BECN2 CCD homodimers. (B) Atomic details of N187–L218 pair in the WT BECN2 CCD structure for each chain. The 2Fo–Fc electron density maps are contoured at 1σ above the mean (blue mesh). (C) Atomic details of L187–L218 pairs for each chain in the N187L BECN2 CCD structure. The 2Fo–Fc electron density maps are contoured at 1σ above the mean (blue mesh).
The N187L CCD homodimer interface has a total buried surface area of 2188 ± 30 Å2, somewhat more than in the WT CCD. Except for the mutated residues, packing at the N187L BECN2 CCD homodimer interface, including the two pairs of inter‐chain surface polar interactions, is similar to WT. N187 in the WT structure [Fig. 3(B)], as well as the leucine at position 187 in the mutant structure [Fig. 3(C)], is clearly defined by electron density. The average buried surface area for the leucines in the N187L CCD homodimer interface from the four chains in the crystallographic asymmetric unit is 94.56 ± 3.06 Å2; significantly more than the 80.44 ± 6.54 Å2 buried for the N187 in the WT CCD interface. In the WT CCD structure, the N187 Cγ packs between side chains of Y215 and L218 from the partner helix, while the polar end of the side chain is solvent‐exposed. In the N187L CCD structure, the L187 Cγ atom packs similarly and the two leucine Cδ atoms make additional hydrophobic interactions with the partner helix via L218, K219, and Y215 side chain atoms as well as the Y215 backbone C atoms. Thus, as expected, the N187L mutation results in increased hydrophobic interactions, leading to tighter homodimerization.
The BECN2 and ATG14 CCD interaction is inversely related to the stability of BECN2 CCD homodimer
Imperfect interactions at the BECN2 homodimer interface that weaken the CCD homodimer likely promote self‐dissociation of the CCD homodimer to facilitate heterodimerization with other CCD containing autophagy proteins such as ATG14. Consistent with this idea, the BECN2 CCD binds tightly to the ATG14 CCD with a binding affinity (Kd) of 0.22 μM (Table 4), which is more than 600‐fold tighter than BECN2 CCD self‐association (Table 2). Notably, the BECN2 CCD:ATG14 CCD interaction is 20‐fold tighter than the BECN1 CCD:ATG14 CCD interaction.18
Table 4.
Summary of Thermodynamic Parameters for Binding of Wild‐Type and Mutant Beclin 2 CCDs to Atg14 CCD
| Beclin 2 CCD | K d (μM) | dH (kJ/mol) | dS (J/K·mol) |
|---|---|---|---|
| Wild type | 0.22 ± 0.04 | −162.2 ± 1.1 | −435.4 ± 2.2 |
| E173L | 0.58 ± 0.14 | −154.3 ± 6.8 | −416.0 ± 21.4 |
| N187L | 0.72 ± 0.11 | −110.5 ± 16.1 | −265.8 ± 57.1 |
| A190L + Y215L | 0.30 ± 0.02 | −120.7 ± 3.9 | −294.0 ± 13.8 |
| A197L + E208L | 1.23 ± 0.06 | −61.7 ± 11.5 | −100.9 ± 40.4 |
| H211L | 0.80 ± 0.18 | −105.7 ± 3.0 | −250.0 ± 12.2 |
| Q222L | 0.79 ± 0.01 | −149.7 ± 0.1 | −402.7 ± 0.2 |
| R243L | 0.54 ± 0.05 | −148.7 ± 0.6 | −396.0 ± 1.6 |
All the mutants created to have more ideal packing interfaces bind more weakly to the ATG14 CCD, although all binding affinities lie within ∼10‐fold of the WT (Table 4). Notably, the trend of increasing self‐association of BECN2 CCD mutants (Table 2) correlates, although not perfectly, with the order of decreasing binding affinity for ATG14. For instance, the A197L + E208L BECN2 CCD mutant, which has no detectable self‐dissociation, displays the weakest interaction, with approximately sixfold lower binding affinity for the ATG14 CCD relative to WT. Similarly, the N187L and H211L mutants, which have significantly tighter self‐association relative to WT, bind to the ATG14 CCD with approximately threefold lower affinity compared to WT. The A190L + Y215L mutant, which self‐associates marginally better than the WT BECN2 CCD, binds to the ATG14 CCD slightly more weakly than does the WT. However, the E173L and Q222L BECN2 CCD mutants that homodimerize very weakly and have noncooperative melting curves also bind 2.5–4‐fold more weakly to the ATG14 CCD than WT BECN2 CCD. This is not entirely surprising because heterodimerization with ATG14 also depends on factors other than BECN2 homodimer dissociation. Perhaps the side chain oxygen and hydrogen atoms of E173 and Q222 are involved in polar interactions with ATG14 residues; therefore, mutating these residues to leucine does not improve binding to the ATG14 CCD. Thus, although all mutations weaken binding of BECN2 CCD to ATG14 CCD, no mutation completely abrogates this interaction. Significantly, all the BECN2 interface mutations impact homodimerization more than heterodimerization.
The BECN2 CCD and ATG14 CCD form a parallel heterodimer
Finally, we used SAXS to probe the structure of the BECN2 CCD:ATG14 CCD heterodimer. To ensure that the SAXS data were collected from a homogeneous sample, SAXS was performed in tandem with size exclusion chromatography (SEC‐SAXS). For this study, the BECN2 CCD was expressed and purified with an 11 kDa N‐terminal SUMO‐His6‐tag; and the ATG14 CCD, as previously described,18 was expressed and purified with a 40 kDa N‐terminal MBP tag, so that the relative orientation of each protein in the complex could be extrapolated by the location of the tags in the low‐resolution envelope. The SEC profile of the SUMO‐His6‐BECN2 CCD:MBP‐ATG14 CCD complex comprises a single peak consistent with an elongated CCD dimer and indicates an absence of aggregated protein (Fig. S3).
The low q‐range region of the Guinier plot is linear in the range of q × R g < 1.3 confirming the absence of aggregation [Fig. 4(A)]. The radius of gyration (R g) of the SUMO‐His6‐BECN2 CCD:MBP‐ATG14 CCD heterodimer estimated from the Guinier plot is 43 Å, which is similar to the R g of 49 Å estimated from the pairwise‐distance distribution function (P(r) plot) [Fig. 4(B)]. The P(r) plot indicates that the BECN2 CCD:MBP‐ATG14 CCD heterodimer is elongated, with the maximum particle dimension (D max) estimated to be 186 Å. The Kratky plot indicates that the heterodimer is well folded [Fig. 4(C)]. The ab initio envelope calculated from the heterodimer SAXS data is “lollipop shaped,” comprising a long rod with a globular head at one end of the rod. The rod‐part of the envelope is somewhat curved and likely corresponds to the heterodimeric CCD, with the head corresponding to MBP and SUMO tags [Fig. 4(D,E)]. Localization of both N‐terminal tags on the same end of the CCD suggests that the BECN2 CCD and ATG14 CCD form a parallel heterodimer.
Figure 4.
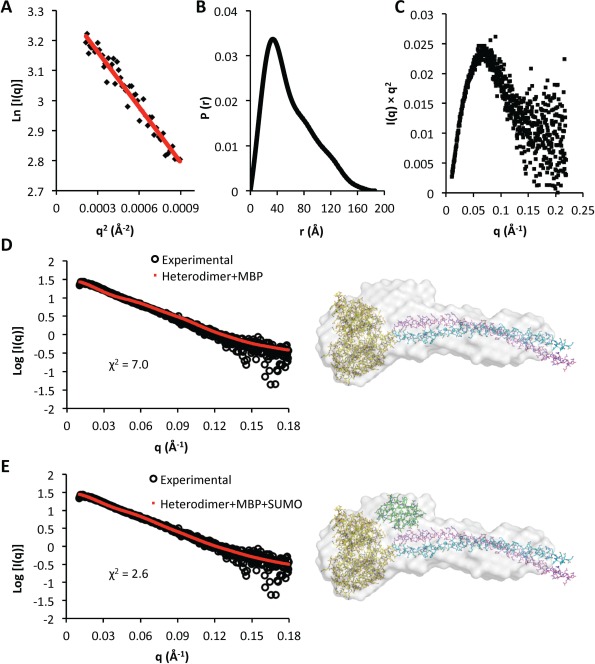
SEC‐SAXS analysis of SUMO‐His6‐BECN2:MBP‐ATG14 CCD heterodimer. (A) Guinier plot for the region s × R g ≤ 1.3. (B) P(r) pairwise distribution. (C) Kratky plot. (D and E) The fits of theoretical scattering profiles (red dots) calculated from the corresponding heterodimer model to the experimental SAXS data (black circles) are shown on the left. The pseudo‐atomic BECN2:MBP‐ATG14 CCD heterodimer models are (D) without SUMO‐His6 and (E) including SUMO‐His6 (right panels). The ab initio calculated molecular envelope is shown in gray. The atomic structures used for constructing the heterodimer model construction are shown in atomic detail with carbons colored according to molecule: MBP, yellow; SUMO, green; BECN2 CCD, violet; and ATG14 CCD, cyan (An interactive view is available in the electronic version of the article).
To further elucidate structural details of the BECN2:ATG14 CCD heterodimer, we built a parallel heterodimer model [Fig. 5(A)]. We first used CCBuilder35 to determine the register of parallel BECN2 CCD and ATG14 CCD sequences that results in optimal packing interactions at the CCD heterodimer interface. Next, we used the Protein Structure Prediction Server ((PS)2)36 to calculate a pseudo‐atomic heterodimer model based on the amino acid sequences of each CCD and the best packing register obtained from CCBuilder. Interestingly, the BECN2:ATG14 CCD model is also curved, with a radius of curvature of 13 nm [Fig. 5(B)], suggesting the heterodimer may be slightly more curved than the BECN2 CCD homodimer. The BECN2:ATG14 CCD heterodimer is less negatively charged compared to the BECN2 CCD homodimer [Fig. 5(B)].
Figure 5.
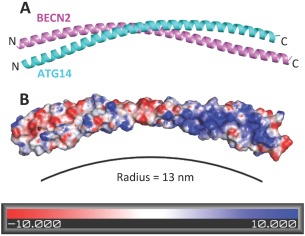
The BECN2:ATG14 CCD heterodimer. (A) The predicted heterodimer model displayed as (A) ribbon with BECN2 in violet and ATG14 in cyan ribbon and (B) molecular surface colored according to electrostatic potential calculated using APBS.
SASREF37 was then used to construct a pseudo‐atomic model comprising MBP and the BECN2:Atg 14 CCD heterodimer, that fits with a χ 2 of 7.0 [Fig. 4(D)] to the experimental SAXS data. The parallel CCD fits the rod part of the “lollipop‐shaped envelope, while MBP is positioned in the head [Fig. 4(D), right panel]. However, this fit leaves unoccupied space in the head. Therefore, SUMO was manually placed into this unoccupied space, which significantly improved agreement with the SAXS data as indicated by a χ 2 of 2.6 [Fig. 4(E)]. MBP and SUMO are therefore located on the same side of the rod‐like part of the ab initio SAXS envelope, since no unoccupied space remains on the other end of the envelope. Thus, the BECN2 CCD and ATG14 CCD form a parallel heterodimer, similar to that suggested for the BECN1:ATG14 CCD.29
The BECN2:ATG14 CCD heterodimer interface
The total surface buried at the heterodimer interface is 1814 Å2, accounting for 21.6% of the BECN2 CCD surface area and also 21.6% of the ATG14 CCD surface area. This buried area is less than the 2070 ± 37 Å2 found in the BECN2 CCD homodimer. Notably, all BECN2 CCD residues at the homodimer interface were positioned at the BECN2:ATG14 CCD heterodimer interface. Only 15 of the 91 ATG14 CCD residues are leucine, valine or isoleucine and in the best‐packed heterodimer model, ten of these residues contribute to the heterodimer interface.
Since the BECN2 CCD and ATG14 CCD form a parallel heterodimer, BECN2 residues at a and d positions of the successive heptad repeats pack against the ATG14 CCD residues at a′ and d′ positions, respectively, resulting in 25 paired interactions (Fig. 6). Given the few hydrophobic residues contributed by ATG14, the heterodimer interface has only ten pairs of acceptable packing interactions, including five ideal pairs, compared to the 12 pairs found in the BECN2 CCD homodimer. This is consistent with the smaller interface buried in the heterodimer model relative to the homodimer. The heterodimer is also stabilized by one pair of inter‐chain polar interactions. However, BECN2:ATG14 CCD heterodimerization is >600‐fold tighter than BECN2 CCD self‐association (Table 4).
Figure 6.
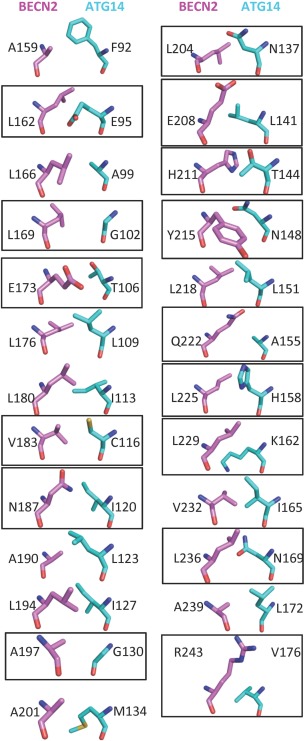
The BECN2:ATG14 CCD heterodimer interface. Interface residues displayed in stick colored as in Figure 1B, with carbons of BECN2 CCD in violet and ATG14 CCD in cyan. Nonideal packing residues are boxed.
The heterodimer model provides a structure‐based explanation for the impact of mutating BECN2 interface residues on heterodimerization with the ATG14 CCD, as assessed by ITC (Table 4). N187L and the A197L + E208L are the two BECN2 CCD mutants that self‐associate most tightly. In the heterodimer model, N187 packs against ATG14 I120. Therefore, mutating N187 to leucine also makes a more hydrophobic L187–I120 heterodimer interface. However, since heterodimerization depends on dissociation of the BECN2 CCD homodimer, the significantly improved homodimerization results in 3.5‐fold lower heterodimerization.
The A197L + E208L mutant does not appear to self‐dissociate. Based on the heterodimer model this double mutation should also result in significantly better hydrophobic packing with ATG14 due to formation of a BECN2 L208:ATG14 L141 pair and a BECN2 L197:ATG14 G130 pair. However, this improved interaction is offset by the substantially decreased A197L + E208L BECN2 CCD homodimer dissociation, such that overall K d for A197L + E208L BECN2:ATG14 CCD heterodimerization is sixfold lower than WT.
The H211L BECN2 CCD mutant also displays improved homodimerization. In the heterodimer model, H211 is involved in polar interactions with ATG14 T144; therefore, mutating H211 to leucine merely results in a nonideal L211–T144 pair. As a result, while the H211L BECN2 CCD self‐associates 10‐fold more strongly than WT, the H211L mutant binds to the ATG14 CCD with approximately four‐fold weaker affinity than the WT.
Both the E173L and Q222L BECN2 CCD form much weaker homodimers compared to WT, but they both also bind to the ATG14 CCD more weakly than WT. In the heterodimer model, E173 packs against ATG14 T106, while Q222 packs against A155. In each case, mutation to leucine does not improve hydrophobic packing at the heterodimer interface. Indeed, the WT E173 side chain may interact better with the ATG14 T106. Both E173 and Q222 may also help stabilize the WT heterodimer structure by polar interactions with surrounding charged or polar residues. Thus, despite the substantially weakened homodimerization, binding of ATG14 CCD to either E173L or Q222L BECN2 CCD is not improved compared to WT due to the absence of improved interactions in the heterodimers.
A190L + Y215L BECN2 CCD self‐dissociation is similar to the WT BECN2 CCD, and the ATG14 CCD also binds to the A190L + Y215L BECN2 CCD with an affinity similar to WT. In the heterodimer model, A190 and Y215 pack against ATG14 L123 and N148, respectively. While mutating A190 to leucine makes an ideal L190–L123 interface pair, mutating Y215 to leucine may not improve the stability of the heterodimer, as the WT heterodimer is likely stabilized by hydrophobic packing of the Y215 side chain against the ATG14 N148 Cα and Cβ and with carbon atoms of surrounding residues combined with hydrogen binding of the Y215 hydroxyl with N148 or with adjacent polar ATG14 residues.
Finally, relative to WT, the R243L BECN2 CCD forms a ∼2.5‐fold weaker homodimer and heterodimer with ATG14. In the heterodimer model, the aliphatic part of the R243 side chain packs against ATG14 V176, while the charged guanidinium group may make electrostatic interactions to surrounding charged ATG14 residues such as ATG14 E177. The loss of these polar interactions in the R243L mutant likely explains the weaker heterodimerization of this mutant.
Discussion
Our detailed structural, biophysical and mutational analysis of BECN2 CCD homodimerization and heterodimerization with ATG14 suggests that the sequence of BECN homologs is optimized to pair with partners such as ATG14 to form functionally active states, rather than to form homodimers, which may represent an autophagy‐inactive reservoir within the cell. Thus, BECN homologs are designed to associate and dissociate in the context of subtle cellular signals.
Unlike canonical CCDs, such as leucine zippers, the BECN2 CCD homodimer is stabilized by a few ideally paired hydrophobic residues, some less well‐packed hydrophobic residues, as well as noncanonical, interchain polar interactions, while being weakened by several nonideal pairings of polar and hydrophobic interface residues. This results in weakly associating, metastable BECN2 homodimers that likely represent an autophagy‐inactive cellular reservoir of BECN2. This homodimer may be further stabilized by interactions with other proteins in the cell.
Indeed, formation of a metastable homodimer appears to be a conserved feature of BECN homologs, as many of the nonideal interface pairs are conserved. Consistent with this hypothesis, structural and biophysical studies show that the BECN1 CCD also exists as a metastable homodimer,17, 18 while cellular studies of the mammalian ortholog, BECN1, indicate that there is a stable cellular reservoir of homodimers, which appears to be further stabilized by binding of BCL2 homologs.38, 39 The weak homodimerization of BECN homologs likely facilitates the dissociation of these homodimers upon induction of autophagy, to allow them to form heterodimers with the CCDs of other autophagy proteins like ATG14 and UVRAG.
BECN homologs are core components of quaternary complexes called PI3KC3 Complex I and Complex II, which also include PI3KC3, p150, and either ATG14 or UVRAG. 28 Å Cryo‐EM reconstructions of Complex I and Complex II indicate that these complexes have similar overall architecture and shape at that resolution.29 A recent 4.4 Å crystal structure of the yeast equivalent of Complex II confirmed results from the Cryo‐EM reconstructions and provided additional information regarding arrangement and interactions of different domains.16 Our SAXS results indicate that BECN2 and ATG14 form a parallel heterodimer, similar to that deduced for BECN1 and ATG14 from the Cryo‐EM reconstruction of Complex I and analogous to the arrangement of BECN and UVRAG homologs in Complex II. However, compared to Complex II, the curvature of the BECN2:ATG14 heterodimer is likely to alter the relative arrangement of membrane‐binding domains of BECN2, PI3KC3 and ATG14 in Complex I. This may influence selection and binding to membranes of different curvature by Complex I and Complex II.
BECN2:ATG14 heterodimer formation is driven by the higher affinities of the BECN2 CCD for the ATG14 CCD rather than for another BECN2 molecule, corroborating earlier qualitative assessments of binding.9 Our heterodimer model indicates that this higher affinity is dictated by better pairing of residues at the heterodimer interface as some polar BECN2 residues are paired with polar ATG14 residues rather than with hydrophobic residues, resulting in better interactions. Strikingly, some of these polar pairs are conserved among ATG14 and BECN homologs, supporting the idea that these polar residues are important for BECN:ATG14 CCD heterodimer formation.
These conclusions are corroborated by our experiments assessing the impact of BECN2 mutagenesis on thermal stability of the homodimer as well as affinity of homodimerization and heterodimerization. Mutating the nonideal BECN2 interface residues to ideal leucine–leucine or leucine–valine pairs largely improves CCD helicity and homodimer stability and self‐association, but adversely impacts binding to ATG14. However, binding to ATG14 is not completely abrogated, especially as some mutations also improve interactions with ATG14. Exceptions to improved homodimerization upon mutagenesis of BECN2 CCD nonideal residues to leucine/valine are primarily due to steric conflicts created within the homodimer upon mutagenesis, and in one case, replacement of polar residues that are stabilized by inter‐ and/or intrachain interactions. Thus, the sequence of BECN homologs is optimized to prevent tight self‐association, yet enable somewhat tighter association with specific partners such as ATG14, while simultaneously preventing overly tight association with these partners, thereby allowing for dynamic exchange of partners for different functions.
Despite the overall similarity in the structure, interactions and function of the BECN1 and BECN2 CCDs, there are important differences. For example, selected single BECN1 CCD mutations of charged interface residues to leucine completely abrogate binding to the ATG14 CCD.17 More recently, it was shown that mutating the BECN1 CCD hydrophobic interface residues to alanine dramatically impairs interaction between full‐length BECN1 and ATG14 and significantly diminishes starvation‐induced autophagy.18 In contrast, none of the mutations we made to the BECN2 CCD abrogate binding to the ATG14 CCD. Therefore, our study indicates that the interaction between the ATG14 CCD and BECN2 CCD is more tolerant to point mutations in BECN2, relative to the self‐association of BECN2; as well as compared to the impact of analogous point mutations in BECN1 on either BECN1 homodimerization or interaction with ATG14. This is consistent with our observations of tighter binding of the BECN2 and ATG14 CCDs and the previously reported stronger interaction between full‐length proteins in the cell.9 Therefore, ATG14 may bind BECN2 more persistently than BECN1, which may result in a steadier upregulation of autophagy. Thus, the differential interactions of the two mammalian BECN paralogs likely enables their different biological functions.
Materials and Methods
Experimental methods are described in Supplementary Information.
Supporting information
Supporting Information
Acknowledgments
The authors declare no conflict of interest. This work was supported by the following grants: NIH NINDS RO3 NS090939 (PI: SCS), NIGMS R15 GM122035 (PI: SCS), NIGMS R15 GM113227 (PI: CLC) and NIAID U19 AI199725 (PI: BL), a National Science Foundation grant MCB‐1413525 (PI: SCS), and NSF and North Dakota EPSCoR grant II‐1355466 doctoral dissertation awards for MS (PI: SCS). Work performed at Bio‐CAT was supported by NIH NIGMS 9P41 GM103622 and use of the Pilatus 3 1M detector funded by NIH NIGMS 1S10OD018090‐01. This work incorporates research conducted at the NE‐CAT beamlines, which are funded by NIH NIGMS P41 GM103403, and includes use of the Pilatus 6M detector on 24‐ID‐C beam line funded by NIH‐ORIP HEI S10 RR029205. This research used resources of the APS, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract no. DE‐AC02‐06CH11357.
References
- 1. Levine B, Klionsky DJ (2004) Development by self‐digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6:463–477. [DOI] [PubMed] [Google Scholar]
- 2. Mizushima N (2007) Autophagy: process and function. Genes Dev 21:2861–2873. [DOI] [PubMed] [Google Scholar]
- 3. Yorimitsu T, Klionsky DJ (2005) Autophagy: molecular machinery for self‐eating. Cell Death Differ 12:1542–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rubinsztein DC, Difiglia M, Heintz N, Nixon RA, Qin ZH, Ravikumar B, Stefanis L, Tolkovsky A (2005) Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy 1:11–22. [DOI] [PubMed] [Google Scholar]
- 5. Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mizushima N, Levine B (2010) Autophagy in mammalian development and differentiation. Nat Cell Biol 12:823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rubinsztein DC, Mariño G, Kroemer G (2011) Autophagy and aging. Cell 146:682–695. [DOI] [PubMed] [Google Scholar]
- 8. Levine B, Mizushima N, Virgin HW (2011) Autophagy in immunity and inflammation. Nature 469:323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He C, Wei Y, Sun K, Li B, Dong X, Zou Z, Liu Y, Kinch LN, Khan S, Sinha S, Xavier RJ, Grishin NV, Xiao G, Eskelinen E‐L, Scherer PE, Whistler JL, Levine B. (2013) Beclin 2 functions in autophagy, degradation of g protein‐coupled receptors, and metabolism. Cell 154:1085–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B (1998) Protection against fatal sindbis virus encephalitis by beclin 1, a novel bcl‐2‐interacting protein. J Virol 72:8586–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dong X, Cheng A, Zou Z, Yang Y‐S, Sumpter RM, Jr. Huang C‐L, Bhagat G, Virgin HW, Lira SA, Levine B (2016) Endolysosomal trafficking of viral g protein‐coupled receptor functions in innate immunity and control of viral oncogenesis. Proc Natl Acad Sci USA 113:2994–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mei Y, Glover K, Su M, Sinha SC (2016) Conformational flexibility of becn1: essential to its key role in autophagy and beyond. Protein Sci. 25:1767–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mei Y, Su M, Soni G, Salem S, Colbert CL, Sinha S (2014) Intrinsically disordered regions in autophagy proteins. Proteins 82:565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee EF, Perugini MA, Pettikiriarachchi A, Evangelista M, Keizer DW, Yao S, Fairlie WD (2016) The becn1 n‐terminal domain is intrinsically disordered. Autophagy 12:460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mei Y, Ramanathan A, Glover K, Stanley C, Sanishvili R, Chakravarthy S, Yang Z, Colbert CL, Sinha SC (2016) Conformational flexibility enables function of a becn1 region essential for starvation‐mediated autophagy. Biochemistry 55:1945–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, Masson GR, Johnson C, Steyaert J, Ktistakis NT, Williams RL. (2015) Structure and flexibility of the endosomal vps34 complex reveals the basis of its function on membranes. Science 350:178–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X, He L, Che KH, Funderburk SF, Pan L, Pan N, Zhang M, Yue Z, Zhao Y (2012) Imperfect interface of beclin1 coiled‐coil domain regulates homodimer and heterodimer formation with atg14l and uvrag. Nat Commun 3:662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mei Y, Su M, Sanishvili R, Chakravarthy S, Colbert CL, Sinha SC (2016) Identification of becn1 and atg14 coiled‐coil interface residues important for starvation‐induced autophagy. Biochemistry 55:4239–−4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Noda NN, Kobayashi T, Adachi W, Fujioka Y, Ohsumi Y, Inagaki F (2012) Structure of the novel c‐terminal domain of vacuolar protein sorting 30/autophagy‐related protein 6 and its specific role in autophagy. J Biol Chem 287:16256–16266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang W, Choi W, Hu W, Mi N, Guo Q, Ma M, Liu M, Tian Y, Lu P, Wang F‐L, Deng H, Liu L, Gao N, Yu L, Shi Y. (2012) Crystal structure and biochemical analyses reveal beclin 1 as a novel membrane binding protein. Cell Res 22:473–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oberstein A, Jeffrey PD, Shi Y (2007) Crystal structure of the bcl‐xl‐beclin 1 peptide complex: beclin 1 is a novel bh3 only protein. J Biol Chem 282:13123–13132. [DOI] [PubMed] [Google Scholar]
- 22. Feng W, Huang S, Wu H, Zhang M (2007) Molecular basis of bcl‐xl's target recognition versatility revealed by the structure of bcl‐xl in complex with the bh3 domain of beclin‐1. J Mol Biol 372:223–235. [DOI] [PubMed] [Google Scholar]
- 23. Ku B, Woo J‐S, Liang C, Lee K‐H, Hong H‐S, Xiaofei E, Kim K‐S, Jung JU, Oh B‐H (2008) Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral bcl‐2 of murine γ‐herpesvirus 68. PLoS Pathog 4:e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sinha S, Colbert CL, Becker N, Wei Y, Levine B (2008) Molecular basis of the regulation of beclin 1‐dependent autophagy by the γ‐herpesvirus 68 bcl‐2 homolog m11. Autophagy 4:989–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sinha S, Levine B (2009) The autophagy effector beclin 1: A novel bh3‐only protein. Oncogene 27:S137–S148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q (2008) Identification of barkor as a mammalian autophagy‐specific factor for beclin 1 and class iii phosphatidylinositol 3‐kinase. Proc Natl Acad Sci USA 105:19211–19216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama‐Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T. (2009) Two beclin 1‐binding proteins, atg14l and rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 11:385–396. [DOI] [PubMed] [Google Scholar]
- 28. Itakura E, Kishi C, Inoue K, Mizushima N (2008) Beclin 1 forms two distinct phosphatidylinositol 3‐kinase complexes with mammalian atg14 and uvrag. Mol Biol Cell 19:5360–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baskaran S, Carlson L‐A, Stjepanovic G, Young LN, Kim DJ, Grob P, Stanley RE, Nogales E, Hurley JH (2014) Architecture and dynamics of the autophagic phosphatidylinositol 3‐kinase complex. eLife 3:e05115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fimia G, Stoykova A, Romagnoli A, Giunta L, Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. (2007) Ambra1 regulates autophagy and development of the nervous system. Nature 447:1121–1125. [DOI] [PubMed] [Google Scholar]
- 31. Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B (2005) Bcl‐2 antiapoptotic proteins inhibit beclin 1‐dependent autophagy. Cell 122:927–939. [DOI] [PubMed] [Google Scholar]
- 32. Su M, Mei Y, Sanishvili R, Levine B, Colbert CL, Sinha S (2014) Targeting γ‐herpesvirus 68 bcl‐2‐mediated down‐regulation of autophagy. J Biol Chem 289:8029–8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mason JM, Arndt KM (2004) Coiled coil domains: stability, specificity, and biological implications. ChemBiochem 5:170–176. [DOI] [PubMed] [Google Scholar]
- 34. Sanchez JG, Okreglicka K, Chandrasekaran V, Welker JM, Sundquist WI, Pornillos O (2014) The tripartite motif coiled‐coil is an elongated antiparallel hairpin dimer. Proc Natl Acad Sci USA 111:2494–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wood CW, Bruning M, Ibarra AA, Bartlett GJ, Thomson AR, Sessions RB, Brady RL, Woolfson DN (2014) Ccbuilder: an interactive web‐based tool for building, designing and assessing coiled‐coil protein assemblies. Bioinformatics 30:3029–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang TT, Hwang JK, Chen CH, Chu CS, Lee CW, Chen CC (2015) (ps)2: protein structure prediction server version 3.0. Nucleic Acids Res 43:W338–W342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petoukhov MV, Svergun DI (2005) Global rigid body modeling of macromolecular complexes against small‐angle scattering data. Biophys J 89:1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Noble C, Dong J, Manser E, Song H (2008) Bcl‐xl and uvrag cause a monomer–dimer switch in beclin1. J Biol Chem 283:26274–26282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adi‐Harel S, Erlich S, Schmukler E, Cohen‐Kedar S, Segev O, Mizrachy L, Hirsch JA, Pinkas‐Kramarski R (2010) Beclin 1 self‐association is independent of autophagy induction by amino acid deprivation and rapamycin treatment. J Cell Biochem 110:1262–1271. [DOI] [PubMed] [Google Scholar]
- 40. Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98:10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. DeLano WL. ( 2002) The pymol molecular graphics system. San Carlos, CA, USA: DeLano Scientific. [Google Scholar]
- 42. Schrodinger, LLC. (2015) The pymol molecular graphics system, version 1.8, forthcoming.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


